Peng et al. (1) question the evidence for early chicken domestication in Northern China (2) based on ancient DNA sequences obtained from archaeological bones. They point out that the sequences used in ref. 2 contain the primer sequences, and therefore haplotype recombinations were introduced. Although they are correct that the primer sequences were unintentionally included in the analyzed sequences, a reanalysis of the data reduced to the fragment between the primers shows that all conclusions drawn in the original publication (2) are also supported by the revised dataset.
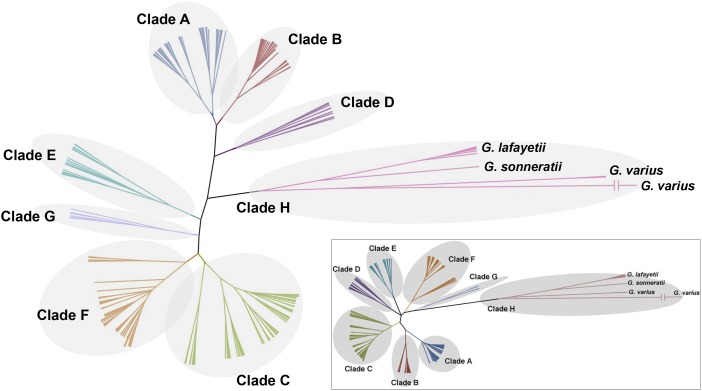
By removing primer sequences from the analyzed control region fragment, the length of the sequences shrinks from 326 to 285 bp. Peng et al. claim that for these 285 bp, many haplotypes cannot be assigned to a haplogroup with confidence. However, they overlooked that we did not use previous haplogroup definitions based on a limited number of sites, but rather, we based haplogroup assignment on phylogenetic results from Bayesian tree reconstruction and network analyses. We redid these analyses, and the structure of the tree stays unchanged, leaving the phylogenetic structure of eight divergent clades (clades A–H) intact (Fig. 1), although the 1,019 sequences analyzed now represent 259 rather than 293 haplotypes (in other words, 34 haplotypes were lost; Table 1). The haplogroup assignments do not change, except for the previous haplotype F2, which is now grouped with E1 and E2 (Table 1). However, these all represent Kenya indigenous chickens, which are of no relevance to the conclusions drawn in ref. 2. Thus, the potential artificial recombination at scored mutation site 246 does not impact our main results (3).
Fig. 1.
Unrooted Bayesian consensus phylogenetic tree of 259 unique haplotypes for the revised 285-bp fragment. (Inset) Original Bayesian tree from the 293 haplotypes for the original 326-bp fragment in ref. 2.
Table 1.
Comparison of the haplotype distributions and haplogroup assignments by removing primer sequences
| Items | A | B | C | D | E | F | G | H | |
| Species | An | 3 (3)/3 (3) | 5 (5)/4 (4) | 1 (1)/1 (1) | |||||
| Dm | 49 (83)/39 (66) | 26 (45)/23 (43) | 33 (80)/29 (78) | 31 (51)/26 (45) | 31 (87)/26 (78) | ||||
| RJF | 3 (3)/3 (3) | 2 (2)/1 (1) | 33 (45)/33 (45) | 18 (18)/18 (18) | 4 (5)/4 (5) | 18 (19)/15 (16) | 6 (6)/6 (6) | ||
| Gs | 13 (20)/12 (20) | ||||||||
| An and RJF | 1 (5)/1 (5) | ||||||||
| Dm and RJF | 2 (159)/2 (166) | 1 (46)/2 (49) | 5 (59)/4 (31) | 3 (6)/3 (13) | 1 (26)/— (—) | ||||
| Dm and Gs | 1 (10)/1 (11) | ||||||||
| An, Dm and RJF | 1 (95)/1 (105) | 1(31) | 1 (6)/— (—) | ||||||
| An, Dm, RJF and Gs | 1 (134)/1 (176) | ||||||||
| Total | 58 (343)/48 (343) | 29 (93)/26 (93) | 77 (194)/72 (194) | 18 (18)/18 (18) | 39 (72)/34 (74) | 53 (273)/43 (271) | 6 (6)/6 (6) | 13 (20)/12 (20) | |
| Incorporated haplotypes | A2, A3, A15, A36, A54 | B1, B27 | C3, C54, C74 | — | E1, E2, F2 | F3, F25, F27, F32, F38, F40, F47, F49 | — | H9, H12 | |
| A10, A46, A53 | B2, B29 | C4, C66 | E18, E39 | F11, F33 | |||||
| A11, A32 | B5, B10 | C5, C69 | E24, E36 | F45, F46 | |||||
| A13, A14 | C49, C57 | E26, E27 | |||||||
| A27, A40 | E30, E31 | ||||||||
| A30, A31 | |||||||||
The data are shown as previous number of haplotype (number of sequences)/present number of haplotype (number of sequences). An, ancient samples; Dm, domestic chickens; Gs, G. sonneratii, G. lafayetii, and G. varius; RJF, red jungle fowls.
In addition, Peng et al. point out that short ancient DNA sequences are insufficient to judge the domestic status of early Holocene samples. In fact, we never concluded that the earliest Holocene chicken remains from northern China represent domestic chicken solely on the basis of the phylogenetic position of the ancient DNA sequences. As we point out in our original publication “…it is—based on genetic analyses alone—of course impossible to prove that the chicken bones analyzed represent domestic rather than wild chicken populations” (2). However, the ancient Chinese chicken sequences along with published ancient and modern sequences document genetic continuity among early chicken populations in northern China and also suggest potential contributions to the gene pool of modern domestic chickens. This result is unaffected by reducing the sequence length of the analyzed fragment from 326 to 285 bp. Even if the mtDNA sequences from Nanzhuangtou and Cishan are from wild junglefowl populations, they support the conclusion that chicken domestication would have been possible in northern China at that time. Thus, although Peng et al. (1) are correct that the length of the sequence fragments needs to be corrected to 285 bp, in contrast to their claims, the ancient DNA results together with the archaeological context still support the up to ∼10,000-y-old Gallus bones from Nanzhuangtou and Cishan being the remains of a population ancestral to at least some of modern chicken mtDNA diversity.
Footnotes
The authors declare no conflict of interest.
References
- 1.Peng MS, Shi NN, Yao YG, Zhang YP. Caveats about interpretation of ancient chicken mtDNAs from Northern China. Proc Natl Acad Sci USA. 2015;112:E1970. doi: 10.1073/pnas.1501151112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiang H, et al. Early Holocene chicken domestication in northern China. Proc Natl Acad Sci USA. 2014;111(49):17564–17569. doi: 10.1073/pnas.1411882111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miao YW, et al. Chicken domestication: An updated perspective based on mitochondrial genomes. Heredity (Edinb) 2013;110(3):277–282. doi: 10.1038/hdy.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]