Significance
Bacteria such as Escherichia coli encounter multiple environments and, as with all organisms, must balance growth and survival when conditions are suboptimal. E. coli uses the RpoS sigma factor, a specialized subunit of RNA polymerase, under conditions of stress or starvation to change the transcriptional program to one that promotes survival. We find that a block in central metabolism, via deletion of a gene encoding pyruvate dehydrogenase, leads to a dramatic increase in RpoS in exponential phase cells. This metabolic cue is likely via reduced amounts of acetyl-CoA. The increase in RpoS is mediated by stabilization of RpoS protein and by translational up-regulation of RpoS synthesis and links this stress response to the state of central metabolism.
Keywords: RssB, ClpXP, acetyl CoA, RpoS, pyruvate dehydrogenase
Abstract
RpoS, the stationary phase/stress sigma factor of Escherichia coli, regulates a large cohort of genes important for the cell to deal with suboptimal conditions. Its level increases quickly in the cell in response to many stresses and returns to low levels when growth resumes. Increased RpoS results from increased translation and decreased RpoS degradation. Translation is positively regulated by small RNAs (sRNAs). Protein stability is positively regulated by anti-adaptors, which prevent the RssB adaptor-mediated degradation of RpoS by the ClpXP protease. Inactivation of aceE, a subunit of pyruvate dehydrogenase (PDH), was found to increase levels of RpoS by affecting both translation and protein degradation. The stabilization of RpoS in aceE mutants is dependent on increased transcription and translation of IraP and IraD, two known anti-adaptors. The aceE mutation also leads to a significant increase in rpoS translation. The sRNAs known to positively regulate RpoS are not responsible for the increased translation; sequences around the start codon are sufficient for the induction of translation. PDH synthesizes acetyl-CoA; acetate supplementation allows the cell to synthesize acetyl-CoA by an alternative, less favored pathway, in part dependent upon RpoS. Acetate addition suppressed the effects of the aceE mutant on induction of the anti-adaptors, RpoS stabilization, and rpoS translation. Thus, the bacterial cell responds to lowered levels of acetyl-CoA by inducing RpoS, allowing reprogramming of E. coli metabolism.
RpoS is a central sigma factor in Escherichia coli involved in gene expression in the stationary phase but also under a large number of stress conditions (1). As with many global transcriptional regulators, RpoS is highly regulated at the transcriptional, translational, and posttranslational levels. Under normal growth conditions, the translation of RpoS is blocked by a secondary structure in the 5′ UTR; this is relieved in response to various signals by the action of small RNAs (sRNAs) (reviewed in ref. 1). In addition, RpoS is rapidly degraded during exponential phase by the ClpXP protease. For RpoS proteolysis to occur, it must interact with RssB, an adaptor protein, which allows recognition of RpoS by the protease. Under stress conditions, RpoS is stabilized, allowing it to rapidly accumulate to activate the RpoS regulon.
Mechanisms leading to RpoS stabilization under stress conditions were not understood for many years. However, the discovery of three small proteins, IraP, IraD, and IraM, which are able to interact with RssB to stabilize RpoS, has provided a major insight into the regulatory pathways inhibiting RpoS proteolysis. These proteins, called anti-adaptor proteins, are each induced under a different stress condition (2). IraP, induced via an increase in ppGpp (3), is necessary for stabilization of RpoS during low phosphate growth. IraM, induced via activation of the PhoQ/PhoP two-component system, is similarly critical for RpoS stabilization during low Mg2+ growth (2). IraD induction is not fully understood, but it clearly plays a role both in the transition to stationary phase, via induction of a ppGpp-dependent promoter, and after DNA damage (4, 5). IraM and IraD are also negatively regulated by H-NS (6).
Regulation of RpoS is also mediated by three sRNAs that positively regulate translation. Each of these sRNAs requires the RNA chaperone Hfq for function, and each pairs with the 5′ UTR of RpoS to make the ribosome-binding site accessible. However, the sRNAs have very different induction pathways. DsrA is synthesized at low temperatures (7, 8). RprA is synthesized in response to activation of the Rcs phosphorelay under conditions of cell-surface stress (9). The third sRNA, ArcZ, is negatively regulated by the ArcB/ArcA two-component system under anaerobic conditions; therefore, ArcZ is more abundant under aerobic conditions (10).
Even with these recent advances, much remains to be discovered about when and how RpoS levels are regulated. For example, translational up-regulation at stationary phase has been reported, independent of the upstream hairpin that is the target for sRNAs (11). RpoS-dependent genes are known to help the cell to respond to a large number of stress conditions and metabolic shifts, suggesting a need for increased synthesis of RpoS and its stabilization under many conditions. Here we find that, in exponential phase cells, perturbing central metabolism by decreasing the pool of acetyl-CoA leads to dramatically increased levels of RpoS. This increase integrates two independent regulatory pathways: RpoS stabilization via the production of two of the known anti-adaptors and induction of RpoS translation. These results demonstrate both a previously unexplored signal for the RpoS general stress response and a unique coupling in signaling between translational regulation and protein stabilization.
Results
RpoS Levels Are Increased in an ΔaceE Mutant.
Lycopene is a red pigment with antioxidative properties that can be produced in microorganisms. It is made in E. coli via the isoprenoid pathway, with the final steps carried out by enzymes cloned from other sources such as Erwinia (reviewed in refs. 12 and 13). Research on optimal conditions for lycopene expression in E. coli has included screening various gene knockouts and overexpression strains. Surprisingly, many regulators of RpoS degradation were identified using these screens. In one study, overexpression of the anti-adaptors IraD (yjiD) and IraM (ycgW), as well as AppY [also known to lead to RpoS stabilization (2)] and RpoS itself were each identified as improving lycopene formation, suggesting that increasing RpoS contributes to higher lycopene biosynthesis (14). In that study, as well as related ones, the deletion of the gene encoding RssB, the adaptor for RpoS degradation (called in ref. 15 hnr), also improved lycopene formation. Therefore, we began this study by asking if other mutants that increase lycopene formation also acted by increasing RpoS synthesis or stability. We focused on gdhA, aceE, and fdhF mutants (14–16). The deletion of fdhF and gdhA had no effect on RpoS stability, nor on the level of RpoS at 0′ (Fig. S1A). These gene deletions thus presumably act independently of increased RpoS in the lycopene synthesis pathway.
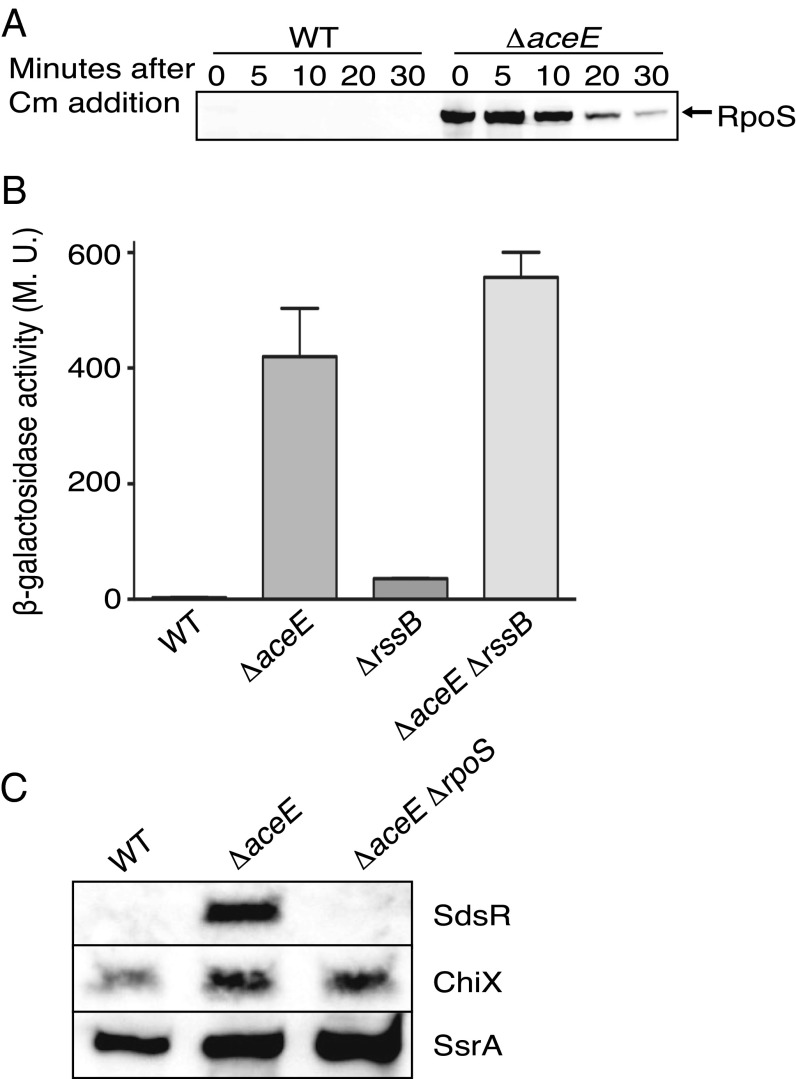
However, a deletion of aceE, encoding one of the subunits of pyruvate dehydrogenase, had a drastic effect on RpoS levels (Fig. 1A). RpoS was barely visible at t = 0 min in a WT strain at the low OD600 of 0.3 at which this experiment was done, but was abundant at 0′ and still present after a chase of 30 min in the ΔaceE background; the half-life of RpoS in midexponential phase changed from 3′ to more than 10′. The high level of RpoS at 0′ suggested that ΔaceE might also increase synthesis of RpoS. This was tested using rpoS-lacZ transcriptional and translational fusions, again measured in exponential phase. There were very modest effects on the transcriptional fusion (Fig. S1B). However, a translational fusion that is also subject to RssB-dependent degradation was almost undetectable in the wild-type background and was increased 140-fold in the aceE background (Fig. 1B). The same experiment was repeated in a strain in which RpoS degradation was blocked by inactivating rssB. Mutating rssB increased the expression of RpoS-Lac 12-fold; however, deleting aceE increased expression of the fusion protein another 15-fold (Fig. 1B). Therefore, there are two additive and independent effects on RpoS of mutating aceE: a 15-fold increase in synthesis and a further 10-fold increase due to stabilization. Because we saw very little increase for the transcriptional fusion, we conclude that the increase in synthesis reflects increased translation.
Fig. 1.
Mutations in aceE lead to increased RpoS levels. (A) WT (MG1655) and ΔaceE (BA334) were grown to an OD600 of ∼0.3, protein synthesis was inhibited by chloramphenicol (Cm) addition, and RpoS levels were analyzed by Western blotting using an anti-RpoS antiserum. (B) Strains containing rpoS-lacZ translational fusions (WT, SG30013; ΔaceE, BA455; ΔrssB, SG30018; ΔrssB ΔaceE, BA679) were grown to an OD600 of ∼0.3 and assayed for β-galactosidase activity. This fusion has the promoters of rpoS and enough of the RpoS ORF to be subject to RssB-dependent degradation. The mean from three replicates is presented, and the standard error of the mean (SEM) is indicated by the error bars. M.U., Miller units. (C) Cells were grown to an OD600 of 0.2 in LB, and samples were removed for RNA analysis. Samples were probed with biotinylated probes for SdsR (Top), ChiX (Middle), or SsrA (Bottom). Strains: WT (NM1100), ΔaceE::kan (BA334), and rpoS::tet ΔaceE::kan (BA530).
If the increased RpoS in the aceE mutant is active, it should be reflected in expression of RpoS-dependent RNAs; this was measured using the RpoS-dependent sRNA SdsR (17). RNA was isolated from wild-type, aceE, and aceE rpoS mutant cells grown to an OD600 of 0.3 and probed for SdsR, as well as for the sRNAs not under RpoS control, ChiX and SsrA (Fig. 1C). SdsR was undetectable in the wild-type cells at this OD, consistent with the expected very low levels of RpoS (Fig. 1A), but was expressed in the aceE mutant in an RpoS-dependent fashion. Therefore, the induced RpoS in the ΔaceE strain is active. As expected, ChiX and SsrA levels were not significantly affected by the aceE mutant (Fig. 1C).
RpoS Stabilization in the aceE Mutant Is Dependent upon Induction of Anti-Adaptors IraP and IraD.
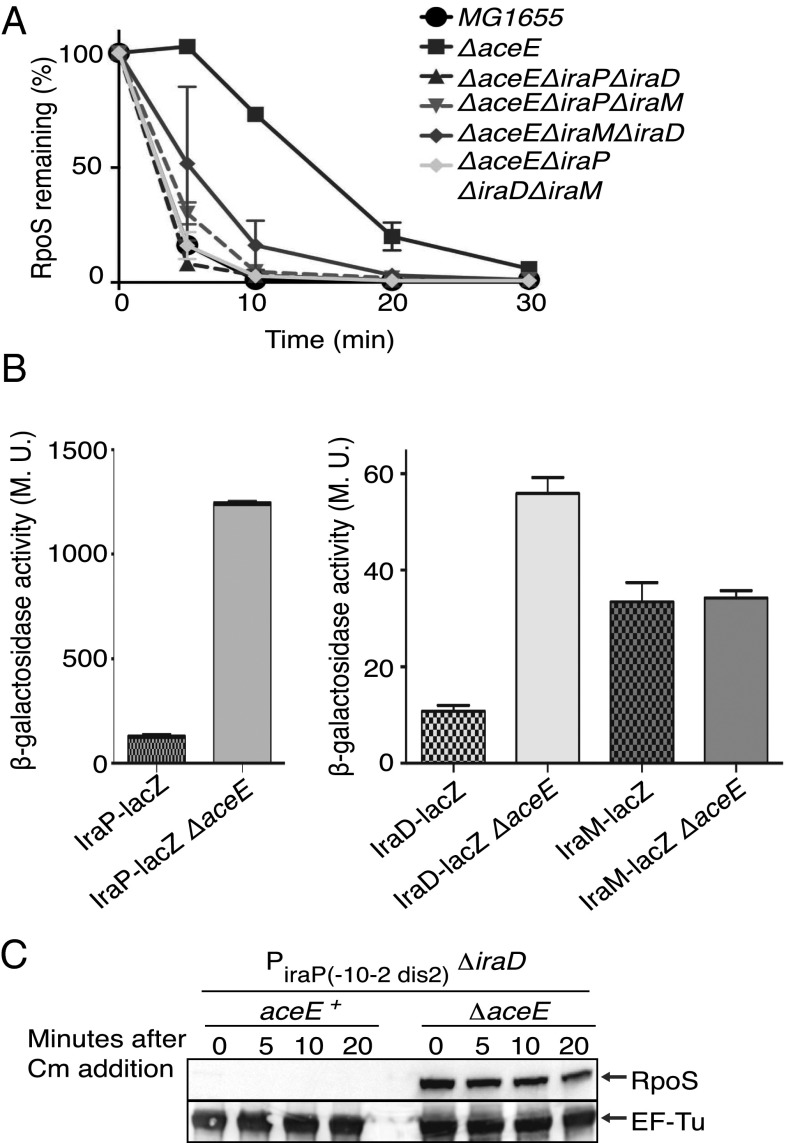
RpoS degradation by the ClpXP protease depends on the adaptor protein RssB. The major mechanism that leads to RpoS stabilization under different stress conditions is inactivation of RssB adaptor activity by anti-adaptor proteins IraP, IraD, and IraM (2, 18, 19). Thus, we investigated if the known anti-adaptors play a role in the stabilization of RpoS in the aceE::kan background. As shown in Fig. 2A, the deletion of all three genes coding for the anti-adaptors restored RpoS degradation in an aceE::kan mutant to the rate seen in the wild-type strain. Single and double mutants of the anti-adaptors in the ΔaceE background suggest that IraP and to a less extent IraD mediate stabilization of RpoS in the ΔaceE strain (Fig. 2A and Fig. S2A). Strains deleted for iraD and iraP or all three anti-adaptors still have a high level of RpoS at the initial time point (0 min) even though RpoS degradation was similar to that seen in the aceE+ host (Fig. S2A), consistent with the increase in translation seen in the aceE mutant (Fig. 1B).
Fig. 2.
RpoS stabilization in an aceE mutant is dependent on the anti-adaptors IraP and IraD. (A) Strains were grown and RpoS levels analyzed by Western blotting as in Fig. 1A. The RpoS level was quantified, with intensity measured at time 0 for each strain set at 100%. The mean from three replicates is presented, and the error bars indicate the SEM. The following strains were used: WT (MG1655); ΔaceE (BA334); and the isogenic derivatives ΔaceE ΔiraP ΔiraD (BA449), ΔaceE ΔiraP ΔiraM (BA447), ΔaceE ΔiraD ΔiraM (BA467), and ΔaceE ΔiraP ΔiraD ΔiraM (BA385). The gels and other mutants are in Fig. S2A. (B) Strains containing transcriptional fusions iraP-lacZ (WT, AB060; ΔaceE, BA407), iraD-lacZ (WT, AB050; ΔaceE, BA411), or iraM-lacZ (WT, AB042; ΔaceE, BA409) were grown in LB to an OD600 of ∼0.3 and assayed for β-galactosidase activity. The mean from three replicates is presented; the SEM is indicated by the error bars. M.U., Miller units. The level of expression of the iraP fusion is significantly higher than that for the other two fusions. (C) Strains were grown as in Fig. 1A, and RpoS levels were analyzed by Western blotting using anti-RpoS antiserum (Upper) or anti–EF-Tu antiserum (Lower). The following strains were used: −10–2 dis2 iraP ΔiraD::tet (BA946) and the isogenic aceE derivative (BA948).
Anti-adaptors act by interacting with the limiting levels of RssB and preventing the interaction of RssB with RpoS (19). Therefore, a stabilization mechanism dependent on anti-adaptor titration of RssB can be bypassed by increasing the levels of RssB. Consistent with the aceE mutant acting via anti-adaptors, increasing the level of RssB also bypassed the aceE effect on stabilization (Fig. S2B).
RssB is an orphan response regulator in E. coli and can be phosphorylated on a conserved aspartate residue, D58. Mutating the D58 residue of RssB to A or P decreases but does not eliminate RpoS degradation in vivo and is still subject to stabilization by the anti-adaptors (2, 18–20). The small-molecule acetyl phosphate can phosphorylate RssB and stimulate degradation in vivo and in vitro (19, 20). Acetyl phosphate is synthesized from acetyl CoA, and one major pathway for acetyl CoA synthesis is dependent upon the product of the aceE gene. Therefore, we examined the dependence of the stabilization by the aceE mutation on RssB phosphorylation. This was tested by comparing RpoS stability in a strain expressing rssBD58P, mutated at the site of phosphorylation, to stability in an rssBD58P ΔaceE double mutant. As expected, the strain with the RssBD58P mutant was not fully functional; the half-life of RpoS was significantly longer than that of the wild-type strain (half-life of 8′, Fig. S2C). However, RpoS was more stable in the aceE::kan strain than in the rssBD58P mutant strain, and the effects of rssBD58P and aceE::kan were additive (half-life in double mutant of 19′). Therefore, the major effect of an aceE mutant in stabilizing RpoS is via the anti-adaptors, and not due to loss of RssB phosphorylation.
IraP and IraD are normally present in the cell at low levels; their synthesis increases enough to stabilize RpoS only under specific stress conditions (2, 4, 18). The fact that IraP and IraD stabilize RpoS in an aceE::kan mutant suggests that their expression is induced in this background.
Induction of the promoters for these anti-adaptors was followed by using transcriptional fusions. The aceE deletion increased the expression of iraP 10-fold and the expression of iraD 5-fold; iraM expression was unchanged by the aceE mutant (Fig. 2B). Both iraP and iraD are positively regulated by ppGpp (3, 5), suggesting that an increase in this alarmone might explain the induction of these anti-adaptors. Direct measurement of ppGpp in the aceE host was not possible because the strain grew very poorly in the 3-(N-morpholino)propansulfonic acid medium needed for the experiment. However, the promoter of iraP itself acts as a measure of ppGpp. It is strongly positively regulated by ppGpp, and mutations in the iraP discriminator abolish the response to ppGpp (3). Consistent with an increase in ppGpp in the aceE mutant, the induction of the iraP transcriptional fusion in an aceE mutant was abolished in a fusion carrying the ppGpp-insensitive discriminator mutant dis2 (changed from AT-rich to GC-rich) (3) (Fig. S3A). The dis2 mutant reduces the basal level of iraP expression significantly, as previously seen (3), but this reduced level was not increased at all in the aceE mutant. Therefore, these results suggest that ppGpp increases in the aceE mutant and in turn induces the promoters of iraP and iraD.
The contribution of ppGpp was further examined in a strain devoid of ppGpp (ppGpp0). In E. coli, RelA mediates ppGpp synthesis during amino acid starvation; SpoT synthesizes but also degrades ppGpp and has been implicated in ppGpp synthesis after various stress treatments (21, 22). The triple mutant (aceE relA spoT) was unable to grow without the addition of acetate (Fig. S3B). The strain was grown in the presence of acetate, acetate washed out, and the stability of RpoS was then examined. Unexpectedly, RpoS was still stable in this strain in the absence of ppGpp (Fig. S3C).
We considered the possibility that, in addition to induction of ppGpp-dependent promoters, there was posttranscriptional induction of the anti-adaptors. This was examined for IraP. Induction of IraP tagged at its C terminus with SPA was examined in wild-type and aceE mutants in both a strain carrying a wild-type iraP promoter and a strain carrying the dis2 iraP promoter mutant, rendering it no longer positively regulated by ppGpp (3). IraP-SPA levels were induced in the aceE mutants even with the dis2 iraP promoter (Fig. S3D). RpoS was partially stabilized even in the dis2 mutant (Fig. S3D). To bypass the low expression of IraP from the dis2 promoter, an additional experiment was done with an up mutation in the −10 region (−10-2), combined with the dis2 mutant (3). In a transcriptional fusion, neither the −10-2 mutant nor the −10-2 dis2 mutant showed further induction in the aceE mutant, confirming the absence of transcriptional induction of IraP from these promoters (Fig. S3E). This same −10-2 dis2 promoter was then used to drive native IraP in isogenic aceE+ and aceE strains; strains were also mutated for iraD so that any stabilization would depend on IraP (Fig. 2C). RpoS was stable in the aceE mutant, but not in the aceE+ parent. Therefore, we conclude that the aceE mutant leads both to ppGpp induction of the iraP promoter and to posttranscriptional induction of IraP, probably by increased translation. This additional induction likely occurs in the ppGpp0 strain as well because RpoS is still stable in that strain.
Translational Up-Regulation of rpoS in aceE Mutants Is Not via the Known sRNA-Dependent Pathway and Is Independent of Most of the rpoS Leader.
RpoS accumulates in the aceE mutant due to increased translation in addition to stabilization of the protein (Fig. 1B, rssB::tet strains, and Fig. S2A, 0′ point). rpoS has a long untranslated leader, and RpoS translation is positively regulated in response to different environmental signals via the action of three different sRNAs; each requires the RNA chaperone Hfq for function (reviewed in ref. 1). Initially, an rpoS-lacZ translational fusion was evaluated for the effects of each of the sRNAs on expression in a ΔaceE mutant (Table S1); an rssB mutant derivative was used to stabilize the fusion protein. Our expectation was that if one or more of the sRNAs were responsible for increased translation in the aceE mutant, deleting the sRNA(s) should abolish the induction. As in previous experiments (10), deletion of dsrA from the aceE+ parent had the biggest effect under these growth conditions, reducing RpoS-LacZ expression significantly. Single mutations in either arcZ or rprA had very little effect. A triple mutant was somewhat more defective than the dsrA deletion alone. However, none of these deletions, including the arcZ dsrA rprA triple mutant, abolished the effect of an aceE mutant. In these experiments, the ratio of aceE−/aceE+ was 4 for the wild-type strain; in the triple mutant the ratio was 20, reflecting the low level of the aceE+ strain. Therefore, the induction of translation of RpoS by aceE is apparently not by the action of any of these Hfq-dependent sRNAs.
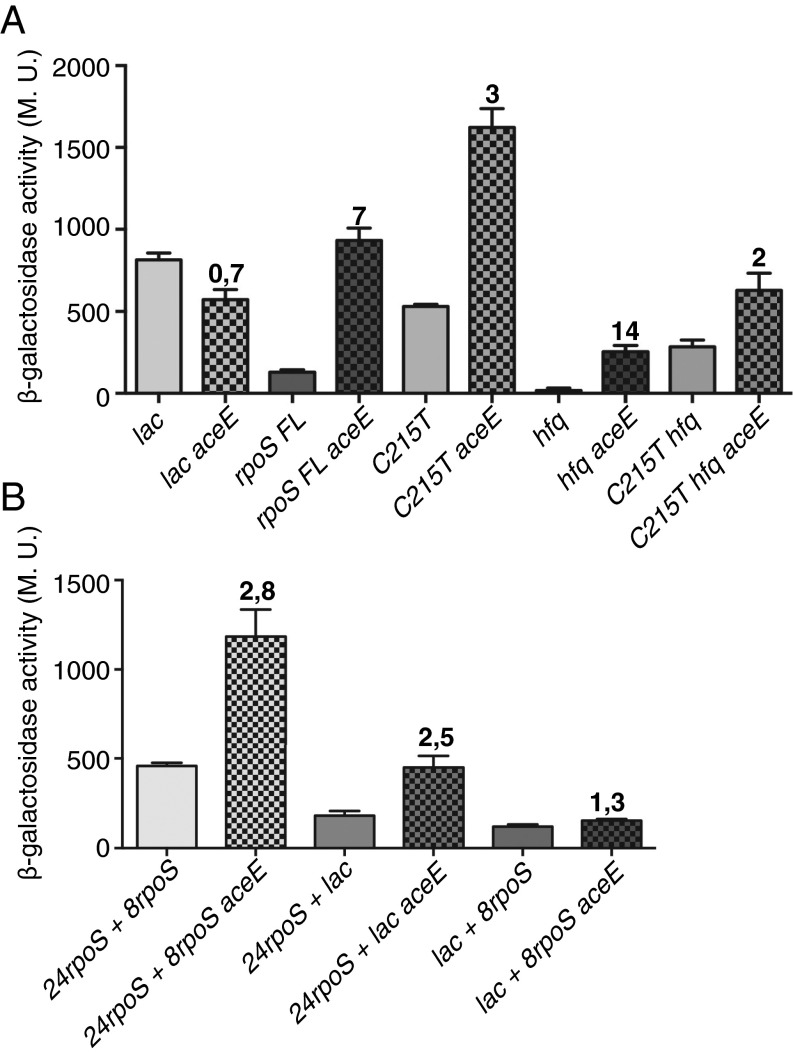
For further analysis of how aceE induces RpoS synthesis, a set of fusions, all expressed from the constitutive Cp17 promoter (23), were constructed and assayed in aceE+ and aceE− backgrounds. The parent fusion carries the full-length leader and 477 nt of the rpoS-coding region, deleting the region necessary for RpoS degradation (24). Thus, levels of RpoS-lac should reflect changes only in mRNA stability and translation. In this strain, an aceE mutant increases expression more than fivefold at OD600 0.3 (Fig. 3A). A control fusion of the promoter to the lac leader and lacZ had a ratio of aceE−/aceE+ of 0.7 (Fig. 3A). A mutation, C125T, which disrupts the inhibitory hairpin and renders translation independent of Hfq (25) (Fig. 3A), increased expression in the wild-type strain by fourfold. An aceE mutant increased expression by an additional threefold. Therefore, by this test the aceE effect is independent of the hairpin, and the hairpin opening is additive with the effect of the aceE mutant (Fig. 3A). We also looked at the effect of an hfq deletion on the Cp17 fusions. The aceE deletion increased expression in the hfq mutant by 14-fold (Fig. 3A). The C125T hfq fusion was increased twofold in the aceE mutant; why this is somewhat less than the C125T fusion is not clear. Overall, these experiments confirm the results in Table S1, ruling out a critical role for sRNAs, Hfq, and the rpoS inhibitory hairpin in the stimulation of RpoS in an aceE mutant.
Fig. 3.
Regions in rpoS needed for translational induction in the aceE mutant. All strains carried an RpoS-LacZ translational fusion driven by a Cp17 promoter; the portion of RpoS present does not include the region necessary for RssB-dependent degradation. Without the promoter there was no expression of lacZ. (A) Cells were grown to an OD600 of 0.3 and were assayed for β-galactosidase. Isogenic pairs of aceE+ and aceE::kan strains were assayed. Strains used were the following: lac (control fusion, with 38-nt leader of lac and lacZ: BA926, BA928); rpoS FL (full-length 567-nt leader of rpoS and 477 nt of rpoS ORF: BA938, BA940); C125T (derivative of rpoS FL with leader mutation C125T, disrupting hairpin: BA942, BA944); hfq (Δhfq derivatives of rpoS FL: BA960, BA984); C125T hfq (Δhfq derivatives of C125T rpoS FL: BA962, 986). (B) Cells were grown and assayed as for A. Strains used were the following: 24rpoS + 8rpoS (BA964, BA966); 24rpoS + lac (BA992, BA996); and lac + 8rpoS (BA994, BA998).
Further fusions were examined to identify the region necessary for the response to the aceE mutant. A fusion carrying the same Cp17 promoter but only 24 nt of the leader and 24 nt of the translated rpoS gene (first eight codons) lacks the inhibitory hairpin and therefore had a higher basal level of expression. The aceE mutant increased this similarly to the open hairpin derivative (Fig. 3B, 24rpoS + 8rpoS, 2.8-fold). These results suggest that the major effect of the aceE mutant on translation/mRNA stability is likely specific to sequences near the sites for the initiation of translation. As a comparison, parallel fusions were constructed expressing the 24 nt of the rpoS leader fused to lacZ at the ATG (24rpoS +lac) and a reciprocal fusion containing the lac leader and eight codons of rpoS (lac + 8rpoS) (Fig. S4). The 24 nt of the rpoS leader were sufficient to give 2.5-fold induction in the aceE mutant, whereas the reciprocal fusion gave no increase (1.3-fold) in the aceE strain (Fig. 3B). Therefore, the region immediately before the ATG is necessary for the effect of ace, and the codons within the ORF do not contain essential information. Because the aceE induction in the full-length fusion (Fig. 3A) was considerably higher (7× rather than 3×), additional information may be contained elsewhere.
Thus, both IraP and RpoS are subject to translational up-regulation in the aceE mutant. This effect is specific; lacZ translation was not similarly up-regulated. The sequences that allow this up-regulation are close to the translational initiation site, at least for RpoS (Fig. 3B).
Metabolic Defects in the aceE Mutant.
aceE encodes the E1 component of pyruvate dehydrogenase (PDH), an enzyme complex that converts pyruvate to acetyl-CoA, critical for the TCA cycle, for fatty acid synthesis and for synthesis of the signaling molecule acetyl-phosphate. The other two subunits of PDH are encoded by aceF and lpd; the lipoamide dehydrogenase (LPD) subunit of PDH, encoded by lpd, is also part of the 2-oxogluatarate dehydrogenase and glycine cleavage system complexes. To examine whether it is the absence of AceE itself or loss of PDH that leads to increased RpoS, mutations in aceF and lpd, as well as a mutation in lipA encoding lipoate synthetase, necessary for all LPD activities, were compared with the aceE mutation for effects on the rpoS-lacZ fusions. Each of these mutations increased the expression of the rpoS-lacZ fusion similarly to that seen for aceE (Fig. S5 A and B), suggesting that loss of PDH activity and thus loss of acetyl-CoA or a downstream product of acetyl-CoA leads to RpoS induction.
One of the major consequences of reduced acetyl-CoA is reduced fatty acid synthesis. It has previously been shown that starvation for fatty acids leads to elevated ppGpp via an interaction of the Acyl carrier protein with SpoT, demonstrating the global regulatory effect of this metabolic change (26). In addition, fabH is synthetically lethal in strains unable to synthesize ppGpp (27), as is aceE (Fig. S3B). Thus, it seemed possible that the inducing signal in the aceE mutant was decreased fatty acid synthesis. A mutation in fabH will block use of acetyl-CoA to make acetoacetyl-ACP, an early step in fatty acid synthesis. The fabH mutation also induced the fusion, although not to the same extent as the mutations in PDH (Fig. S5 A and B). All of the mutations that block PDH activity also grew very slowly; fabH growth was intermediate between the PDH mutants and wild type, correlating with the less dramatic induction of RpoS (Fig. S5C). Thus, a defect in fatty acid synthesis partially mimics the PDH mutant.
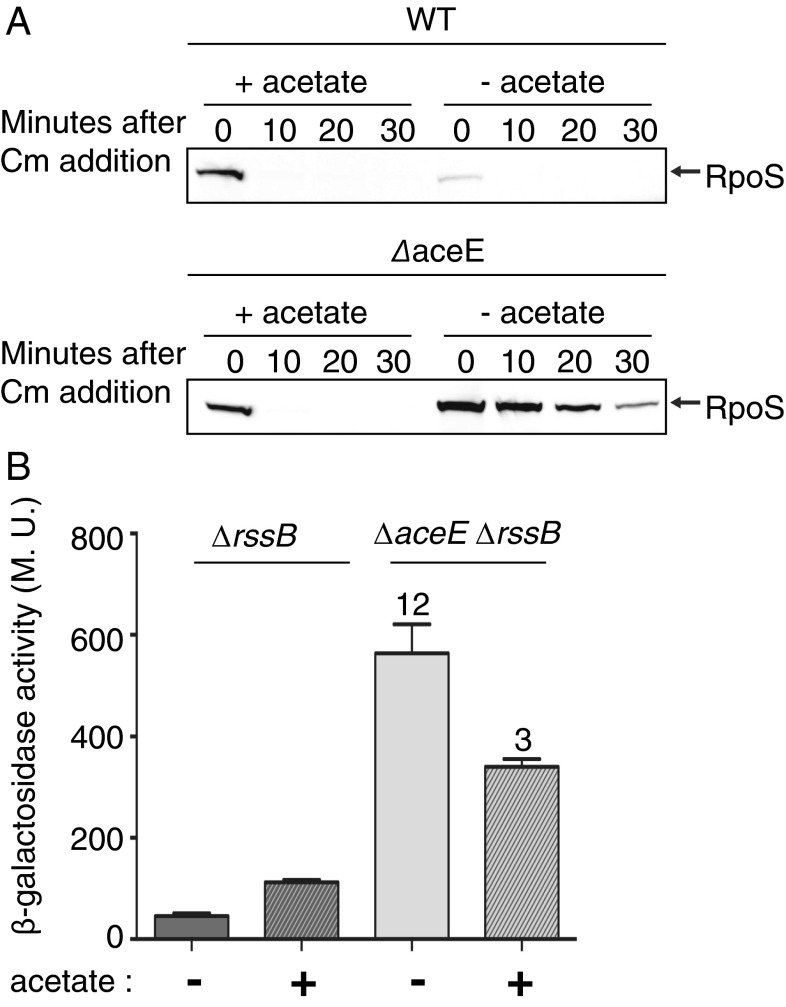
Alternative pathways for synthesis of acetyl-CoA from acetate exist in E. coli. One uses acetyl-CoA synthetase (Acs); a parallel pathway uses Pta and AckA when external acetate levels are high. If lowered levels of acetyl-CoA are responsible for the signals leading to RpoS induction, adding acetate to the medium should improve acetyl-CoA levels and reverse induction of RpoS. As seen in Fig. S6A, acetate improved growth. Addition of acetate also led to reduced stability of RpoS (Fig. 4A) and reduced expression of both iraP and iraD transcriptional fusions (Fig. S6B). In addition, the translational induction of RpoS was partially reversed (Fig. 4B). Addition of other carbon sources that do not directly improve acetyl-CoA synthesis (succinate and oleate) did not mimic the effect of acetate in reversing RpoS stability (Fig. S6 C and D) (28). These results are consistent with a depletion of acetyl-CoA or its products as the major defect in the aceE mutant.
Fig. 4.
Acetate overcomes effect of aceE mutant. (A) WT (MG1655) and ΔaceE (BA334) strains were grown and RpoS levels were analyzed as for Fig. 1A. (B) Strains contain the rpoS-lacZ translational fusion used in Fig. 1B (under the control of rpoS promoters and subject to RssB-dependent degradation). ΔrssB (SG30018) and ΔaceE ΔrssB (BA679) strains were grown at 37° in LB in the absence or presence of acetate (30 mM) and assayed as for Fig. 1B.
Discussion
RpoS is used by bacteria to respond to a variety of starvation conditions and stresses in what has been termed a “general stress response.” Under these conditions, the level of RpoS increases, reflecting stabilization of the protein and increased translation. RpoS-dependent genes encode functions that help the cell repair damage and await better times. Because of the range of RpoS-dependent functions, one RpoS-inducing stress can provide cross-resistance to many others (reviewed in ref. 1). As cells enter stationary phase, some combination of changes (starvation, accumulation of toxic by-products, other changes) lead to higher levels of RpoS. The pathways for inducing RpoS during stationary phase in complex medium have not been fully defined, but include increased translation and decreased degradation.
Here, we find that a mutation in the gene encoding a subunit of PDH leads to induction of RpoS in exponential phase; our studies were primarily in cells mutant for aceE, encoding the E1a subunit of PDH. This increase resulted from decreased RpoS degradation and increased translation. RpoS was stabilized via two anti-adaptor proteins, IraP and IraD. Induction of these anti-adaptors is in part likely due to increased levels of ppGpp, a positive regulator of both the iraP and iraD promoters (3, 5). However, even in the absence of ppGpp (Fig. S3C) or in a cell carrying a ppGpp-independent mutation in the iraP promoter (Fig. S3D), RpoS is stabilized. IraP translation is apparently also up-regulated in the aceE mutant sufficiently to stabilize RpoS, although the basis for this up-regulation is not yet understood.
The second component of induction is high-level translation of RpoS. Surprisingly, the activation of translation was independent of the well-studied small regulatory RNAs that are responsible for translational induction of RpoS translation under other growth conditions (Table S1). Activation of translation was also independent of the long rpoS leader where these sRNAs act; a fusion that contains only the final 24 nt of the rpoS leader was still induced in an aceE mutant (Fig. 3B). Hirsch and Elliott used a similarly short rpoS translational fusion (24 nt of leader and eight codons of RpoS) and observed stationary phase induction (11).
Thus, perturbing the metabolism of pyruvate to acetyl-CoA has uncovered a previously unrecognized signaling pathway for high level induction of RpoS. PDH uses pyruvate, CoASH, and NAD to synthesize acetyl-CoA, a precursor for a variety of cellular functions, including the TCA cycle and fatty acid synthesis. Acetyl-CoA also is the precursor for the signaling molecule acetyl-phosphate, which can also be metabolized to acetate, thus providing a pathway from pyruvate, via PDH, to acetate. An alternative pathway from pyruvate to acetate uses the enzyme PoxB, which is expressed in an RpoS-dependent fashion (29). Although PoxB does not itself synthesize acetyl-CoA, AMP-forming Acs can use the acetate formed by PoxB and synthesize acetyl-CoA (30, 31). Thus, one consequence of the induction of RpoS in PDH-deficient cells is likely to be expression of higher levels of PoxB, increasing the alternative route for acetate and acetyl-CoA synthesis. Another consequence may be that PoxB, unlike PDH, does not convert NAD to NADH.
Although our results are consistent with lack of acetyl-CoA as the primary signal for RpoS induction, inhibition of the fatty acid synthesis pathway in a fabH mutation also leads to an intermediate level of RpoS induction (Fig. S5 A and B). A fabH mutation will block use of acetyl-CoA to make acetoacetyl-ACP, an early step in fatty acid synthesis; therefore, a product beyond FabH may be what is sensed in these mutants.
The role of ppGpp in the aceE stress response is not entirely clear. We, as well as others (R. Harinarayanan and M. Cashel), see synthetic lethality of a strain unable to synthesize ppGpp in the aceE mutant. It has also been reported that fabH is synthetically lethal with a ppGpp null strain (27). Therefore, interference with synthesis of acetyl-CoA or the resulting defect in fatty acid synthesis is likely to induce ppGpp synthesis and may be the lethal event in ppGpp-null cells. Although ppGpp certainly contributes to RpoS induction via induction of the anti-adaptors IraP and IraD in the aceE mutant, our results suggest that additional levels of regulation allow the cell to induce RpoS independently of ppGpp.
As noted above, cells mutant in aceE grow slowly (Fig. S5C), consistent with a general decrease in protein synthesis. If translation is limited in these cells, and RpoS (and IraP) escape this limitation, this would explain the rapid increase in their relative abundance. There are a number of other examples of such selective translation. After cold shock, specific genes escape the general translation slowdown (32). Some ribonuclease toxins encoded by toxin/antitoxin pairs cut at specific sequences in mRNAs, degrading most but not all mRNAs. We suggest that, in the absence of acetyl-CoA or one of its downstream products, RpoS translation is designed to escape, allowing the cell to partially deal with the metabolic defect. The mechanistic basis of this selectivity will be the subject of future research.
Materials and Methods
Media and Growth Conditions.
Cells were grown in Lennox broth (LB) at 37 °C with aeration. When indicated, sodium acetate was added to a final concentration of 30 mM (adjusted to pH 7.0).
Strains and Plasmids.
All strains used are derivatives of MG1655 or DJ480 (MG1655 ΔlacX74) (33); strains were made either by P1 transduction, selecting for the appropriate antibiotic resistance marker, or by lambda Red recombineering (34) as outlined in Table S2 and SI Materials and Methods. Primers are listed in Table S3.
The Cp17 synthetic promoter sequence is from Jensen and Hammer (23); the iraP promoter mutations were described previously (3). The piraD2-lacZ fusion starts at the P2 promoter of iraD at −137 and extends through the entire ORF into the 10th codon of lacZ.
Assay for in Vivo RpoS Degradation.
Cells were grown overnight in LB medium, diluted into fresh LB medium at an OD600 of ∼0.01, and grown to midlogarithmic phase (OD600 of ∼0.3) at 37 °C. Chloramphenicol (200 μg/mL) or tetracylcline (100 μg/mL) was added. Samples were analyzed as described by Battesti et al. (19). Values presented are the mean of at least three independent assays.
Northern Blots.
Cultures were grown to an OD600 of ∼0.3, and RNA was extracted by the hot phenol method (35). RNAs were separated on an acrylamide gel, transferred to a nylon membrane, probed, and developed as described (36). Biotinylated probe sequences are listed in Table S3.
β-Galactosidase Assay.
Triplicate cultures were grown in LB medium at 37 °C and assayed using the assay described by Miller (37). Averaged values are presented.
Supplementary Material
Acknowledgments
We thank M. Cashel, P. Moreau, P. Mandin, and members of our laboratory for comments on the manuscript; E. Bouveret for discussion of the work; M. Cashel and R. Harinarayanan for sharing unpublished results; and F. Barras for providing facilities and support during a portion of this work. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. A.B. was supported in part by the Fondation pour la Recherche Medicale.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504639112/-/DCSupplemental.
References
- 1.Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S. Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol. 2008;68(2):298–313. doi: 10.1111/j.1365-2958.2008.06146.x. [DOI] [PubMed] [Google Scholar]
- 3.Bougdour A, Gottesman S. ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc Natl Acad Sci USA. 2007;104(31):12896–12901. doi: 10.1073/pnas.0705561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merrikh H, Ferrazzoli AE, Bougdour A, Olivier-Mason A, Lovett ST. A DNA damage response in Escherichia coli involving the alternative sigma factor, RpoS. Proc Natl Acad Sci USA. 2009;106(2):611–616. doi: 10.1073/pnas.0803665106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merrikh H, Ferrazzoli AE, Lovett ST. Growth phase and (p)ppGpp control of IraD, a regulator of RpoS stability, in Escherichia coli. J Bacteriol. 2009;191(24):7436–7446. doi: 10.1128/JB.00412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battesti A, Tsegaye YM, Packer DG, Majdalani N, Gottesman S. H-NS regulation of IraD and IraM antiadaptors for control of RpoS degradation. J Bacteriol. 2012;194(10):2470–2478. doi: 10.1128/JB.00132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Repoila F, Gottesman S. Signal transduction cascade for regulation of RpoS: Temperature regulation of DsrA. J Bacteriol. 2001;183(13):4012–4023. doi: 10.1128/JB.183.13.4012-4023.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Repoila F, Gottesman S. Temperature sensing by the dsrA promoter. J Bacteriol. 2003;185(22):6609–6614. doi: 10.1128/JB.185.22.6609-6614.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol. 2002;46(3):813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 10.Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29(18):3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch M, Elliott T. Role of ppGpp in rpoS stationary-phase regulation in Escherichia coli. J Bacteriol. 2002;184(18):5077–5087. doi: 10.1128/JB.184.18.5077-5087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Misawa N, Shimada H. Metabolic engineering for the production of carotenoids in non-carotenogenic bacteria and yeasts. J Biotechnol. 1997;59(3):169–181. doi: 10.1016/s0168-1656(97)00154-5. [DOI] [PubMed] [Google Scholar]
- 13.Immethun CM, Hoynes-O’Connor AG, Balassy A, Moon TS. Microbial production of isoprenoids enabled by synthetic biology. Front Microbiol. 2013;4:75. doi: 10.3389/fmicb.2013.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Y-S, Stephanopoulos G. Multi-dimensional gene target search for improving lycopene biosynthesis in Escherichia coli. Metab Eng. 2007;9(4):337–347. doi: 10.1016/j.ymben.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Alper H, Stephanopoulos G. Uncovering the gene knockout landscape for improved lycopene production in E. coli. Appl Microbiol Biotechnol. 2008;78(5):801–810. doi: 10.1007/s00253-008-1373-x. [DOI] [PubMed] [Google Scholar]
- 16.Alper H, Miyaoku K, Stephanopoulos G. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat Biotechnol. 2005;23(5):612–616. doi: 10.1038/nbt1083. [DOI] [PubMed] [Google Scholar]
- 17.Fröhlich KS, Papenfort K, Berger AA, Vogel J. A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res. 2012;40(8):3623–3640. doi: 10.1093/nar/gkr1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bougdour A, Wickner S, Gottesman S. Modulating RssB activity: IraP, a novel regulator of σ(S) stability in Escherichia coli. Genes Dev. 2006;20(7):884–897. doi: 10.1101/gad.1400306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battesti A, et al. Anti-adaptors provide multiple modes for regulation of the RssB adaptor protein. Genes Dev. 2013;27(24):2722–2735. doi: 10.1101/gad.229617.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouché S, et al. Regulation of RssB-dependent proteolysis in Escherichia coli: A role for acetyl phosphate in a response regulator-controlled process. Mol Microbiol. 1998;27(4):787–795. doi: 10.1046/j.1365-2958.1998.00725.x. [DOI] [PubMed] [Google Scholar]
- 21.Cashel M, Gentry DR, Hernandez VJ, Vinella D. The stringent response. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, K. B. Low J, Magasanik B, et al., editors. Escherichia coli and Salmonella typhimurium. Vol 1. ASM; Washington, DC: 1996. pp. 1458–1496. [Google Scholar]
- 22.Potrykus K, Cashel M. (p)ppGpp: Still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 23.Jensen PR, Hammer K. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol. 1998;64(1):82–87. doi: 10.1128/aem.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stüdemann A, et al. Sequential recognition of two distinct sites in sigma(S) by the proteolytic targeting factor RssB and ClpX. EMBO J. 2003;22(16):4111–4120. doi: 10.1093/emboj/cdg411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown L, Elliott T. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J Bacteriol. 1997;179(3):656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Battesti A, Bouveret E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol. 2006;62(4):1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 27.Yao Z, Davis RM, Kishony R, Kahne D, Ruiz N. Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc Natl Acad Sci USA. 2012;109(38):E2561–E2568. doi: 10.1073/pnas.1209742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iram SH, Cronan JE. The beta-oxidation systems of Escherichia coli and Salmonella enterica are not functionally equivalent. J Bacteriol. 2006;188(2):599–608. doi: 10.1128/JB.188.2.599-608.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang Y-Y, Wang A-Y, Cronan JE., Jr Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS(katF) gene. Mol Microbiol. 1994;11(6):1019–1028. doi: 10.1111/j.1365-2958.1994.tb00380.x. [DOI] [PubMed] [Google Scholar]
- 30.Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69(1):12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumari S, et al. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 2000;182(15):4173–4179. doi: 10.1128/jb.182.15.4173-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gualerzi CO, Giuliodori AM, Pon CL. Transcriptional and post-transcriptional control of cold-shock genes. J Mol Biol. 2003;331(3):527–539. doi: 10.1016/s0022-2836(03)00732-0. [DOI] [PubMed] [Google Scholar]
- 33.Cabrera JE, Jin D-J. Growth phase and growth rate regulation of the rapA gene, encoding the RNA polymerase-associated protein RapA in Escherichia coli. J Bacteriol. 2001;183(20):6126–6134. doi: 10.1128/JB.183.20.6126-6134.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: A homologous recombination-based method of genetic engineering. Nat Protoc. 2009;4(2):206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187(20):6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Lay N, Gottesman S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol Microbiol. 2012;86(3):524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller JH. A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.