Significance
Membrane contact sites, the sites of close physical proximity between intracellular membranes, are emerging as key players in multiple cellular processes. In particular, endoplasmic reticulum (ER)–plasma membrane (PM) contact sites play important roles in calcium homeostasis, signaling, and lipid regulation/exchange. However, the architecture of these contact sites remains poorly understood. Here we study the 3D architecture of ER–PM contact sites at molecular resolution using cryo-electron tomography. We define the structural signature of the ER–PM tethers mediated by the extended synaptotagmins (E-Syts) and show that E-Syts regulate ER–PM distance in a cytosolic Ca2+-dependent manner. These findings provide an important foundation towards elucidating E-Syt function and more generally the mechanisms of cross-talk between the ER and the PM.
Keywords: E-Syt, cryo-electron microscopy, TULIP, phosphoinositides, lipid transfer
Abstract
The close apposition between the endoplasmic reticulum (ER) and the plasma membrane (PM) plays important roles in Ca2+ homeostasis, signaling, and lipid metabolism. The extended synaptotagmins (E-Syts; tricalbins in yeast) are ER-anchored proteins that mediate the tethering of the ER to the PM and are thought to mediate lipid transfer between the two membranes. E-Syt cytoplasmic domains comprise a synaptotagmin-like mitochondrial-lipid–binding protein (SMP) domain followed by five C2 domains in E-Syt1 and three C2 domains in E-Syt2/3. Here, we used cryo-electron tomography to study the 3D architecture of E-Syt–mediated ER–PM contacts at molecular resolution. In vitrified frozen-hydrated mammalian cells overexpressing individual E-Syts, in which E-Syt–dependent contacts were by far the predominant contacts, ER–PM distance (19–22 nm) correlated with the amino acid length of the cytosolic region of E-Syts (i.e., the number of C2 domains). Elevation of cytosolic Ca2+ shortened the ER–PM distance at E-Syt1–dependent contacts sites. E-Syt–mediated contacts displayed a characteristic electron-dense layer between the ER and the PM. These features were strikingly different from those observed in cells exposed to conditions that induce contacts mediated by the stromal interaction molecule 1 (STIM1) and the Ca2+ channel Orai1 as well as store operated Ca2+ entry. In these cells the gap between the ER and the PM was spanned by filamentous structures perpendicular to the membranes. Our results define specific ultrastructural features of E-Syt–dependent ER–PM contacts and reveal their structural plasticity, which may impact on the cross-talk between the ER and the PM and the functions of E-Syts in lipid transport between the two bilayers.
The endoplasmic reticulum (ER) consists of a complex network of tubules and cisternae that extends throughout the cell and forms close appositions (“contact sites”) with other membranous organelles and with the plasma membrane (PM) (1, 2). The best characterized function of ER–PM contacts is in Ca2+ homeostasis, as ER–PM contacts mediate the excitation–contraction coupling in muscle and store-operated Ca2+ entry (SOCE) in all metazoan cells. In SOCE, upon depletion of Ca2+ in the ER, the ER protein stromal interaction molecule 1 (STIM1) oligomerizes, binds, and activates Orai1 Ca2+ channels at the PM to drive influx of extracellular Ca2+, thereby allowing homeostatic regulation of ER Ca2+ levels (3, 4). However, growing evidence suggests that ER–PM contacts also play more general roles, including signaling (5, 6) and the regulation of both lipid metabolism and transport between bilayers (1, 7–17).
We recently have shown that the three mammalian extended synaptotagmins (E-Syts), homologs of the yeast tricalbins (18, 19), act as ER–PM tethers (20). E-Syts are ER-anchored proteins (via an N-terminal hairpin) containing a synaptotagmin-like mitochondrial-lipid–binding protein (SMP) domain followed by five (E-Syt1) or three (E-Syt2/3) C2 domains. SMP domains are present in proteins that localize at various organelle contact sites, where they have been implicated in lipid transfer between bilayers (19, 21–23). As shown by a recent structural and mass spectrometry study of E-Syt2, the SMP domain dimerizes to form a ∼90-Å-long cylinder traversed by a longitudinally oriented channel lined with hydrophobic residues that harbor glycerophospholipids (23). C2 domains typically are membrane-binding modules whose interaction with the bilayer often is potentiated by elevations in cytosolic Ca2+ and/or facilitated by the presence of acidic phospholipids. Accordingly, the property of the E-Syts to tether the ER to the PM is mediated by C2 domain-dependent interactions with phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] in the PM, and in the case of E-Syt1 is additionally regulated by the elevation of cytosolic Ca2+ (11, 20). Because E-Syt1 interacts with both E-Syt2 and E-Syt3, E-Syt1 acts as a Ca2+-dependent regulator of E-Syt–mediated ER–PM tethering.
The structural features of E-Syt–dependent ER–PM contacts and how the E-Syts and other contact-resident proteins are organized between the bilayers remain unknown. Here we have investigated ER–PM contact architecture in close-to-native conditions using cryo-electron tomography (cryo-ET), a method that enables 3D visualization at molecular resolution of fully hydrated, unstained biological structures in situ, within optimally preserved vitrified cells (24). ER–PM contacts predominantly formed and populated by E-Syt1, E-Syt3, or STIM1 were induced by the transient overexpression of these proteins in COS-7 cells coupled to pharmacological treatments. We found that E-Syt–mediated ER–PM contacts show an electron-dense layer between the two apposed membranes, whereas contacts in which STIM1-mediated tethering to the PM predominates show filaments linking the two membranes. At E-Syt–mediated contacts the ER–PM distance correlates with the length of the cytosolic portion of the specific overexpressed E-Syt isoform and is regulated further by cytosolic Ca2+ in the case of E-Syt1.
Results
ER Morphology at the Cell Edge.
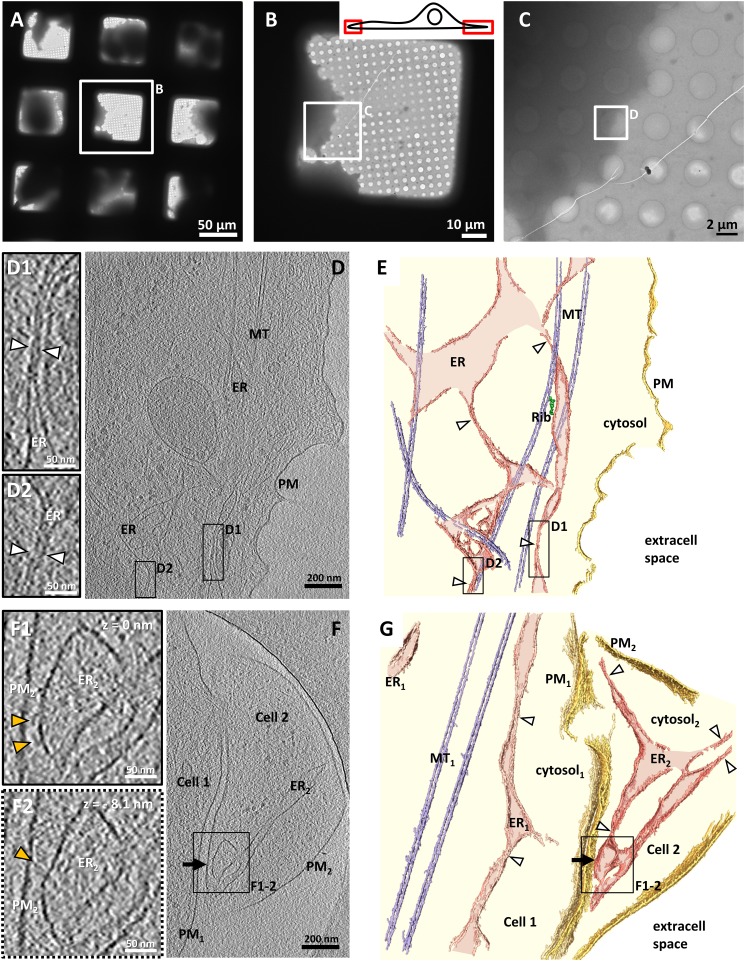
We first characterized peripheral ER morphology in untransfected COS-7 cells by cryo-ET. Cells were grown and subsequently vitrified on EM grids coated with a holey carbon support (Fig. 1A), and thin regions at the edge of the cells were targeted for cryo-ET (Fig. 1 B and C). These regions often contained ER at a certain distance from the PM and in most cases associated with microtubules (Fig. 1 D and E). The ER consisted of irregularly shaped tubular structures, with larger segments connected to each other by extremely narrow tubules (∼15 nm in diameter, measured between the cytosolic leaflets of both membranes; white arrowheads in Fig. 1 D1, D2, E, and G) (25). Similar morphology of the peripheral ER was observed at the edge of Ptk2 cells (Fig. S1 A and B), indicating that this feature is common to other mammalian cells.
Fig. 1.
ER morphology at the edge of untransfected COS-7 cells. (A–C) Images at increasing levels of magnification of cells vitrified on an EM grid coated with a holey carbon support. (B, Inset) Cartoon illustrating the regions of the cell accessible for cryo-ET without need of thinning procedures. (D) Tomographic slice of the region selected in C. In all images, ER, PM, and MT denote endoplasmic reticulum, plasma membrane, and microtubules, respectively. (D1 and D2) Higher-magnification tomographic slices of the regions labeled in D showing narrow ER tubules (white arrowheads). In all figures, the line style of the frame indicates whether the enlarged area in a subpanel was extracted from the tomographic slice shown in the main panel (solid line) or a different tomographic slice (dotted line). (E) 3D rendering of the tomogram shown in D. For all renderings, extracellular space (extracell space, white), cytosol (light yellow), PM (gold), ER (pink), ER lumen (light pink), microtubules (MT, light blue) and ER-bound ribosomes (Rib, green) are indicated. White arrowheads denote narrow ER tubules. (F) Tomographic slice of two cells. The cell on the right (Cell 2) contains a native ER–PM contact (black arrow). (F1 and F2) Higher-magnification tomographic slices of the ER–PM contact in the region labeled in F. The slices depict the same region in x and y at different z heights (z = 0 nm and z = −8.1 nm, respectively). Structures tethered to the ER are indicated by yellow arrowheads. (G) 3D rendering of the tomogram shown in F. D, D1, D2, F, F1, and F2 show 2.7-nm-thick tomographic slices.
Native ER–PM contacts were observed, although only rarely, at the edge of naive cells (black arrow in Fig. 1 F and G and Movie S1). They were formed by wide ER compartments connected to the rest of the ER by narrow tubules (white arrowheads in Fig. 1G). The ER–PM distance at these sites was 25.4 ± 1.6 nm (mean ± SEM) measured between the cytosolic leaflet of the membranes for all contacts within each tomogram (n = 3 tomograms), and electron-dense structures of variable morphology were found tethering the two membranes (yellow arrowheads in Fig. 1 F1 and F2 and Movie S1).
To gain insights into the molecular organization of E-Syt–dependent ER–PM contacts, we examined COS-7 cells in which overexpression of Myc-tagged E-Syt constructs had induced massive formation of such contacts (20). We focused our structural analysis on E-Syt1 and E-Syt3, because, although E-Syt2 and E-Syt3 share similar properties (20), E-Syt2 was less effective in inducing ER–PM contacts in the thin regions at the edge of COS-7 cells amenable to direct cryo-ET imaging despite being expressed at similar levels as E-Syt3 (Fig. S2A).
E-Syt3–Mediated ER–PM Contacts.
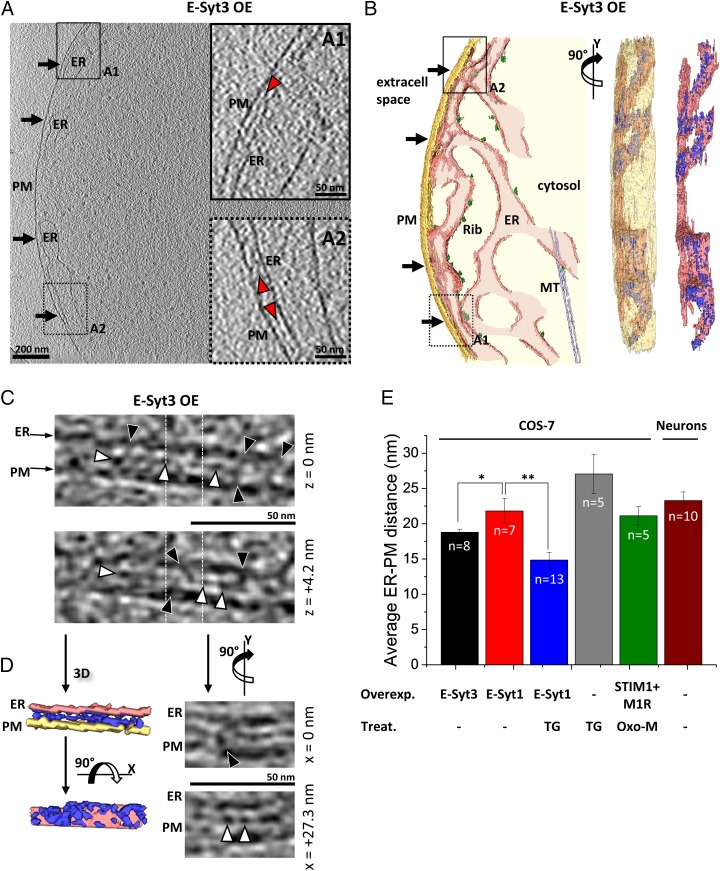
In Myc-E-Syt3–expressing cells the abundance of contacts was obvious, so Myc-E-Syt3–transfected cells could be distinguished readily in EM micrographs by the accumulation of ER at the cell edge (black arrows in Fig. 2 A and B and Movie S2). At such contacts, which are mediated by the PM binding of the C-terminal C2 (C2C) domain of E-Syt3 (20), the ER–PM distance was 18.8 ± 0.4 nm (mean ± SEM, n = 8 tomograms) (Fig. 2E), and a prominent electron-dense layer, significantly denser than the surrounding cytosol and henceforth referred to as “intermediate density” (red arrowheads in Fig. 2 A1 and A2; P < 0.001 by paired t test, Fig. S3A), was observed between the membranes in most of the membrane apposition area. The intermediate density was located 7.6 ± 0.4 nm (mean ± SEM) away from the ER membrane (measured from the cytosolic leaflet of the ER membrane to the center of the density). A higher-resolution cryo-ET dataset recorded using a direct electron detector revealed that the intermediate density consisted of interconnected globular structures (∼5 nm in diameter) (white arrowheads in Fig. 2 C and D and Movie S3), often linked to the ER and to the PM by filamentous bridges (black arrowheads in Fig. 2 C and D and Movie S3). A few of these bridges also seemed to be connected to large extracellular or ER luminal densities (Fig. S1C). Altogether, at E-Syt3–mediated contacts the distance between the ER and the PM was spanned by a dense protein array that adopted a well-defined molecular arrangement.
Fig. 2.
E-Syt3–mediated ER–PM contacts in COS-7 cells. (A) Tomographic slice of ER–PM contacts (black arrows) in a cell overexpressing Myc-E-Syt3 (E-Syt3 OE). (A1 and A2) Higher-magnification tomographic slices of the ER–PM contact in the regions labeled in A. The intermediate density is indicated by red arrowheads. The style of the line framing the subpanel indicates whether the enlarged area was extracted from the tomographic slice in the main panel (solid line) or from a different slice (dotted line). (B) 3D rendering of the tomogram shown in A in a top view (Left), an en face view with a semitransparent PM (90° rotation along the y axis) (Center), and an en face view showing only the ER and all densities present at the ER–PM contacts up to an arbitrary grayscale intensity threshold (dark blue) (Right). (C) Higher-resolution data recorded with a direct electron detector. The tomographic slices depict the same region in x and y at different z heights [z = 0 nm (Upper) and z = +4.2 nm (Lower)]. Globular densities and bridges between the ER and the PM are indicated by white and black arrowheads, respectively. Vertical white dotted lines mark the positions in which orthogonal tomographic slices are shown in D, Right. (D, Left) 3D renderings of the area shown in C depicting the densities (dark blue) present between the ER and the PM up to an arbitrary threshold over 11 tomographic slices (z = −10.5 nm to z = 10.5 nm) in a view perpendicular to the membrane (Upper Left) and an en face view of the ER membrane with a 90° rotation along the x axis (Lower Left). (Right) Orthogonal tomographic slices at the positions marked by white dotted lines in C (90° rotation along the y axis). The tomographic slices depict the same region in y and z at different x positions [x = 0 nm (Upper) and x = +27.3 nm (Lower)]. A, A1, and A2 show 2.7-nm-thick tomographic slices. C and D show 2.1-nm-thick tomographic slices. (E) Average ER–PM distances. Error bars indicate SEM. Overexpressed proteins (Overexpr.) are Myc-E-Syt3 (E-Syt3), Myc-E-Syt1 (E-Syt1), and mRFP-STIM1 plus the muscarinic receptor M1R (STIM1 + M1R). −, no overexpression. Pharmacological treatments (Treat.) are TG and Oxo-M. −, no treatment. The number of tomograms for each condition (n) is indicated within the bars. *P < 0.05 and **P < 0.01 by Student’s t test.
Greater ER–PM Distance at E-Syt1–Mediated Contacts.
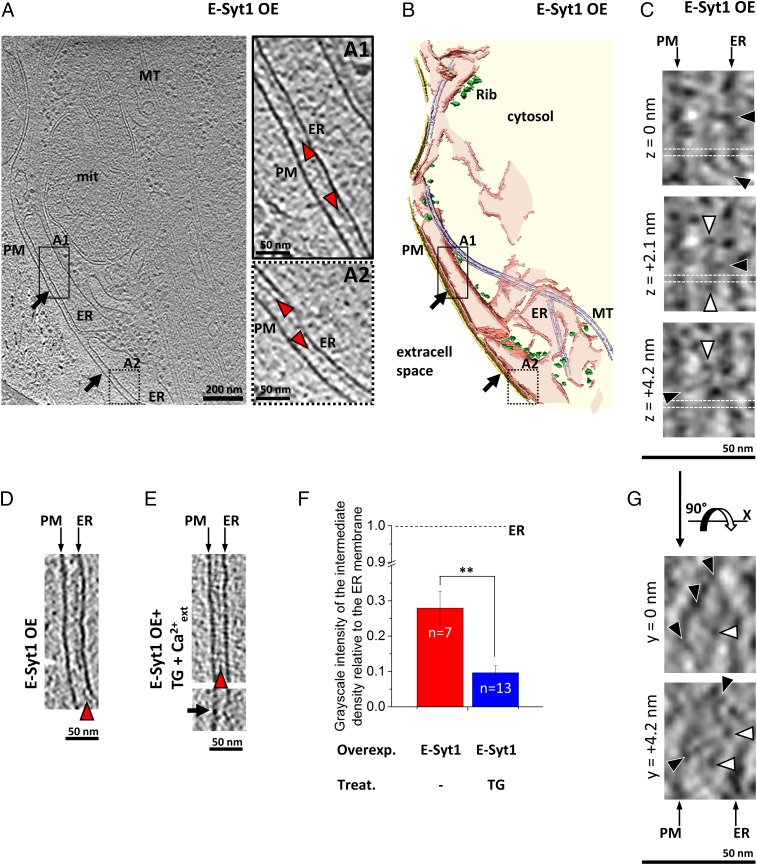
E-Syt1 localizes throughout the entire ER at resting Ca2+ levels in COS-7 cells, but it concentrates at ER–PM appositions upon the elevation of cytosolic Ca2+ (20). Such recruitment critically requires Ca2+ binding by its central C2C domain. Nevertheless, we observed large ER–PM contacts in some Myc-E-Sy1–expressing cells (Fig. 3 A and B). At such contacts, the average ER–PM distance was 21.8 ± 1.8 nm (mean ± SEM, n = 7 tomograms), significantly greater than at E-Syt3–mediated contacts (P < 0.05 by t test) (Fig. 2E) and substantially more variable (error bars in Fig. 2E; compare Fig. S3 A and B), suggesting that E-Syt1 has a higher degree of structural plasticity.
Fig. 3.
E-Syt1–mediated ER–PM contacts in COS-7 cells. (A) Tomographic slice of ER–PM contacts (black arrows) in a cell overexpressing Myc-E-Syt1 (E-Syt1 OE). (A1 and A2) Higher-magnification tomographic slices of the ER–PM contact in the boxed regions in A. The intermediate density between the ER and the PM is indicated by red arrowheads. The style of the line framing the subpanel indicates whether the enlarged area was extracted from the tomographic slice in the main panel (solid line) or from a different slice (dotted line). (B) 3D rendering of the tomogram shown in A. (C) Higher-resolution tomographic slices from data recorded with a direct electron detector. The slices depict the same region in x and y at different z heights (z = 0 nm, z = +2.1 nm, and z = +4.2 nm). Globular densities and bridges between the ER and the PM are indicated by white and black arrowheads, respectively. Horizontal white dotted lines mark the positions in which orthogonal tomographic slices are shown in G. (D) ER–PM contact in a Myc-E-Syt1–transfected cell without further treatments (E-Syt1 OE). (E) ER–PM contacts in two Myc-E-Syt1–transfected cells treated with TG to elevate cytosolic Ca2+ (E-Syt1 OE + TG +Ca2+ext). Note that the distance between the ER and the PM is shorter than in D. In the lower image, a black arrow points to an area where the ER–PM distance is ∼8 nm. (F) Average grayscale intensity of the intermediate density relative to the ER membrane in Myc-E-Syt1 (E-Syt1)-overexpressing cells (a value of 1 would indicate an intermediate density as electron dense as the ER membrane). Error bars indicate SEM. Overexpressed protein (Overexpr.): Myc-E-Syt1 (E-Syt1). Pharmacological treatment (Treat.): TG or no treatment (−). The number of tomograms for each condition (n) is indicated within the bars. Note that the intermediate density was significantly less pronounced for Myc-E-Syt1–transfected cells treated with TG in the presence of extracellular Ca2+. **P < 0.01 by Student’s t test. (G) Orthogonal tomographic slices at the positions marked by white dotted lines in C (90° rotation along the x axis). The tomographic slices depict the same region in x and z at different y positions (y = 0 nm and y = +4.2 nm). A, A1, A2, D, and E show 2.7-nm-thick tomographic slices. C and G show 2.1-nm-thick tomographic slices.
The increased distance of the ER from the PM at Myc-E-Syt1–mediated contacts could be explained by the additional C2 domains of E-Syt1 relative to E-Syt3 (Fig. S2B) if the tethering function of E-Syt1 at resting Ca2+ levels was mediated by the binding to the PM of the C-terminal C2 domain of E-Syt1 (C2E), which shares strong similarity to the C2C domain of E-Syt2/3. Like these two other C2 domains, which mediate the binding of E-Syt2/3 to the PM, it contains a conserved basic patch that is implicated in binding acidic phospholipids (Fig. S2C). Additionally, unlike full-length E-Syt1 (Fig. S4 A and B and see below), an E-Syt1 mutant lacking the C2E domain (E-Syt1 ΔC2E) failed to translocate to the PM upon increase in cytosolic Ca2+ (Fig. S4 C and D). This finding indicates that both the C2C domain, which is required for Ca2+-dependent recruitment to the PM (20), and the C2E domain cooperate in the formation of E-Syt1–mediated ER–PM contacts.
Like Myc-E-Syt3–mediated contacts, Myc-E-Syt1–mediated contacts showed an intermediate density between the ER and the PM (red arrowheads in Fig. 3 A1, A2, and D) that was significantly denser than the surrounding cytosol (P < 0.01 by paired t test) (Fig. S3B). Both the average distance between this intermediate density and the ER membrane (Fig. S3 B and G) and its architecture, as shown by a higher-resolution dataset (Fig. 3 C and G), were similar to that observed in Myc-E-Syt3–mediated contacts (but note the increased variability in Fig. S3B). Given that the cytosolic domains of the E-Syts differ in their C-terminal region but are closely related to each other in their N-terminal region (Fig. S2B), it is likely that this N-terminal region, i.e., the SMP domain plus the C2A and C2B domains, is involved in the formation of the intermediate density.
E-Syt1 Regulates ER–PM Distance in a Ca2+-Dependent Manner.
Elevations in cytosolic Ca2+ concentration lead to an increase in E-Syt1–mediated ER–PM contacts via the Ca2+-dependent PM binding of the E-Syt1 C2C domain (20). To explore the role of cytosolic Ca2+ in the structural plasticity of E-Syt1–mediated ER–PM contacts, we stimulated Myc-E-Syt1–transfected COS-7 cells with thapsigargin (TG), a potent sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) inhibitor that activates SOCE by blocking ER Ca2+ reuptake and thus inducing Ca2+ depletion from the ER. Cells were incubated in Ca2+-free buffer containing 2 µM TG for 10 min to induce depletion of Ca2+ within the ER, then were switched to Ca2+-containing (2 mM) buffer, and were frozen immediately for cryo-ET analysis. This procedure resulted in a massive Ca2+ influx upon the readdition of extracellular Ca2+ and induced a dramatic synchronized increase of E-Syt1–mediated ER–PM contacts as monitored by live imaging (Fig. S4 A and B) (Olof Idevall-Hagren, personal communication). Even though these conditions also induce the recruitment of STIM1 to ER–PM contacts (26), overexpressed E-Syt1 was likely the dominating tether in the absence of STIM1 overexpression. Accordingly, an intermediate density was observed between the ER and the PM (Fig. 3E, Upper, and Fig. S3C), similar to that observed in the contacts formed upon Myc-E-Syt3 (Fig. 2 A, C, and D) or Myc-E-Syt1 (Fig. 3 A, C, D, and G) overexpression at resting cytosolic Ca2+. However, the ER–PM distance was reduced significantly, by more than 30%, to 14.8 ± 1.1 nm (mean ± SEM, n = 13 tomograms; P < 0.01 by t test) (Figs. 2E and 3E) in comparison with untreated Myc-E-Syt1–transfected cells (in which E-Syt1–mediated contacts predominate), with occasional focal closer contacts below 10 nm (Fig. 3E, Lower). Although the average distance between the intermediate density and the ER membrane was unchanged (Fig. 3E, Upper, and Fig. S3G), this structure became significantly less prominent on average (P < 0.01 by t test) (Fig. 3F). Interestingly, the variability in both the distance between the ER and the PM and the distance between the ER and the intermediate density was reduced substantially compared with E-Syt1–mediated contacts at resting Ca2+ levels (Fig. S3C and compare error bars in Fig. 2E). These data suggest that the elevation in cytosolic Ca2+ resulted in a molecular rearrangement of E-Syt1, which in turn led to the shortening of ER–PM distance, weakening of the intermediate density, and reduced structural variability.
E-Syt–Mediated ER–PM Contacts Are Structurally Distinct from Those Mediated by STIM1.
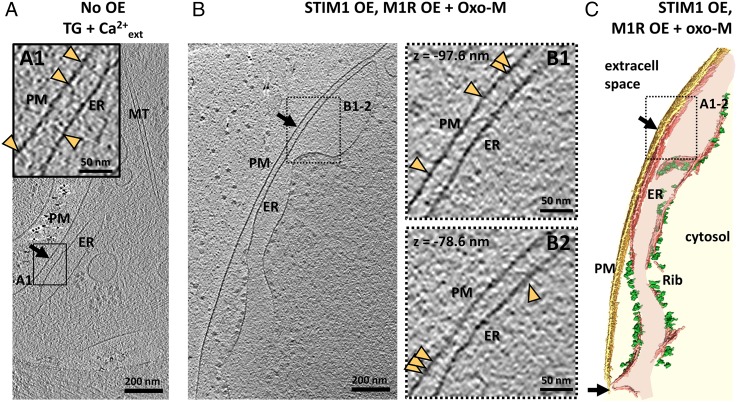
We next compared the structure of E-Syt–dependent contacts with those mediated by STIM1. Upon reduction of ER luminal Ca2+, STIM proteins tether the ER to the PM by interacting with PM Orai Ca2+ channels, thereby activating SOCE (3). Two approaches were used to study STIM1–Orai contacts in COS-7 cells. First, untransfected cells were incubated with 2 µM TG in Ca2+-containing buffer before freezing, i.e., a condition leading to the loss of Ca2+ from the ER because of the TG-induced block of the ER SERCA pump. Contacts formed under this condition (Fig. 4A) were different from E-Syt–mediated contacts: The ER–PM distance was 27.1 ± 2.8 nm (mean ± SEM, n = 5 tomograms) (Fig. 2E), and filaments linked the ER and the PM (yellow arrowheads in Fig. 4A1) with no obvious intermediate density such as observed in E-Syt–transfected cells (Fig. S3D).
Fig. 4.
STIM1-mediated ER–PM contacts in COS-7 cells. (A) Tomographic slice of an ER–PM contact (black arrow) in an untransfected cell treated with 2 µM TG in Ca2+-containing buffer for 15 min immediately before freezing (TG +Ca2+ext). (A1) Higher-magnification tomographic slice of the ER–PM contact in the region labeled in A. Filaments emanating from the ER and the PM are indicated by yellow arrowheads. (B) Tomographic slice of an ER–PM contact (black arrow) in a cell cotransfected with mRFP-STIM1 and M1R and treated with 10 μM Oxo-M for 400 s before freezing (STIM1 OE, M1R OE + oxo-M). (B1 and B2) Higher-magnification tomographic slices of the ER–PM contact in the region labeled in A. Filaments bridging the ER and the PM are indicated by yellow arrowheads. The dotted lines framing B1 and B2 indicate that the enlarged areas were extracted from tomographic slices other than that shown in the main panel (z = −97.6 nm and z = −78.6 nm, taking the slice in the main panel as 0 nm). (C) 3D rendering of the tomogram shown in B. A, A1, B, B1, and B2, show 2.7-nm-thick tomographic slices.
Second, COS-7 cells cotransfected with mRFP-STIM1 and the muscarinic receptor M1 were stimulated with 10 μM of the muscarinic receptor agonist oxotremorine M (Oxo-M) for 400 s before freezing. The activation of the muscarinic receptor stimulates phospholipase C leading to the hydrolysis of PM PI(4, 5)P2 and release of inositol triphosphate (IP3), thus resulting in the reduction of ER Ca2+ stores through the activation of IP3 receptors on the ER (27). In the absence of E-Syt overexpression, these conditions led to the formation of predominantly STIM1-mediated ER–PM contacts (Fig. S5). Accordingly, the intermediate density typical of E-Syt–mediated contacts was not observed at these ER–PM contacts (Fig. 4 B and C and Fig. S3E). Instead, the gap between the ER and the PM (21.1 ± 1.3 nm; mean ± SEM, n = 5 tomograms) (Fig. 2E) was spanned by numerous filaments roughly perpendicular to the ER and the PM (yellow arrowheads in Fig. 4 B1 and B2), similar to the filaments observed in untransfected cells stimulated with 2 µM TG (see above). This distance was smaller than that observed in cells in which STIM1 contacts were generated in TG-treated untransfected cells, but this finding was not investigated further. One possibility is that the difference may be related to the decrease in PI(4,5)P2 which accompanies the increase in cytosolic Ca2+ in Oxo-M–stimulated cells but not in TG-treated cells.
ER–PM Contacts in Neurons.
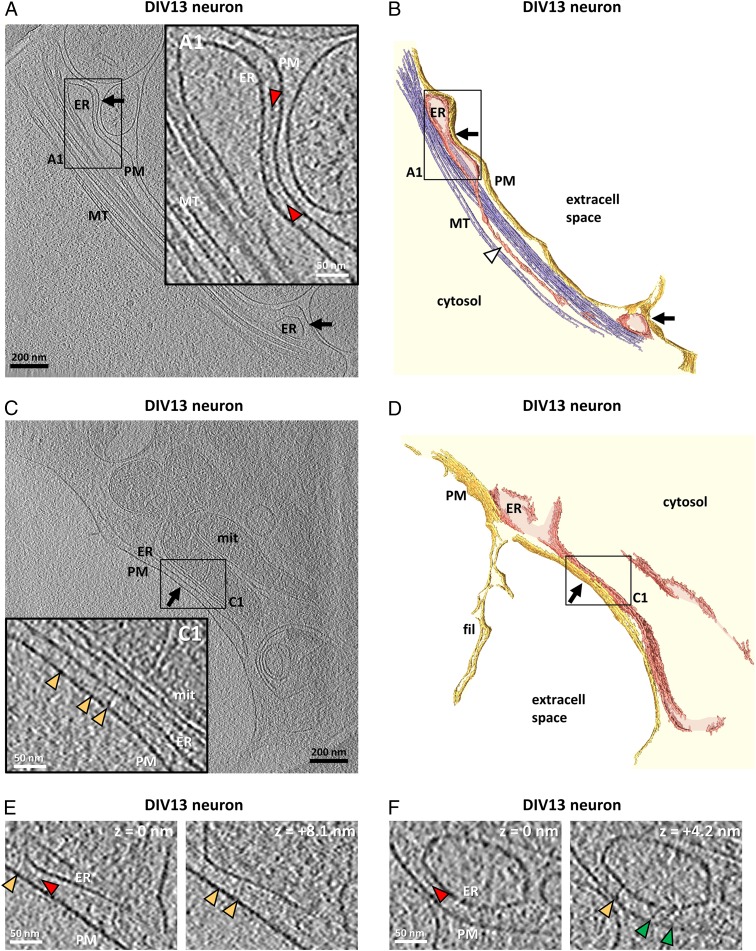
ER–PM contacts are observed frequently in neurons (25, 28–31). Thus, to complement our overexpression experiments, we analyzed native ER–PM contacts in untransfected neurons cultured on EM grids. We focused our analysis on neuronal processes, which were generally thin enough for direct cryo-ET imaging. The overall ER morphology in these regions was comparable to that observed in the periphery of COS-7 (Fig. 1 D–G) and Ptk2 cells (Fig. S1 A and B), with very narrow ER tubules interconnecting wider compartments (white arrowhead in Fig. 5B). However, ER–PM contacts were found more frequently in neurons. On average, the ER–PM distance at the contact sites was 23.3 ± 1.2 nm (mean ± SEM, n = 10 tomograms), i.e., in the range of contacts observed in COS-7 cells (Fig. 2E). In some neuronal ER–PM contacts (black arrows in Fig. 5 A and B and Movie S4), an electron-dense layer was observed ∼8 nm away from the ER membrane (red arrowheads in Fig. 5A1), reminiscent to the intermediate density observed upon overexpression of E-Syts in COS-7 cells (compare with Figs. 2 and 3). In other contacts (black arrow in Fig. 5 C and D), filamentous structures linked the ER to the PM (yellow arrowheads in Fig. 5C1), reminiscent of the structures found at STIM1-mediated ER–PM contacts (Fig. 4). Occasionally, contacts showed both filaments and an intermediate density (Fig. 5 E and F) or tethers of an unidentified nature (green arrowheads in Fig. 5F). These findings support the existence in neurons of contacts mediated by the E-Syts and also of some contacts mediated by STIM1 but also indicate that additional, structurally distinct contact architectures coexist.
Fig. 5.
Native ER-PM contacts in untransfected DIV13 cultured neurons. (A) Tomographic slice of ER–PM contacts (black arrows) in a cultured neuron. (A1) Higher-magnification tomographic slice of the ER–PM contact in the region labeled in A. The intermediate density between the ER and the PM is indicated by red arrowheads. (B) 3D rendering of the tomogram shown in A. A narrow ER tubule is indicated by a white arrowhead. (C) Tomographic slice of an ER–PM contact (black arrow) in another neuron. (C1) Higher-magnification tomographic slice of the ER–PM contact in the region boxed in C. Note that the ER simultaneously establishes a contact with a neighboring mitochondrion (mit). The ER and the outer mitochondrial membranes are at a distance of ∼8 nm (measured between the cytosolic leaflets of both membranes). The filaments bridging the ER and the PM are indicated by yellow arrowheads. The solid lines framing the boxed areas in A1 and C1 indicate that the enlarged views were extracted from the same tomographic slice as the corresponding main panels. (D) 3D rendering of the tomogram shown in C. fil, filopodium. (E and F) Neuronal ER–PM contacts showing filaments (yellow arrowheads), an intermediate density (red arrowheads), and tethers of different morphology (green arrowheads). The slices depict the same regions in x and y at different z heights [z = 0 nm and z = +8.1 (E); z = 0 nm and z = +4.2 nm (F)]. A, A1, C, C1, E, and F show 2.7-nm-thick tomographic slices.
Discussion
In this study we used cryo-ET to analyze the 3D architecture of ER–PM contacts within vitrified mammalian cells. Taking advantage of overexpression and pharmacological treatments, we visualized in situ ER–PM contacts expected to be mediated primarily by the E-Syts.
At putative E-Syt–mediated contacts we observed an electron-dense layer, the intermediate density, ∼8 nm away from the ER membrane (E-Syt conditions in Fig. S6). The similar morphology of this density in E-Syt1– and E-Syt3–mediated contacts suggests that the portions of the cytosolic domains that are most similar in these two proteins (the SMP and C2A and C2B domains) play a role in the formation of such density. The intermediate density was not observed at ER–PM contacts expected to be mediated primarily by STIM1, which, in agreement with previous structural models (32), showed filaments linking the ER to the PM (Fig. S6, Upper Right).
Although E-Syt1 recruitment to the PM was shown to be stimulated by elevations in cytosolic Ca2+ (20), a finding that we have confirmed here, we now have observed that some E-Syt1–dependent contacts occurred even at resting Ca2+ levels in our cells and under our experimental conditions. This result is consistent with the strong similarity of the C2E domain of E-Syt1 to the C2C domains of E-Syt2 and E-Syt3, which are known to mediate the constitutive interaction of these two proteins with the PM. Perhaps the generally low levels of association of E-Syt1 with the PM at resting cytosolic Ca2+ concentration are caused by inhibitory interactions that may partially mask the basic patch of the C2E domain. Furthermore, at resting Ca2+ levels, the distance between the ER and the PM was larger and more variable in contacts mediated by E-Syt1 than in those mediated by E-Syt3 (compare E-Syt3 OE and E-Syt1 OE in Fig. S6), in good agreement with the greater amino acid length of the cytosolic portion of E-Syt1 relative to E-Syt3. In response to strong elevations of cytosolic Ca2+, the recruitment of E-Syt1 to the cell cortex correlated with a reduction of the length of E-Syt1–dependent ER to PM tethers indicating a dynamic conformational change (shortening) of the cytosolic portion of E-Syt1. Modulation of ER–PM distance by tethering molecules also was previously reported by other studies (11, 31). Because we previously have shown that the C2C domain of E-Syt1 is required for the binding of E-Syt1 to the PM in a cytosolic Ca2+-dependent manner (20), we speculate that the reduction of ER–PM distance in E-Syt1–mediated contacts upon elevation of cytosolic Ca2+ could be accounted by the interaction of its C2C domain with the PM. This shortening from C2E- to C2C-mediated PM binding could be consistent with the magnitude of the change in the ER–PM distance that we quantified.
The property of E-Syts to harbor phospholipids in their SMP domains (23), the localization of several SMP domain-containing proteins at membrane contact sites (19), and the similarity of SMP modules to modules whose known function is to exchange lipids (21, 23) strongly suggest that E-Syts have a role in lipid exchange between membranes. Furthermore, the peculiar elongated structure of the 9-nm-long SMP domain obligate dimer, with a deep hydrophobic groove that spans the entire domain from tip to tip and nests the acyl groups of phospholipids, had suggested the possibility that lipid exchange may be achieved by direct flow of lipids from one bilayer to the other via the SMP domain bridge [the tunnel model of lipid exchange (23)]. This model cannot be excluded, because we observed very few focal contacts with ER–PM distances less than 10 nm. However, the larger ER–PM distances observed at the great majority of E-Syt–dependent contacts are more consistent with the shuttle model (23), in which lipids first are extracted by the SMP domain at one membrane and then are delivered to the other membrane by a translocation of the SMP domain kept in place by C2 domain-dependent interactions. Cytosolic Ca2+ may regulate such exchanges, because all E-Syts exhibit Ca2+-dependent properties: Although the ER–PM tethering function is strongly regulated by Ca2+ only in the case of E-Syt1 (because the properties of its C2C domain; see ref. 20), the C2A domains of all three E-Syts have conserved Ca2+-binding pockets that promote Ca2+-dependent bilayer binding (33, 34). This property may account for a conformational change of their cytosolic domains, which results in phospholipid extraction from membranes and transfer. The weakening of the intermediate density observed by cryo-ET after increase of cytosolic Ca2+ supports a flexible and dynamic nature of the cytosolic region of E-Syt1.
We complemented our overexpression study with the investigation of native ER–PM contacts in untransfected neurons. We found that at least some native ER–PM contacts in neurons share key features with the contacts mediated by E-Syts. Some neuronal ER–PM contacts showed an electron-dense layer ∼8 nm away from the ER membrane, similar to E-Syts–mediated contacts (Fig. S6, Lower Left), whereas others showed filamentous structures reminiscent of STIM1-mediated contacts and/or tethers of unidentified nature (Fig. S6, Lower Right). These findings suggest that neuronal ER–PM contacts are dynamic and might be populated by different molecules, and point to additional tethering molecules beyond E-Syts and STIM1.
Materials and Methods
Antibodies and Chemicals.
Sources of reagents were as follows: anti–E-Syt1 (HPA016858; Sigma-Aldrich); anti–E-Syt2 (HPA002132; Sigma-Aldrich); anti–E-Syt3 (HPA039200; Sigma-Aldrich); anti-Myc 9E10 (sc-40; Santa Cruz Biotechnology); and anti-actin (69100; MPBiosciences). Oxo-M and atropine were obtained from Sigma-Aldrich, and TG was obtained from Life Technologies.
Plasmids.
The following reagents were kind gifts: mRFP-Sec61β from T. Rapoport, Harvard University, Cambridge, MA (35); mRFP-STIM1 from B. Baird, Cornell University, Ithaca, NY (36); and M1 muscarinic acetylcholine receptor (M1R) from B. Hille, University of Washington, Seattle (27). The generation of Myc-E-Syt1, Myc-E-Syt2 (Myc-E-Syt2S was used in this study), Myc-E-Syt3, and EGFP-E-Syt1 was described in ref. 20.
Cloning of EGFP-E-Syt1ΔC2E.
cDNAs of E-Syt1 were amplified by PCR. In all PCR reactions, PfuUltra II DNA polymerase (Agilent) was used. The primers used were
5′ SalI_E-Syt1: ATTCTGCAGTCGACATGGAGCGATCTCCAGGAGAGGGCCC
3′ SacII_E-Syt1_C2E_De: GGATCCCGGGCCCGCGGCTAAGGCCCGGCTGGAGCCTCAAG
PCR products were ligated between SalI and SacII in the pEGFP-C1 vector (Clontech) to generate EGFP-E-Syt1ΔC2E.
Cell Culture and Transfection.
COS-7 cells were cultured in Dulbecco's modified essential Eagle medium (DMEM) (Life Technologies) supplemented with 10% (vol/vol) FBS at 37 °C and 5% CO2. Neuronal culture was prepared as previously described (37). Transfection of plasmids was carried out with Lipofectamine 2000 (Life Technologies), according to the manufacturer’s instructions.
Live-Cell Imaging.
For imaging experiments, cells were plated on 35-mm glass-bottomed dishes at low density (MatTek Corp.). Live-cell imaging was carried out 1 d after transfection. Total internal reflection fluorescence (TIRF) microscopy was performed on a setup built around a Nikon Ti-E microscope equipped with a 60× 1.49-NA objective. Excitation light was provided by 488-nm (for GFP) and 561-nm (mRFP) DPSS lasers coupled to the TIRF illuminator through an optical fiber. The output from the lasers was controlled by an acousto-optic tunable filter, and fluorescence was detected with an EM-CCD camera (Andor DU-887). Acquisition was controlled by Andor iQ software. Images were sampled at 0.20 Hz with exposure times in the 100- to 500-ms range. TIRF microscopy was carried out at 37 °C.
To induce SOCE in COS-7 cells expressing either EGFP-E-Syt1 or EGFP-E-Syt1 ΔC2E, the cells were washed twice and incubated with Ca2+-free buffer [140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 10 mM Hepes, 10 mM glucose, and 3 mM EGTA, (pH 7.4)] for 10 min in the presence of 2 μM TG, and Ca2+ buffer [140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 10 mM Hepes, 10 mM glucose, and 2 mM CaCl2 (pH 7.4)] was added back after ER Ca2+ store depletion. Cells were imaged with a TIRF microscope. In a parallel experiment, COS-7 cells expressing Myc-E-Syt1 were frozen for cryo-ET immediately after Ca2+ add-back with the same protocol.
To induce the formation of STIM1-mediated ER–PM contacts in COS-7 cells coexpressing mRFP-STIM1 and M1R, the cells incubated in Ca2+ buffer were stimulated with 10 μM Oxo-M. Cells were imaged with a TIRF microscope. In a parallel experiment, COS-7 cells coexpressing mRFP-STIM1 and M1R were frozen for cryo-ET 400 s after the addition of 10 uM of Oxo-M.
Biochemical Analysis of E-Syts.
COS-7 cells expressing Myc-tagged E-Syts were lysed in SDS lysis buffer [10 mM Tris⋅HCl, 150 mM NaCl, 2% SDS (pH 8.0)]. Cell lysates were incubated at 60 °C for 20 min followed by incubation at 70 °C for 10 min. Benzonase Nuclease (Novagen) was added, and the samples were incubated further at room temperature for 20 min. Cell lysates were loaded and separated in 8% SDS/PAGE gel, and immunoblotting was carried out as described in ref. 20.
Sequence Analysis.
Sequence alignment was carried out by Clustal W2 (EMBL-EBI). The following protein sequences were used: the C2B domain of Syt1 (amino acids 274–382), the C2E domain of E-Syt1 (amino acids 972–1081), the C2C domain of E-Syt2 (amino acids 759–868), and the C2C domain of E-Syt3 (amino acids 755–864).
Cell Plating and Vitrification.
Holey carbon EM grids (Quantifoil) were incubated with 70% ethanol for 10 min and were washed extensively with water. Grids were incubated at 37 °C in DMEM supplemented with 10% FBS for 2–3 h, and then approximately 75,000 COS-7 cells were plated per grid. Cells were grown for 48 h at 37 °C and 5% CO2 before vitrification. Plasmids were transfected 24 h before vitrification using Lipofectamine 2000 (Life Technologies). Neuronal cultures were prepared as previously described (37), and cells were cultured for 13 d before vitrification.
When indicated, 2 µM TG (15–20 min) or 10 μM Oxo-M (400 s) was applied immediately before vitrification (all at room temperature). Grids were mounted in a vitrification device (manual plunger, FEI Vitrobot Mark III or Leica EM GP). A 3-μL drop of a concentrated solution of 10-nm BSA-gold tracers (Aurion) resuspended in the solution in which the cells were incubated was deposited on the EM grid and incubated for 10 s. Grids were blotted with Whatman filter paper (GE Healthcare Life Sciences), plunged into liquid ethane cooled at liquid nitrogen temperature, and stored in liquid nitrogen until imaging.
Cryo-ET.
All data except those in Figs. 2 C and D and 3 C and G and Fig. S1C were recorded as follows: Vitrified EM grids were mounted on a model 626 specimen transfer holder (Gatan) cooled below −150 °C and inserted into a Tecnai F20 microscope (FEI). The microscope was equipped with a field-emission gun operated at 200 kV and a 4 × 4 k CCD camera (Gatan). Tilt series were collected using SerialEM low-dose acquisition scheme (38), typically from −60° to 60° with 2° angular increments. Pixel size was 0.9 nm at the specimen level, and the defocus was set to −8 μm. For data in Fig. S1 A and B, the pixel size at the specimen level was 1.2 nm, and the defocus was set to −10 μm. The total dose was kept below 150 e-/Å2.
The data in Figs. 2 C and D and 3 C and G and Fig. S1C were recorded as follows: Vitrified EM grids were inserted into a Polara G2 microscope (FEI) and kept below −150 °C at all times. The microscope was equipped with a field-emission gun operated at 300 kV, a postcolumn energy filter, and a K2 Summit direct electron detector (Gatan). For tilt series, zero-loss images were recorded in dose fractionation mode using the SerialEM low-dose acquisition scheme (38), typically from −60° to 60° with 2° angular increments. Pixel size was 0.5 nm at the specimen level, and the defocus was set to −5 μm. The total dose was kept below 150 e-/Å2.
Raw frames from the direct electron detector were aligned using in-house software. For both microscopes, tilt series were aligned using gold beads as fiducial markers, reconstructed and denoised by anisotropic nonlinear diffusion filtering using the software package IMOD (39). CCD data were binned three times (final voxel size, 2.7 nm) and reconstructed by weighted back-projection with analytical weighting. Data in Fig. S1 A and B were binned twice for reconstruction (final voxel size, 2.4 nm). For the direct electron detector, integrated aligned projections were binned four times using an anti-aliasing filter (final voxel size, 2.1 nm) and reconstructed by simultaneous iterative reconstruction. All tomographic images are displayed using the interpolation tool of IMOD.
Image Segmentation and Statistical Analysis.
The ER and PM were manually segmented in Amira (FEI). In tomograms containing multiple ER–PM contact sites, all contact sites were pooled for subsequent analysis. For the analysis of grayscale intensity, in each tomogram the pooled contact was divided into one-pixel-thick layers parallel to the PM using a previously described Pyto package (40). For ER–PM distances (Fig. 2E) and grayscale intensity profiles (Fig. S3), averages were calculated weighting the contribution of each tomogram by the size of the ER–PM contacts it contained (ER area at the contact site). For the quantification of the grayscale intensity of the intermediate density relative to the ER membrane (Fig. 3F), the positions of the ER membrane and the intermediate density were identified in every tomogram as the first and second minima, respectively, of the average grayscale intensity profiles (Fig. S3). Then, the ratio of grayscale intensity of the intermediate density and the ER membrane was calculated for every tomogram. The average ratios are displayed in Fig. 3F. The dynamic thresholding module of the Pyto package mentioned above was used to segment the densities present between the ER and the PM in Fig. 2 B and D, Left, and Movies S1, S2, and S4. Student's t test was used for statistical analysis of these values, which appeared to be normally distributed. The confidence values are indicated in the graphs: *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Frank Wilson for outstanding technical support; Xinran Liu, Hideki Shigematsu, and Stacy Wilson for help with microscopy experiments; Mirko Messa for help and advice with neuronal cultures; Olof Idevall-Hagren for sharing unpublished results and methodology; the Yale Center for Cellular and Molecular Imaging; Dmitry Tegunov for the direct electron detector frame alignment software; and Eri Sakata for critical reading of the manuscript. This work was supported in part by National Institutes of Health (NIH) Grants R37NS36251, DK082700, and DK45735 (to P.D.C.) and by fellowships from the Alexander von Humboldt Foundation (to R.F.-B.) and the Uehara Memorial Foundation and the Japan Society for the Promotion of Science (to Y.S.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 4837.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503191112/-/DCSupplemental.
References
- 1.Stefan CJ, Manford AG, Emr SD. ER-PM connections: sites of information transfer and inter-organelle communication. Curr Opin Cell Biol. 2013;25(4):434–442. doi: 10.1016/j.ceb.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helle SC, et al. Organization and function of membrane contact sites. Biochim Biophys Acta. 2013;1833(11):2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 3.Carrasco S, Meyer T. STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu Rev Biochem. 2011;80(1):973–1000. doi: 10.1146/annurev-biochem-061609-165311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogan PG, Lewis RS, Rao A. Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Annu Rev Immunol. 2010;28(1):491–533. doi: 10.1146/annurev.immunol.021908.132550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderie I, Schulz I, Schmid A. Direct interaction between ER membrane-bound PTP1B and its plasma membrane-anchored targets. Cell Signal. 2007;19(3):582–592. doi: 10.1016/j.cellsig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Haj FG, et al. Regulation of signaling at regions of cell-cell contact by endoplasmic reticulum-bound protein-tyrosine phosphatase 1B. PLoS ONE. 2012;7(5):e36633. doi: 10.1371/journal.pone.0036633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pichler H, et al. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur J Biochem. 2001;268(8):2351–2361. doi: 10.1046/j.1432-1327.2001.02116.x. [DOI] [PubMed] [Google Scholar]
- 8.Stefan CJ, et al. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144(3):389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 9.Maeda K, et al. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature. 2013;501(7466):257–261. doi: 10.1038/nature12430. [DOI] [PubMed] [Google Scholar]
- 10.Tavassoli S, et al. Plasma membrane—endoplasmic reticulum contact sites regulate phosphatidylcholine synthesis. EMBO Rep. 2013;14(5):434–440. doi: 10.1038/embor.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang CL, et al. Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Reports. 2013;5(3):813–825. doi: 10.1016/j.celrep.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 12.Levine T, Loewen C. Inter-organelle membrane contact sites: through a glass, darkly. Curr Opin Cell Biol. 2006;18(4):371–378. doi: 10.1016/j.ceb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Beh CT, McMaster CR, Kozminski KG, Menon AK. A detour for yeast oxysterol binding proteins. J Biol Chem. 2012;287(14):11481–11488. doi: 10.1074/jbc.R111.338400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Filseck JM, Mesmin B, Bigay J, Antonny B, Drin G. Building lipid ‘PIPelines’ throughout the cell by ORP/Osh proteins. Biochem Soc Trans. 2014;42(5):1465–1470. doi: 10.1042/BST20140143. [DOI] [PubMed] [Google Scholar]
- 15.Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010;11(10):739–750. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- 16.Holthuis JC, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510(7503):48–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- 17.Holthuis JC, Levine TP. Lipid traffic: floppy drives and a superhighway. Nat Rev Mol Cell Biol. 2005;6(3):209–220. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- 18.Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell. 2012;23(6):1129–1140. doi: 10.1016/j.devcel.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Toulmay A, Prinz WA. A conserved membrane-binding domain targets proteins to organelle contact sites. J Cell Sci. 2012;125(Pt 1):49–58. doi: 10.1242/jcs.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giordano F, et al. PI(4,5)P(2)-dependent and Ca2+-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 2013;153(7):1494–1509. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopec KO, Alva V, Lupas AN. Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria. Bioinformatics. 2010;26(16):1927–1931. doi: 10.1093/bioinformatics/btq326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325(5939):477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schauder CM, et al. Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature. 2014;510(7506):552–555. doi: 10.1038/nature13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lučič V, Rigort A, Baumeister W. Cryo-electron tomography: the challenge of doing structural biology in situ. J Cell Biol. 2013;202(3):407–419. doi: 10.1083/jcb.201304193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metuzals J, Chang D, Hammar K, Reese TS. Organization of the cortical endoplasmic reticulum in the squid giant axon. J Neurocytol. 1997;26(8):529–539. doi: 10.1023/a:1015482407202. [DOI] [PubMed] [Google Scholar]
- 26.Orci L, et al. STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc Natl Acad Sci USA. 2009;106(46):19358–19362. doi: 10.1073/pnas.0911280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suh BC, Horowitz LF, Hirdes W, Mackie K, Hille B. Regulation of KCNQ2/KCNQ3 current by G protein cycling: the kinetics of receptor-mediated signaling by Gq. J Gen Physiol. 2004;123(6):663–683. doi: 10.1085/jgp.200409029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi M, et al. Cell- and stimulus-dependent heterogeneity of synaptic vesicle endocytic recycling mechanisms revealed by studies of dynamin 1-null neurons. Proc Natl Acad Sci USA. 2008;105(6):2175–2180. doi: 10.1073/pnas.0712171105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenbluth J. Subsurface cisterns and their relationship to the neuronal plasma membrane. J Cell Biol. 1962;13(3):405–421. doi: 10.1083/jcb.13.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spacek J, Harris KM. Three-dimensional organization of smooth endoplasmic reticulum in hippocampal CA1 dendrites and dendritic spines of the immature and mature rat. J Neurosci. 1997;17(1):190–203. doi: 10.1523/JNEUROSCI.17-01-00190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petkovic M, et al. The SNARE Sec22b has a non-fusogenic function in plasma membrane expansion. Nat Cell Biol. 2014;16(5):434–444. doi: 10.1038/ncb2937. [DOI] [PubMed] [Google Scholar]
- 32.Soboloff J, Rothberg BS, Madesh M, Gill DL. STIM proteins: dynamic calcium signal transducers. Nat Rev Mol Cell Biol. 2012;13(9):549–565. doi: 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min SW, Chang WP, Südhof TC. E-Syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proc Natl Acad Sci USA. 2007;104(10):3823–3828. doi: 10.1073/pnas.0611725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J, et al. Structure and Ca2+-binding properties of the tandem C2 domains of E-Syt2. Structure. 2014;22(2):269–280. doi: 10.1016/j.str.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibata Y, et al. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem. 2008;283(27):18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calloway N, Vig M, Kinet JP, Holowka D, Baird B. Molecular clustering of STIM1 with Orai1/CRACM1 at the plasma membrane depends dynamically on depletion of Ca2+ stores and on electrostatic interactions. Mol Biol Cell. 2009;20(1):389–399. doi: 10.1091/mbc.E07-11-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raimondi A, et al. Overlapping role of dynamin isoforms in synaptic vesicle endocytosis. Neuron. 2011;70(6):1100–1114. doi: 10.1016/j.neuron.2011.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 40.Fernández-Busnadiego R, et al. Quantitative analysis of the native presynaptic cytomatrix by cryoelectron tomography. J Cell Biol. 2010;188(1):145–156. doi: 10.1083/jcb.200908082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.