Significance
Variation is the raw material for evolution, but studies have yet to unify the effects of intrinsic genetic and developmental interactions that influence variation with the extrinsic factors that shape organismal diversity on a large scale. Analysis of phenotypic integration can bridge different facets of evolutionary study and address fundamental questions on developmental and evolutionary dynamics through deep time. Our study of Late Pleistocene saber-toothed cats and dire wolves demonstrates that developmental integration channeled increasing amounts of morphological variation through 27,000 years of climate change, validating results from laboratory-based studies with the natural experiments captured in the fossil record.
Keywords: phenotypic integration, modularity, macroevolution, carnivorans, Late Pleistocene
Abstract
Variation is the raw material for natural selection, but the factors shaping variation are still poorly understood. Genetic and developmental interactions can direct variation, but there has been little synthesis of these effects with the extrinsic factors that can shape biodiversity over large scales. The study of phenotypic integration and modularity has the capacity to unify these aspects of evolutionary study by estimating genetic and developmental interactions through the quantitative analysis of morphology, allowing for combined assessment of intrinsic and extrinsic effects. Data from the fossil record in particular are central to our understanding of phenotypic integration and modularity because they provide the only information on deep-time developmental and evolutionary dynamics, including trends in trait relationships and their role in shaping organismal diversity. Here, we demonstrate the important perspective on phenotypic integration provided by the fossil record with a study of Smilodon fatalis (saber-toothed cats) and Canis dirus (dire wolves). We quantified temporal trends in size, variance, phenotypic integration, and direct developmental integration (fluctuating asymmetry) through 27,000 y of Late Pleistocene climate change. Both S. fatalis and C. dirus showed a gradual decrease in magnitude of phenotypic integration and an increase in variance and the correlation between fluctuating asymmetry and overall integration through time, suggesting that developmental integration mediated morphological response to environmental change in the later populations of these species. These results are consistent with experimental studies and represent, to our knowledge, the first deep-time validation of the importance of developmental integration in stabilizing morphological evolution through periods of environmental change.
How do extrinsic and intrinsic forces interact to shape the diversity of life? Macroevolutionary analyses of biodiversity typically focus on factors such as competition, environmental change, or mass extinctions, which can affect individual or multiple lineages over long time periods. In contrast, microevolutionary studies of diversity are usually concerned with the genetic and developmental changes that shape population-level variation, the raw material of natural selection. This divergence in focus can be attributed in large part to differences in data resolution. Population- or even species-level variation is often difficult to estimate in extinct or rare taxa whereas natural changes in extrinsic factors rarely occur on the time scales represented by large samples of living taxa. The study of morphological trait interactions, or phenotypic integration (1–4), provides an almost unique opportunity to unify these different scales, and foci, of evolutionary biology into combined analyses of organismal diversity that are equally applicable to Drosophila and dinosaurs.
Phenotypic integration is based on the intuitive idea that organisms are made up of interrelated traits and that the relationships among these traits are not equal (1, 2). Rather, genetic, developmental, and functional interactions result in some traits covarying strongly with each other while others have little covariance at all (Fig. 1). Moreover, the organization of traits often can be characterized as modular, in that traits form groups that have strong interactions within each group, but relatively weak interactions between groups (5, 6). The study of these trait relationships, or their integration, and their frequent division into modules has typically focused on model organisms with detailed genealogies and ontogenies (3–9), which allows for distinguishing among different levels and sources of integration, from genetic and developmental to evolutionary integration (10) (SI Text). However, through quantitative analysis of phenotypic trait covariances, genetic and developmental changes can be reconstructed in taxa for which molecular data will never be available, such as fossils (1, 11).
Fig. 1.
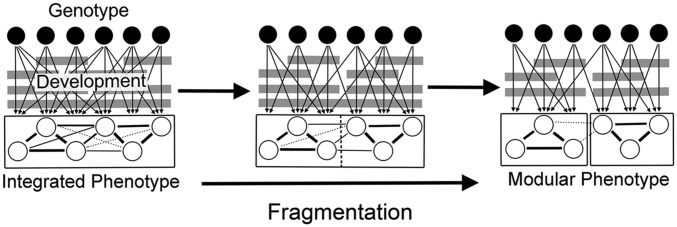
Genetic pleiotropy and developmental interactions contribute to the integration of phenotypic traits, which are often observed to form distinct modules, sets of highly integrated traits with strong correlations (solid lines) within modules and relatively weaker (thin solid lines and dotted lines) and/or fewer correlations between modules. It is hypothesized that fragmentation of integrated traits through time leads to the evolution of modular phenotypes. Modified from refs. 6 and 9.
Much recent work has focused on characterizing large-scale patterns of trait relationships through comparative studies of extant and fossil taxa (11–23), in some cases demonstrating shifts in phenotypic integration related to changes in development (24–26), function (27–29), and environment (30–33). Although focused overwhelmingly on model organisms, these studies provide a foundation for understanding how phenotypic integration changes through ontogenetic and evolutionary time and how it relates to myriad factors shaping morphological evolution. However, much work remains to be done in this field, and paleontological data will be critical to incorporate in future studies to reconstruct the evolution of phenotypic integration and establish its role in shaping organismal diversity.
The importance of fossil data to the study of phenotypic integration can be illustrated in a few key points. First, fossils are the only source of data for most of organismal variation. Understanding the patterns and processes that shape morphological evolution and phenotypic integration requires an accurate estimate of all variation in overall form and in phenotypic integration through time, not only the small proportion that is represented by living taxa. Second, known correspondences between shifts in developmental patterns and shifts in trait integration allow for the inference of development in fossil taxa through examination of phenotypic integration. Direct evidence on the timing of evolutionary changes in development in turn allows for more precise understanding of the selection pressures driving the developmental transitions that define many large clades. Lastly, fossils provide direct evidence of evolutionary responses to large-scale extrinsic factors, such as environmental change (34–36), allowing for combined studies of intrinsic and extrinsic influences on organismal evolution.
There are many hypotheses concerning phenotypic integration that paleontological data are uniquely suited to address, but perceptions of sample size restrictions and a long focus on experimental analyses have left the following questions, by and large, unanswered: When did observed shifts in patterns of phenotypic integration, and associated developmental transitions, occur in the evolution of major clades? Does phenotypic integration mediate organismal response to environmental change? Does the influence of developmental interactions on variation increase through time/clade history? Is phenotypic integration more labile during radiations or early in clade evolution? Are there persistent trends in phenotypic integration and modularity? Does modularity increase evolvability and how has this shaped organismal diversity? Here, we discuss these topics in further detail, highlighting some of the provocative yet preliminary studies that approach this field from a deep-time perspective.
Reconstructing Deep-Time Evolutionary Development Through Analysis of Phenotypic Integration in Fossils
Recent studies linking shifts in phenotypic integration to differences in development among major clades have raised the exciting prospect of reconstructing developmental patterns from adult fossil specimens, potentially illuminating the often obscure biology of stem representatives for living clades (24–26). For example, the three major clades of extant mammals, placentals, marsupials, and monotremes, are characterized by markedly different developmental strategies that are reflected in well-studied heterochronies in limb ossification. Monotremes (platypuses and echidnas) differ from almost all other tetrapods in ossifying distal limb elements first, and progressing toward the most proximal bones, rather than the typical proximal-to-distal direction (37). Adult monotremes also show strong covariation in shape between the serial homologs of the limbs (e.g., humerus covaries most strongly with the femur), with little integration within limbs (e.g., within the forelimb) (24). In contrast, marsupials, pouched mammals with short intrauterine gestation periods and a requisite crawl to the pouch where highly altricial young undergo most of their development, show the opposite pattern. Fore- and hindlimbs show strong integration within each limb, but weak or no integration across the limbs (24, 26, 38). Different again are the placental mammals, which show strong within-limb and between-limb integration, reflecting both functional associations and serial homology (28). These differences in integration as measured from adult morphology correspond with heterochronic shifts in ossification (25) and in gene-expression patterns (39).
These straightforward links among phenotypic integration, skeletal development, and gene expression in limb buds demonstrate the remarkable potential of fossil data on phenotypic integration to elucidate the evolution of development. Data on limb integration in early mammal fossils could establish the ancestral developmental condition for mammals and the timing of development shifts leading to the major mammalian clades, as demonstrated by some promising analyses exploring related questions with fossil data. A recent analysis of evolutionary integration in the mammalian shoulder girdle compared pooled samples of marsupials, placentals, and nonmammalian synapsids, the extinct mammalian stem group, to show that placentals and synapsids share a common pattern of (relatively weak) scapular integration, but that marsupials have increased their within-scapular integration, perhaps alongside their increased within-forelimb integration, early in their evolution (40). Future study of integration in limb elements of synapsids may demonstrate that marsupials and monotremes show a derived, more modular condition whereas placentals have retained an ancestral pattern of integration across the appendicular skeleton.
There are limited studies of phenotypic integration in fossil vertebrates other than mammals, but one recent study demonstrated that ichthyosaurs show strong integration across serial homologs (humerus and femur), like monotremes, despite strongly divergent sizes and presumably functions for the fore- and hindlimbs in these secondarily aquatic reptiles (41). In contrast, all three groups of tetrapods that have evolved flapping flight (pterosaurs, birds, and bats) show dissociation of serial homologs of the fore- and hindlimb, reflecting the functional divergence of these structures (27). It is likely that either a pattern of strong integration between serial homologs alone (as in monotremes and ichthyosaurs) or a fully-integrated pattern with strong correlations among serial homologs and within limbs (as in placentals) is ancestral to tetrapods, but further data, particularly from taxa that occurred early in the terrestrialization of vertebrates, is needed to establish evolutionary trends in postcranial integration. Given the divergent developmental strategies and morphologies of many extant clades, data on phenotypic integration from transitional fossils may also provide the only direct evidence on the timing and selective pressures underlying the evolution of reproductive strategies across vertebrates.
Of course, linking observed patterns of phenotypic integration to specific developmental interactions is not always straightforward. Many layers of developmental patterning may obscure each other, termed the “palimpsest” problem (42). Indeed, there seems to be little to no relationship between cranial phenotypic modules and coordination of skeletal heterochronies (25, 43–45), unlike in the postcranial skeleton. Nonetheless, quantitative approaches can estimate the developmental contribution to phenotypic integration in fossil taxa, even without identifying a specific developmental driver, by measuring fluctuating asymmetry (FA). Simply, whereas genetic and environmental perturbations affect symmetric traits similarly, developmental errors lead to random, nondirectional asymmetries in the morphology, thereby revealing patterns of direct developmental interactions among traits (22, 46–49). By quantifying FA, one can test whether the patterns of direct developmental interactions correspond to major components of variation across individuals and among taxa, as would be expected if developmental integration is a significant constraint on evolutionary variation. For example, recent studies of extant taxa ranging from skulls of domesticated and wild carnivorans to compound leaves in plants have demonstrated a significant correspondence between FA and overall variation at all levels (22, 23), suggesting that developmental integration is a significant influence on morphological evolution.
Studies of FA hold great promise for revealing developmental and evolutionary dynamics in long-extinct taxa (16–19). A recent analysis of three species of trilobites from early in their Cambrian diversification used comparisons of FA and overall variation to estimate how developmental interactions shaped variation between species. The results suggested that developmental integration was relatively labile and had little correlation with directions of variation between species (18), in contrast to the studies of extant taxa (22, 23). This analysis also demonstrates one of the key benefits of examining fossil data—the ability to directly sample taxa from early in a clade’s history, when developmental canalization might be less pronounced than in more established clades. Whether this pattern is specific to trilobites or more generally representative of early periods of clade evolution and radiation requires much further study with deep-time data.
Studies of fluctuating asymmetry also suggest that developmental integration may play an important role in channeling variation through periods of environmental change. An interesting series of laboratory experiments examined the effects of artificially induced environmental stress on developmental integration and variation in shrew mandibles, demonstrating that FA and variation rose in stressed population but that this increased variation was channeled along the same directions as variation between species (32, 47). Whether these experimental studies are representative of evolutionary responses to changes in the natural environment is difficult to assess, but data from the fossil record are ideally suited to further test the interactions among developmental integration, phenotypic integration, morphological variation, and environmental change over large time scales.
Resolving Macroevolutionary Trends in Phenotypic Integration and Modularity
It is hypothesized that modularity has increased, and integration decreased, through time to maintain or increase evolvability (6), the ability of organisms to evolve in response to changing environments and selection pressures (23, 50) despite changes in complexity (51, 52). The basis for this prediction is that strong integration of traits is thought to be a major control on phenotypic variation, and ultimately, phenotypic evolution (Fig. 2). Left unchecked, increasing complexity of the genetic effects, developmental pathways, and functional mechanisms that drive trait integration may be expected to impose overwhelming constraints on variation, thereby limiting the evolution of morphological diversity (6). Increasing modularity has been proposed as a mechanism for offsetting cumulative genetic pleiotropy and developmental canalization, allowing the fragmentation of larger integrated units into smaller modules that maintain the necessary interactions among traits within each module but allow the newly fragmented units to vary independently of one other (Fig. 1).
Fig. 2.
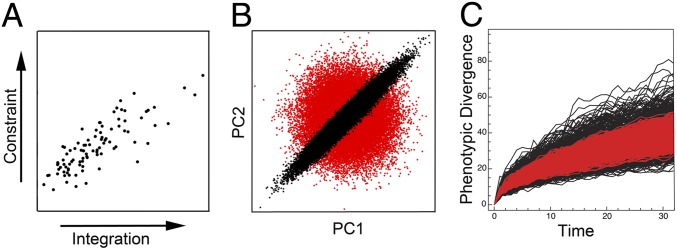
Phenotypic integration shapes morphological variation. (A) Greater integration of traits constrains the response to selection to a direction that is concordant with that of maximum variation, shown here for simulations based on empirical data from mammalian crania. Constraint is measured as the vector correlation between the response vector and the first principal component of each matrix. Integration is measured as the relative eigenvalue standard deviation (SD) of each matrix. (B) Integrated structures (black dots) repartition variance along preferred directions of change, such that, over time, they will explore fewer directions of morphospace than unintegrated structures (red dots), but will achieve a great range of shapes in those preferred directions. (C) This bias in direction of evolutionary change will ultimately produce both more and less divergent morphologies in an integrated structure (black lineages) than expected from an unintegrated one (red lineages). Modified from ref. 63.
There is currently little data to support this hypothesis of increasing modularity because phylogenetic comparisons have identified shifts in modularity but have, as of yet, rarely provided any robust evidence as to their polarity. It is almost certain that modularity has increased from the first simple organisms to most of those making up the modern biota. However, a similar argument may be made for complexity; yet, rigorous analyses have failed to identify a significant trend toward increasing complexity across the tree of life (51). Indeed, a review of the few relevant macroevolutionary studies suggests that it is unlikely that there are simple trends toward decreased phenotypic integration, and increased modularity, through time. For example, multiple studies of phenotypic integration in the mammalian cranium have supported a common pattern in most living marsupials and placentals (11, 15), and, where fossil mammals have been examined, most show similar patterns as their extant relatives (11). One interesting exception is that of extinct saber-toothed cats (Smilodon fatalis), in which patterns of cranial integration diverge from those of extant and extinct conical-toothed cats (53) by fragmentation of the anterior face into two new modules (11) (SI Text), possibly due to strong sexual selection on canine size (54) or the simple biomechanical requirements of accommodating these extreme structures. The polarity of this shift can be unambiguously resolved and supports the hypothesis that increased modularity evolved through fragmentation of trait relationships driven by divergent requirements of previously integrated traits (5). The lack of integration of the fore- and hindlimbs in marsupials and various flying tetrapods noted above likely represent multiple additional examples of evolutionary fragmentation of modules, but further data on limb integration across living and extinct tetrapods is needed to polarize these shifts.
Floral evolution may offer another example of increased modularity through evolutionary time. Although there are no analyses of fossil plants yet, there is a long history of studies of phenotypic integration and modularity in plants, starting with Berg’s description of “correlation pleiades” in flowering plants (2). Indeed, the distinct separation of functions in flowering plant structures is perhaps the clearest example of functional and variational modularity in organisms that exists in the modern world, with many quantitative studies demonstrating that floral traits are almost entirely decoupled from vegetative traits (29, 30, 55). A recent comparative study that compiled data from plants, vertebrates, and invertebrates found that the lowest levels of integration across all taxa were observed within floral elements and between floral and vegetative elements (56). Presumably, the dissociation between floral and vegetative elements is a derived condition that evolved as floral morphology and function diverged from that of its vegetative, and probably highly integrated, ancestral form. Because the fossil record provides primary data on the early evolution of angiosperms, it is hoped that future work will incorporate fossils to elucidate the evolution of the extreme modularity observed in flowering plants.
In contrast to evidence for fragmentation of traits into new modules through vertebrate and plant evolution, data from the rich fossil record of marine invertebrates do not support increased modularity, and decreased integration, through time. A recent study of evolutionary integration in crinoids used data from over 1,000 species that span over 400 million years to demonstrate that magnitude of integration does not change significantly through much of crinoid evolutionary history. The only significant change observed was a step-wise shift toward increased integration separating Paleozoic and post-Paleozoic taxa (16), in contrast to hypotheses of trends toward increased modularity and decreased integration through time. Unfortunately, there is no similar work with which to compare this study yet, but the temporal perspective available in the dense fossil record for many marine invertebrate clades is ideally suited to assessing trends in integration and modularity, and further work along these lines is sorely needed.
Evolvability and Modularity: The Macroevolutionary Consequences of Phenotypic Integration
As noted above, a great deal of discussion has concerned what, if any, significance phenotypic integration has for morphological evolution (2, 5, 6, 8, 57–61), and resolving the impact of trait interactions on morphological variation has the potential to fundamentally change models of evolution. Many studies have tested for correlations between developmental integration, variational integration, and overall variation across taxa to assess the influence of developmental interactions on morphological evolution (18, 22, 23). Others have simulated the potential effects of trait integration on the ability of populations to respond to selection (50, 62). Combining simulations with empirical data has shown that phenotypic integration does influence the direction of interspecific variation (50). For example, simulations using empirically derived covariance matrices from mammal crania have also shown that high integration is associated with lower ability to respond to selection (62), with both the magnitude and direction of response to selection mediated by phenotypic integration (Fig. 2A) (63).
Simulations can also estimate how phenotypic integration may manifest its influence on larger time scales. By directing the response to selection and repartitioning variance along preferred trajectories, phenotypic integration essentially constrains evolutionary change in some directions and facilitates it in others (9). Perhaps counterintuitively, this effect may actually increase the range of morphospace occupation for an integrated structure, due to the evolution of more extreme morphologies, as well as more convergence, along preferred axes, compared with an unintegrated one (Fig. 2B). Over time, the effect of phenotypic integration will produce taxa that are both more divergent and less divergent in morphology than would be otherwise expected (Fig. 2C) (63).
Fewer studies have tried to directly estimate the relationship between integration and morphological variation or disparity in empirical datasets (16, 64). Analyses of morphological disparity and integration have compared trait variances in strongly and weakly integrated traits across a few clades of mammals, suggesting that strong integration may constrain trait variation across taxa although its effect is relatively weak (64). Interestingly, this weak effect on disparity may not translate simply to evolutionary rates because some of the most strongly integrated cranial traits showed limited disparity but high rates of evolution in an analysis across carnivorans (63). These results present one possible hypothesis: Phenotypic integration may constrain morphological variation to certain “preferred” directions but has little influence on the speed of changes within the allowed space, a pattern that reflects previous studies finding that evolution is more bounded by constraints than predicted (65).
In contrast to the results for mammals, a recent study of 58 species of extant mantis shrimp compared evolutionary rates across taxa with markedly different patterns of segment modularity, demonstrating that taxa with greater modularity showed higher rates of evolution (20). Different again were the results from the comparative analysis described above of over 1,000 fossil crinoids in which no temporal concordance between morphological disparity and degree of phenotypic integration was found (16). Rather more interestingly, the results suggested that changes in the structure of integration, rather than its overall magnitude, have constrained the diversity dynamics of crinoids through the Phanerozoic (16). Combined, these studies suggest that phenotypic integration, and, more specifically, changes in patterns of integration and modularity do influence trait variation, but far more work is required to fully understand how integration and modularity have shaped organismal diversity through time.
From the Laboratory to the Rock Record: How Phenotypic Integration Mediated 27,000 y of Morphological Evolution in Late Pleistocene Carnivorans
Incorporating fossil data into studies of phenotypic integration will ultimately allow for analysis of the genetic, developmental, and functional drivers of trait variation in concert with data on large-scale external influences on diversity, such as mass extinctions, climate, and ecological interactions. Combined, these data can provide a more comprehensive understanding of the factors shaping morphological diversity through time. Although there is yet little direct study bridging these topics in deep time, experimental studies of extant taxa provide testable hypotheses for examining how phenotypic integration and environmental change may interact on a macroevolutionary scale. Here, we build on these studies to assess cranial integration, modularity, fluctuating asymmetry, and variance in Late Pleistocene carnivorans S. fatalis (sabertoothed cat) and Canis dirus (dire wolf). Using a 3D landmark dataset of specimens from four dated pits spanning ∼27,000 years, we demonstrate the potential insights on these topics available through the analysis of fossil data.
Results
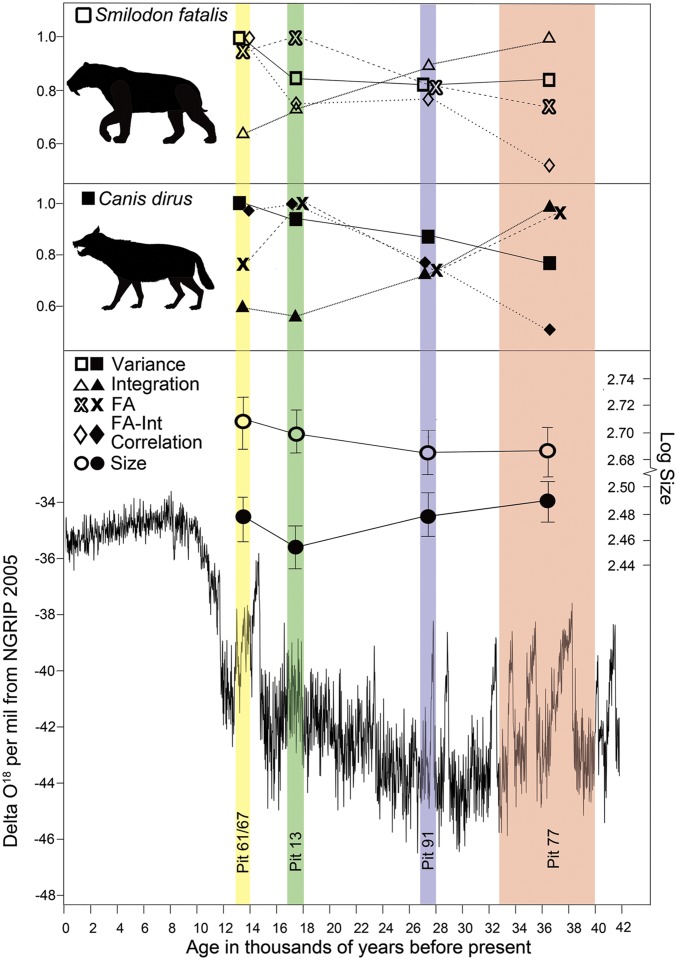
Canonical variates analysis demonstrated that all pit samples of S. fatalis and C. dirus were significantly different in shape (P ≤ 0.01), with the exception of the two oldest samples of S. fatalis, pit 77 and pit 91 (P = 0.092). In addition to changes in cranial shape, S. fatalis increased in size, measured as centroid size (Table S1 and Fig. 3), from the oldest pits (pit 77 and pit 91) to the youngest pits (pit 13 and pit 61/67), although only pit 61/67 specimens were significantly larger than those from pit 77 and pit 91 (P = 0.011 and 0.0031, respectively). In contrast, only C. dirus pit 13 differed significantly in size from other pits, with a significant decrease in body size in that sample (all P < 0.005). Interindividual variance increased through time in both species, with pit 61/67 significantly greater in variance than all other pits in S. fatalis (P < 0.001), and both pit 13 and pit 61/67 significantly greater in variance (P < 0.001) than the two older pits in C. dirus, although this observed increase in C. dirus may in part be due to the low sample size for pit 77 (Table S1 and Fig. 3). Fluctuating asymmetry similarly increases from the older samples to the younger ones in S. fatalis although it is slightly higher in pit 13 than the youngest pit, 61/67. Magnitude of fluctuating asymmetry varies more through time in C. dirus; it is highest in pit 13, as in S. fatalis, but also high in the poorly sampled oldest pit, pit 77, and lower in pits 91 and 61/67 (Table S1 and Fig. 3).
Fig. 3.
Temporal patterns in log centroid size, interindividual variance, FA, phenotypic integration, and the correlation between fluctuating asymmetry and overall integration for S. fatalis and C. dirus from the Rancho La Brea tar pits. Variance, FA, integration, and correlation are scaled relative to their maximum values. Unscaled values in Table S1. Data on pit ages from refs. 35 and 36. Delta O18 curve from ref. 67. Open symbols indicate S. fatalis; closed symbols indicate C. dirus. Silhouettes from phylopic.org.
Overall phenotypic integration, measured as relative eigenvalue SD (66), decreased consistently through time in both S. fatalis and C. dirus (Table S1 and Fig. 3). FA was not significantly correlated with overall integration in the oldest sample for S. fatalis (pit 77) or the two oldest samples for C. dirus (pit 77 and pit 91) but was significantly correlated (P < 0.001) with overall integration in all other samples. All pitwise comparisons of overall integration and of FA were significant (all P < 0.01) (Table S2).
Discussion
We analyzed integration, modularity, and variance in two contemporaneous Late Pleistocene carnivorans that survived multiple glacial-interglacial cycles before their extinction ∼10,000 years ago. Although uncertainties in precise dates limit extrapolation to specific climatic events (35, 36), our sample spans multiple episodes of climate change (Fig. 3), through which both S. fatalis and C. dirus display increasing variance over time. The highest levels of FA in C. dirus were observed in the penultimate sample, pit 13, which also shows the highest incidence of tooth fracture and wear and, presumably, the highest nutritional stress for this species (34). FA is also highest in S. fatalis from pit 13 although only marginally more than in pit 61/67, and tooth damage is similarly high and near-equal in these two populations (34).
Fluctuating asymmetry was increasingly correlated with phenotypic integration within each pit sample, demonstrating that the increased variance was channeled by developmental interactions along existing preferred directions of shape change. FA and overall phenotypic integration were also significantly correlated across pit samples. Thus, both size and shape changed significantly, variance increased, and magnitude of integration decreased through the last years of S. fatalis and C. dirus, but the patterns of phenotypic and developmental integration do not seem to have changed substantially and instead channeled these changes along existing directions of variation. These results are also consistent with repeated patterns of shape evolution observed in S. fatalis mandibular morphology (35) and with neotenic changes observed in C. dirus cranial morphology (36) through the same interval.
This study of Late Pleistocene carnivorans provides, to our knowledge, the first evidence from the fossil record validating experimental analyses of the relationships among fluctuating asymmetry, phenotypic integration, variance, and environmental change or stress (32, 47). Although attributing these responses to specific environmental events is complicated by uncertainties in the precise dating of pits, extensive tooth wear and fracturing suggest that both C. dirus and S. fatalis may have experienced higher levels of nutritional stress in the Late Pleistocene than is observed in modern carnivorans. The concordance of temporal patterns for most attributes measured here (phenotypic integration, variance, FA/integration correlation) in both C. dirus and S. fatalis and the congruent peaks in FA and tooth damage in C. dirus support the suggestion that Late Pleistocene carnivorans exhibit shifts in FA and variance in response to extrinsic forces (e.g., climate change). Moreover, direct developmental interactions, measured through FA, increasingly mediated the morphological response to environmental change in both species.
Conclusions
Phenotypic integration and modularity are fundamental concepts in evolutionary biology and provide a powerful link between genetics, evolutionary developmental biology, and paleobiology. The vast majority of work on phenotypic integration and modularity has been conducted in model organisms, but recent decades have seen a rapid expansion of comparative analyses assessing the conservation of integration and modules across clades and even through deep time. Fossil data allow for the expansion of this field into exciting new areas, from elucidating developmental patterns in long-extinct taxa and reconstructing developmental and evolutionary dynamics through time to polarizing and assessing trends in phenotypic integration and modularity and rigorously examining the influence of trait interactions in shaping the evolution of diversity. The few studies of phenotypic integration that have dipped into deep time reveal the great potential for incorporating fossil data into this field. We anticipate that future work will increasingly exploit the vast record of past life to address fundamental questions surrounding the evolution and significance of phenotypic integration and modularity.
Materials and Methods
Our dataset is composed of 3D landmark data (Table S3) from undeformed crania of 97 specimens of S. fatalis and 83 specimens of C. dirus from the Rancho La Brea tar pits. Specimens were recovered from four pits ranging from ∼40,000 to ∼13,000 years ago (Table S1 and Fig. 3) (35, 36). Canonical variates analysis (CVA) was used to determine whether cranial shape significantly discriminated pit samples of S. fatalis and C. dirus, with significant differences assessed using Procrustes distances. Repeatability-adjusted matrix correlations were generated for each pitwise sample for overall phenotypic integration and for FA, as well as for comparing overall integration and FA within each pit sample. Variance was measured as total Procrustes distance from mean shape for each pit sample. Integration was measured as relative eigenvalue SD of the trait covariance matrix (66). Additional details on methods are provided in SI Text.
Supplementary Material
Acknowledgments
We thank Neil Shubin and David Jablonski for the invitation to contribute to this timely volume. We thank P. D. Polly, J. Finarelli, and B. Van Valkenburgh for relevant discussions. We thank J. Harris, A. Farrell, S. Cox, and G. Takeuchi at the Page Museum for access to specimens.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403667112/-/DCSupplemental.
References
- 1.Olson EC, Miller RL. Morphological Integration. Univ of Chicago Press; Chicago: 1958. [Google Scholar]
- 2.Berg RL. The ecological significance of corelation pleiades. Evolution. 1960;14:171–180. [Google Scholar]
- 3.Pigliucci M, Preston K. Phenotypic Integration. Oxford Univ Press; Oxford: 2004. [Google Scholar]
- 4.Schlosser G, Wagner GP, editors. Modularity in Development and Evolution. Univ of Chicago Press; Chicago: 2004. [Google Scholar]
- 5.Wagner GP. Homologues, natural kinds and the evolution of modularity. Am Zool. 1996;36(1):36–43. [Google Scholar]
- 6.Wagner GP, Altenberg L. Perspective: Complex adaptations and the evolution of evolvability. Evolution. 1996;50(3):967–976. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 7.Wagner GP, Mezey JG. The role of genetic architecture contraints in the origin of variational modularity. In: Schlosser G, Wagner GP, editors. Modularity in Development and Evolution. Univ of Chicago Press; Chicago: 2004. pp. 338–358. [Google Scholar]
- 8.Arnold SJ. Constraints on phenotypic evolution. Am Nat. 1992;140(Suppl 1):S85–S107. doi: 10.1086/285398. [DOI] [PubMed] [Google Scholar]
- 9.Klingenberg CP. Integration, modules, and development: Molecules to morphology to evolution. In: Pigliucci M, Preston K, editors. Phenotypic Integration. Oxford Univ Press; Oxford: 2004. pp. 213–230. [Google Scholar]
- 10.Klingenberg CP. Studying morphological integration and modularity at multiple levels: Concepts and analysis. Philos Trans R Soc Lond B Biol Sci. 2014;369(1649):20130249. doi: 10.1098/rstb.2013.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goswami A. Cranial modularity shifts during mammalian evolution. Am Nat. 2006;168(2):270–280. doi: 10.1086/505758. [DOI] [PubMed] [Google Scholar]
- 12.Armbruster WS, Pélabon C, Bolstad GH, Hansen TF. Integrated phenotypes: Understanding trait covariation in plants and animals. Philos Trans R Soc Lond B Biol Sci. 2014;369(1649):20130245. doi: 10.1098/rstb.2013.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klingenberg CP. Cranial integration and modularity: Insights into evolution and development from morphometric data. Hystrix. 2013;24:43–58. [Google Scholar]
- 14.Polly PD, Head JJ, Cohn MJ. Testing modularity and dissociation: The evolution of regional proportions in snakes. In: Zelditch ML, editor. Beyond Heterochrony: The Evolution Of Development. Wiley-Liss; New York: 2001. pp. 305–335. [Google Scholar]
- 15.Porto A, de Oliveira FB, Shirai L, De Conto V, Marroig G. The evolution of modularity in the mammalian skull I: Morphological integration patterns and magnitudes. Evol Biol. 2009;36:118–135. [Google Scholar]
- 16.Gerber S. On the relationship between the macroevolutionary trajectories of morphological integration and morphological disparity. PLoS ONE. 2013;8(5):e63913. doi: 10.1371/journal.pone.0063913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber S, Hopkins MJ. Mosaic heterochrony and evolutionary modularity: The trilobite genus Zacanthopsis as a case study. Evolution. 2011;65(11):3241–3252. doi: 10.1111/j.1558-5646.2011.01363.x. [DOI] [PubMed] [Google Scholar]
- 18.Webster M, Zelditch ML. Evolutionary lability of integration in Cambrian ptychopariod trilobites. Evol Biol. 2011;38:144–162. [Google Scholar]
- 19.Webster M, Zelditch ML. Modularity of a Cambrian ptychoparioid trilobite cranidium. Evol Dev. 2011;13(1):96–109. doi: 10.1111/j.1525-142X.2010.00459.x. [DOI] [PubMed] [Google Scholar]
- 20.Claverie T, Patek SN. Modularity and rates of evolutionary change in a power-amplified prey capture system. Evolution. 2013;67(11):3191–3207. doi: 10.1111/evo.12185. [DOI] [PubMed] [Google Scholar]
- 21.Zelditch ML, Sheets HD, Fink WL. The spatial complexity and evolutionary dynamics of growth. In: Zelditch ML, editor. Beyond Heterochrony: The Evolution of Development. Wiley-Liss; New York: 2001. pp. 145–194. [Google Scholar]
- 22.Drake AG, Klingenberg CP. Large-scale diversification of skull shape in domestic dogs: Disparity and modularity. Am Nat. 2010;175(3):289–301. doi: 10.1086/650372. [DOI] [PubMed] [Google Scholar]
- 23.Klingenberg CP, Duttke S, Whelan S, Kim M. Developmental plasticity, morphological variation and evolvability: A multilevel analysis of morphometric integration in the shape of compound leaves. J Evol Biol. 2012;25(1):115–129. doi: 10.1111/j.1420-9101.2011.02410.x. [DOI] [PubMed] [Google Scholar]
- 24.Bennett CV, Goswami A. Does reproductive strategy drive limb integration in marsupials and monotremes? Mamm Biol. 2011;76:79–83. [Google Scholar]
- 25.Goswami A, Weisbecker V, Sánchez-Villagra MR. Developmental modularity and the marsupial-placental dichotomy. J Exp Zoolog B Mol Dev Evol. 2009;312B(3):186–195. doi: 10.1002/jez.b.21283. [DOI] [PubMed] [Google Scholar]
- 26.Kelly EM, Sears KE. Reduced integration in marsupial limbs and the implications for mammalian evolution. Biol J Linn Soc Lond. 2011;102:22–36. [Google Scholar]
- 27.Bell E, Andres B, Goswami A. Integration and dissociation of limb elements in flying vertebrates: A comparison of pterosaurs, birds and bats. J Evol Biol. 2011;24(12):2586–2599. doi: 10.1111/j.1420-9101.2011.02381.x. [DOI] [PubMed] [Google Scholar]
- 28.Young NM, Hallgrímsson B. Serial homology and the evolution of mammalian limb covariation structure. Evolution. 2005;59(12):2691–2704. [PubMed] [Google Scholar]
- 29.Armbruster WS, Pélabon C, Hansen TF, Mulder CPH. Floral integration, modularity, and accuracy: Distinguishing complex adaptations from genetic constraints. In: Pigliucci M, Preston K, editors. Phenotypic Integration. Oxford Univ Press; Oxford: 2004. pp. 23–49. [Google Scholar]
- 30.Murren CJ. Phenotypic integration in plants. Plant Species Biol. 2002;17:89–99. [Google Scholar]
- 31.Pigliucci M. Phenotypic integration in a model organism. In: Pigliucci M, Preston K, editors. Phenotypic Integration. Oxford Univ Press; Oxford: 2004. pp. 155–175. [Google Scholar]
- 32.Badyaev AV, Foresman KR. Evolution of morphological integration. I. Functional units channel stress-induced variation in shrew mandibles. Am Nat. 2004;163(6):868–879. doi: 10.1086/386551. [DOI] [PubMed] [Google Scholar]
- 33.Badyaev AV, Foresman KR, Young RL. Evolution of morphological integration: Developmental accommodation of stress-induced variation. Am Nat. 2005;166(3):382–395. doi: 10.1086/432559. [DOI] [PubMed] [Google Scholar]
- 34.Binder WJ, Valkenburgh BV. A comparison of tooth wear and breakage in Rancho La Brea sabertooth cats and dire wolves across time. J Vertebr Paleontol. 2010;30:255–261. [Google Scholar]
- 35.Meachen JA, O’Keefe FR, Sadleir RW. Evolution in the sabre-tooth cat, Smilodon fatalis, in response to Pleistocene climate change. J Evol Biol. 2014;27(4):714–723. doi: 10.1111/jeb.12340. [DOI] [PubMed] [Google Scholar]
- 36.O'Keefe FR, Binder WJ, Frost SR, Sadlier RW, Van Valkenburgh B. Cranial morphometrics of the dire wolf, Canis dirus, at Rancho La Brea: Temporal variability and its links to nutrient stress and climate. Palaeontol Electronica. 2014 17(1):17.1.17A. [Google Scholar]
- 37.Weisbecker V. Monotreme ossification sequences and the riddle of mammalian skeletal development. Evolution. 2011;65(5):1323–1335. doi: 10.1111/j.1558-5646.2011.01234.x. [DOI] [PubMed] [Google Scholar]
- 38.Weisbecker V, Goswami A, Wroe S, Sánchez-Villagra MR. Ossification heterochrony in the therian postcranial skeleton and the marsupial-placental dichotomy. Evolution. 2008;62(8):2027–2041. doi: 10.1111/j.1558-5646.2008.00424.x. [DOI] [PubMed] [Google Scholar]
- 39.Sears KE, Doroba C, Cao X, Xie D, Zhong S. Molecluar determinants of marsupial integration and constraint. In: Asher RJ, Mueller J, editors. From Clone to Bone: The Synergy of Morphological and Molecular Tools in Palaeobiology. Cambridge Univ Press; Cambridge, UK: 2012. [Google Scholar]
- 40.Sears KE, Bianchi C, Powers L, Beck AL. Integration of the mammalian shoulder girdle within populations and over evolutionary time. J Evol Biol. 2013;26(7):1536–1548. doi: 10.1111/jeb.12160. [DOI] [PubMed] [Google Scholar]
- 41.Maxwell EE, Dececchi TA. Ontogenetic and stratigraphic influence on observed phenotypic integration in the limb skeleton of a fossil tetrapod. Paleobiology. 2012;39:123–134. [Google Scholar]
- 42.Hallgrímsson B, et al. Deciphering the palimpsest: Studying the relationship between morphological integration and phenotypic covariation. Evol Biol. 2009;36(4):355–376. doi: 10.1007/s11692-009-9076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goswami A. Cranial modularity and sequence heterochrony in mammals. Evol Dev. 2007;9(3):290–298. doi: 10.1111/j.1525-142X.2007.00161.x. [DOI] [PubMed] [Google Scholar]
- 44.Koyabu D, et al. Heterochrony and developmental modularity of cranial osteogenesis in lipotyphlan mammals. Evodevo. 2011;2:21. doi: 10.1186/2041-9139-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson LAB. Cranial suture closure patterns in Sciuridae: Heterochrony and modularity. J Mamm Evol. 2013;21(2):257–268. [Google Scholar]
- 46.Klingenberg CP, McIntyre GS. Geometric morphometrics of developmental instability: Analyzing patterns of fluctuating asymmetry with Procrustes methods. Evolution. 1998;52:1363–1375. doi: 10.1111/j.1558-5646.1998.tb02018.x. [DOI] [PubMed] [Google Scholar]
- 47.Badyaev AV, Foresman KR, Fernandes MV. Stress and developmental stability: Vegetation removal causes increased fluctuating asymmetry in shrews. Ecology. 2000;81(2):336–345. [Google Scholar]
- 48.Goswami A, Polly PD. Methods for studying morphological integration and modularity. In: Alroy J, Hunt EG, editors. Quantitative Methods in Paleobiology. Paleontological Society Special Publications; Ithaca, NY: 2010. pp. 213–243. [Google Scholar]
- 49.Hallgrímsson B, Willmore K, Hall BK. Canalization, developmental stability, and morphological integration in primate limbs. Am J Phys Anthropol. 2002;45(Suppl 35):131–158. doi: 10.1002/ajpa.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen TF, Houle D. Measuring and comparing evolvability and constraint in multivariate characters. J Evol Biol. 2008;21(5):1201–1219. doi: 10.1111/j.1420-9101.2008.01573.x. [DOI] [PubMed] [Google Scholar]
- 51.Marcot JD, McShea DW. Increasing hierarchical complexity throughout the history of life: Phylogenetic tests of trend mechanisms. Paleobiology. 2007;33:182–200. [Google Scholar]
- 52.Estevbe-Altava B, Marugan-Lobon J, Botella H, Rasskin-Gutman D. Structural constraints in the evolution of tetrapod skull complexity: Williston's Law revisited using network models. Evol Biol. 2013;40:209–219. [Google Scholar]
- 53.Meloro C, Slater GJ. Covariation in the skull modules of cats: The challenge of growing saber-like canines. J Vertebr Paleontol. 2012;32:677–685. [Google Scholar]
- 54.Randau M, Carbone C, Turvey ST. Canine evolution in sabretoothed carnivores: Natural selection or sexual selection? PLoS ONE. 2013;8(8):e72868. doi: 10.1371/journal.pone.0072868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pelabon C, Armbruster WS, Hansen T. Experiment evidence for the Berg hypothesis: Vegetative traits are more sensitive than pollination traits to environmental variation. Funct Ecol. 2011;25:247–257. [Google Scholar]
- 56.Conner JK, Cooper IA, La Rosa RJ, Pérez SG, Royer AM. Patterns of phenotypic correlations among morphological traits across plants and animals. Philos Trans R Soc Lond B Biol Sci. 2014;369(1649):20130246. doi: 10.1098/rstb.2013.0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lande R. The genetic covariance between characters maintained by pleiotropic mutations. Genetics. 1980;94(1):203–215. doi: 10.1093/genetics/94.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner GP. The influence of variation and of developmental constraints on the rate of multivariate phenotypic evolution. J Evol Biol. 1988;1:45–66. [Google Scholar]
- 59.Wagner GP. Advances in Artificial Life: Third European Conference on Artificial Life. Springer; Berlin: 1995. Adaptation and the modular design of organisms; pp. 317–328. [Google Scholar]
- 60.Schluter D. Adaptive radiation along genetic lines of least resistance. Evolution. 1996;50:1766–1774. doi: 10.1111/j.1558-5646.1996.tb03563.x. [DOI] [PubMed] [Google Scholar]
- 61.Eble G. The macroevolution of phenotypic integration. In: Pigliucci M, Preston K, editors. Phenotypic Integration. Oxford Univ Press; Oxford: 2004. pp. 253–273. [Google Scholar]
- 62.Marroig G, Shirai L, Porto A, de Oliveira FB, De Conto V. The evolution of modularity in the mammalian skull II: Evolutionary consequences. Evol Biol. 2009;36:136–148. [Google Scholar]
- 63.Goswami A, Smaers JB, Soligo C, Polly PD. The macroevolutionary consequences of phenotypic integration: From development to deep time. Philos Trans R Soc Lond B Biol Sci. 2014;369(1649):20130254. doi: 10.1098/rstb.2013.0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goswami A, Polly PD. The influence of modularity on cranial morphological disparity in Carnivora and Primates (Mammalia) PLoS ONE. 2010;5(3):e9517. doi: 10.1371/journal.pone.0009517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunt G. Measuring rates of phenotypic evolution and the inseparability of tempo and mode. Paleobiology. 2012;38:351–373. [Google Scholar]
- 66.Pavlicev M, Cheverud JM, Wagner GP. Measuring morphological integration using eigenvalue variance. Evol Biol. 2009;36:157–170. [Google Scholar]
- 67.Andersen KK, et al. North Greenland Ice Core Project members High-resolution record of Northern Hemisphere climate extending into the last interglacial period. Nature. 2004;431(7005):147–151. doi: 10.1038/nature02805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.