Significance
Estrogen receptor β (ERβ) is thought to be the predominant nuclear receptor that regulates estrogen signaling in a wide variety of tissues. In several circumstances, it can oppose the effects of ERα. Here, we generated a novel ERβ exon 3-deleted mouse model (ERβ-Δex3) in which the first zinc finger of the DNA binding domain is missing but the remaining C-terminus part of the receptor is present. We observed that mutant male mice were normal but females were anovulatory. Thus, this work demonstrates that most of the physiological functions of ERβ do not involve estrogen response element binding.
Keywords: mouse model, nuclear receptors, targeted disruption, ventral prostate, AP-1
Abstract
In 1998, an estrogen receptor β (ERβ) knockout (KO) mouse was created by interrupting the gene at the DNA binding domain (DBD) with a neocassette. The mutant females were subfertile and there were abnormalities in the brain, prostate, lung, colon, and immune system. In 2008, another ERβ mutant mouse was generated by deleting ERβ exon 3 which encodes the first zinc finger in the DBD. The female mice of this strain were unable to ovulate but were otherwise normal. The differences in the phenotypes of the two KO strains, have led to questions about the physiological function of ERβ. In the present study, we created an ERβ exon 3-deleted mouse (ERβ-Δex3) and confirmed that the only observable defect was anovulation. Despite the two in-frame stop codons introduced by splicing between exons 2 and 4, an ERβ protein was expressed in nuclei of prostate epithelial cells. Using two different anti-ERβ antibodies, we showed that an in-frame ligand binding domain and C terminus were present in the ERβ-Δex3 protein. Moreover, with nuclear extracts from ERβ-Δex3 prostates, there was an ERβ-dependent retardation of migration of activator protein-1 response elements in EMSA. Unlike the original knockout mouse, expression of Ki67, androgen receptor, and Dachshund-1 in prostate epithelium was not altered in the ERβ-Δex3 mouse. We conclude that very little of ERβ transcriptional activity depends on binding to classical estrogen response elements (EREs).
The first estrogen receptor β knockout (ERβ−/−) mouse was generated in 1998 in the laboratory of Oliver Smithies and revealed a key role for this receptor in female and male reproduction (1). This mouse was characterized by a reduced ability to ovulate, incomplete differentiation of the epithelium in the mammary gland and prostate, defective migration of cortical neurons during fetal development (2, 3), age-dependent myeloid leukemia (4), and severe abnormalities in the lungs (5, 6) and colon (7). Aging male ERβ−/− mice developed prostate hyperplasia and prostatic intraepithelial neoplasia (PIN), a premalignant stage of adenocarcinoma (8). ERβ is also expressed in normal human prostate and in prostate cancer of Gleason grades up to 3 + 3 but its expression decreases as cancer progresses (9–11). The loss of ERβ expression during the progression from high-grade PIN to cancer suggests that ERβ may act as a tumor suppressor. The physiological functions of ERβ deduced from the original ERβ−/− mouse have been questioned (12) and suggested to be due to the presence of the neocassette, used to interrupt translation of the gene at the DNA binding domain (DBD). Indeed, a different ERβ mutant mouse created by deletion of exon 3 (encoding the DNA recognition sequence), showed no abnormalities other than sterility in both males and females (12).
For many years, the transcriptional action of estrogen receptors was thought to be mediated only through binding to specific estrogen response elements (EREs) on DNA. It is now known that one-third of the categorized human 17β-estradiol–responsive genes are transcribed via indirect ER–DNA association through protein–protein interactions with several transcription factors (13). Both ERα and ERβ can exert transcriptional regulation by tethering to other transcription factors such as c-Fos/c-Jun (activator protein-1, AP-1), Sp1, or NF-κB without themselves binding to EREs on DNA (13–18). Genome landscaping has revealed that the non–ERE-dependent mode of transcription is the preferred pathway used by ERβ to regulate transcription of its target genes. Remarkably, only 5% of ERβ-interacting regions include only EREs or ERE half-sites, whereas ∼60% contain AP-1–like binding regions together with ERE-like sites (19). In vitro studies have consistently revealed that ERβ is a weaker activator of classical EREs than ERα (20, 21). Initially, these data were interpreted as evidence that ERβ was not of much physiological relevance, whereas they actually highlighted the fact that both ERα and ERβ mediate transcription through two distinct pathways, with ERβ using predominantly non-ERE pathways.
Because ERβ−/− mouse lines generated in different laboratories have such different phenotypes, the physiological functions of ERβ remain debated. In the present study, we generated a novel exon 3-deleted mouse (ERβ-Δex3) and show that most of the physiological functions of ERβ were preserved in this mutant mouse.
Results
Generation of Floxed ERβ and ERβ-Δex3 Mice.
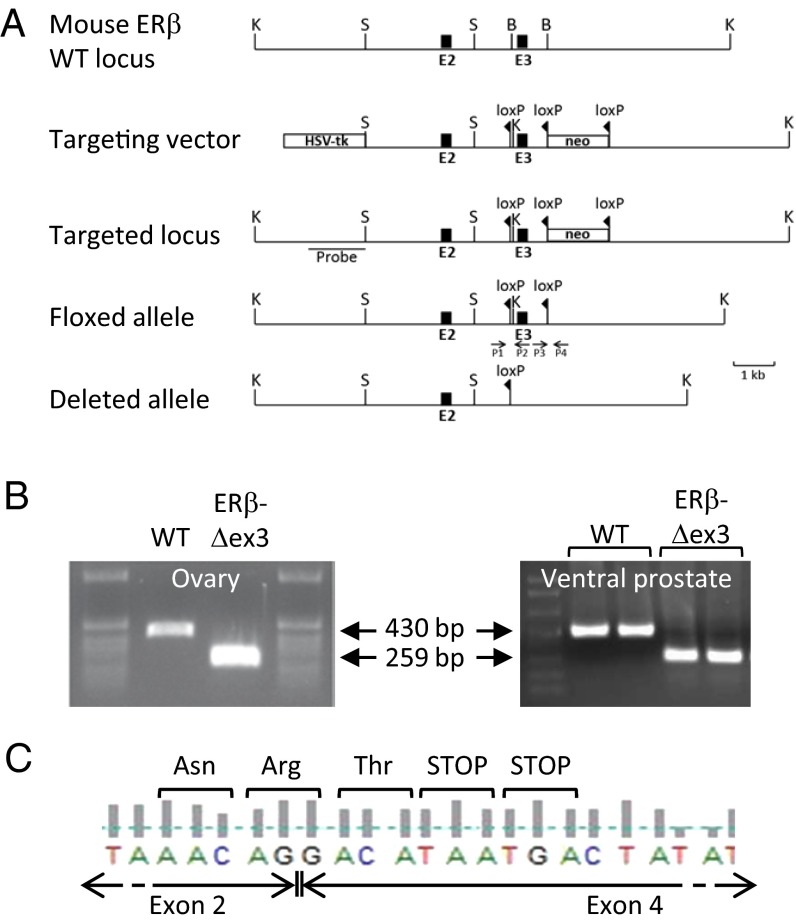
To generate ERβ-Δex3 mice, we used the Cre/loxP recombination system to target exon 3. Exon 3 of the ERβ gene encodes the first zinc finger in the DBD (ERE recognition finger) and removal of this exon should have resulted in a frame shift in the coding region after splicing from exon 2 to exon 4. The targeting vector was designed to introduce a loxP site in intron 2 and a loxP flanked neomycin cassette in intron 3 (Fig. 1A). Mice with a deleted ERβ allele were generated by crossing ERβ floxed mice with transgenic CMV-Cre or Rosa-Cre deleter mice. Breeding with both strains of Cre-deleter mice produced similar results; therefore the resultant mutant mice were named ERβ-exon 3 deleted (ERβ-Δex3) mice. An extended protocol for the generation of the ERβ-Δex3 mutant mice and genotyping data are described in SI Materials and Methods and Fig. S1).
Fig. 1.
Targeted disruption of the mouse ERβ gene. Structure of the WT ERβ allele, targeting vector, targeted locus, floxed allele, and deleted allele after Cre-recombination are shown with the KpnI (K), BamHI (B), and SalI (S) restriction sites. PCR genotyping primers (P1, P2, P3, and P4) are indicated by arrows (A). RT-PCR analysis of ERβ total RNA from WT and ERβ-Δex3 mice (B). WT ovaries and ventral prostate expressed a 430 bp ERβ mRNA, whereas a shorter transcript (259 bp) lacking exon 3 was detected in the ERβ-Δex3 mice. DNA sequencing analysis of cDNA extracted from mutant ovaries and ventral prostate indicates that splicing between exons 2 and 4 occurs in ERβ-Δex3 mice and generates a frame shift in the reading frame (C). This frame shift results in the creation of two in-frame stop codons.
Verification of Exon 3 Deletion.
To verify that exon 3 of the ERβ gene was deleted in mutant mice, we analyzed DNA and confirmed that exon 3 was deleted (Fig. S1 C and D). RNA was extracted from ovaries and ventral prostates (VPs) and analyzed by RT-PCR using primers located in exons 2 and 5. ERβ mRNA was present in the ovary and VP from wild-type (WT) and ERβ-Δex3 mice (Fig. 1B) and sequencing of the amplified product in mutant mouse revealed that it lacked exon 3 and that splicing had occurred between exons 2 and 4 (Fig. 1C). Because of the two in-frame stop codons present at the beginning of exon 4, the predicted translated protein was expected to be composed of 121 amino acids from ERβ and one amino acid from the frame shift in exon 4 and should lack both the DBD and the ligand binding domain (LBD) of ERβ. Similarly, an additional RT-PCR with a primer overlapping the stop codon in exon 9 together with the exon 2 primer detected only a single band. These data together with subsequent sequencing of amplification products suggest that no alternative splicing has occurred. Therefore, determination of cDNA sequences, as well as RT-PCR experiments with different sets of primers confirmed deletion of exon 3 and showed that no alternative splicing had occurred and no transcript variants were produced in this mouse.
Ovarian Dysfunction in ERβ-Δex3 Mouse.
The ERβ-Δex3 female mice are infertile. The overall appearance of the ovaries and uteri were not different from those of WT mice (Fig. S2). However, histological analysis of ovarian cross-sections revealed absence of corpora lutea, an increase in the number of atretic follicles with no detectable increase in the number of growing follicles, and no viable preovulatory follicles (Fig. S2). Thus, the infertility of the ERβ-Δex3 mouse appears to be due to a lack of late follicular maturation and ovulation.
Ventral Prostates of the ERβ-Δex3 Mouse.
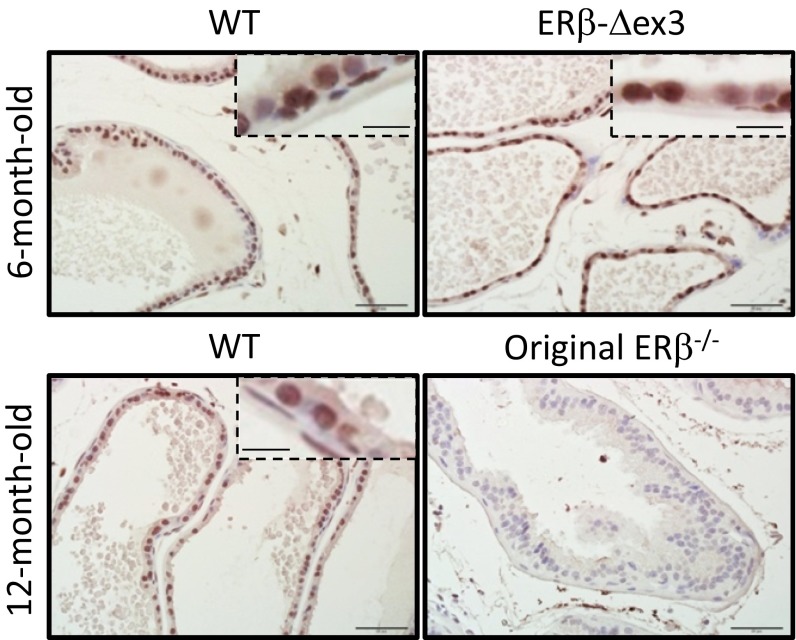
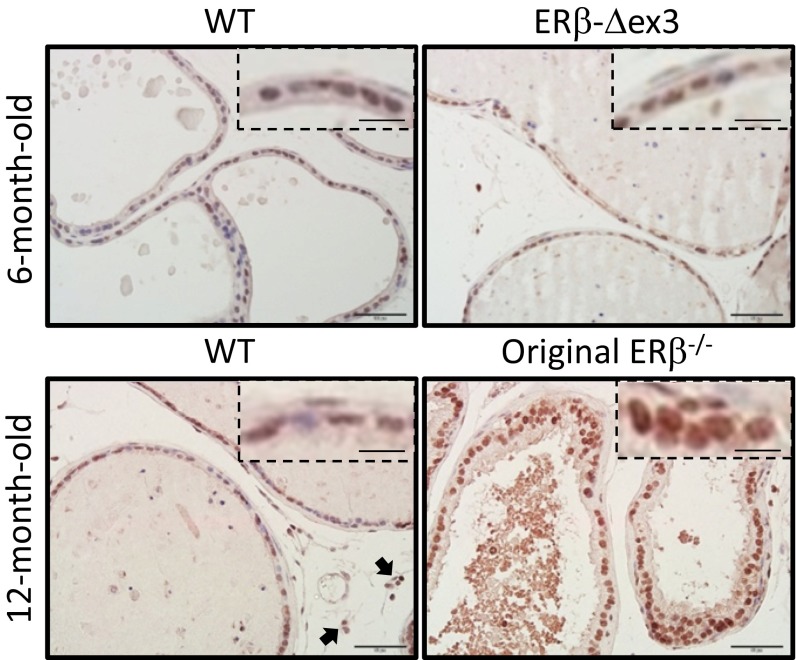
Although ovarian defects in the ERβ-Δex3 mouse were similar to those in the original ERβ−/− mouse, their ventral prostate appears to be unaffected by loss of ERβ exon 3. Because gene expression, morphology, and proliferation were affected in the ventral prostate of the original ERβ−/− mouse, the reason for this difference was further investigated. By immunohistochemical analysis, ERβ was localized in the nuclei of the epithelial, basal, and stromal cells of the ventral prostate of WT and ERβ-Δex3 mice (Fig. 2). Expression of ERβ was similar whether CMV-Cre or Rosa-Cre were used to delete exon 3 (Fig. 2). With the same ERβ 503 antibody, no signal was detected in the prostatic epithelium of original ERβ−/− mice.
Fig. 2.
Localization of ERβ in the WT and ERβ-Δex3 mouse prostates. Ventral prostates of WT, ERβ-Δex3, and original ERβ−/− mice aged 6 or 12 mo were stained for ERβ. ERβ staining is mostly localized in the nuclei of the epithelial cells. Some basal and stromal cells are also positive. Epithelial cells of the WT and ERβ-Δex3 VP present a strong signal. There is no significant difference in the staining between the WT and the mutant mice. Furthermore, there is no detectable staining in the original ERβ−/− mouse prostate. For each picture, a close-up view of the epithelial cell layer has been included. (Scale bars, 50 µm or 12.5 µm for the close-up view.)
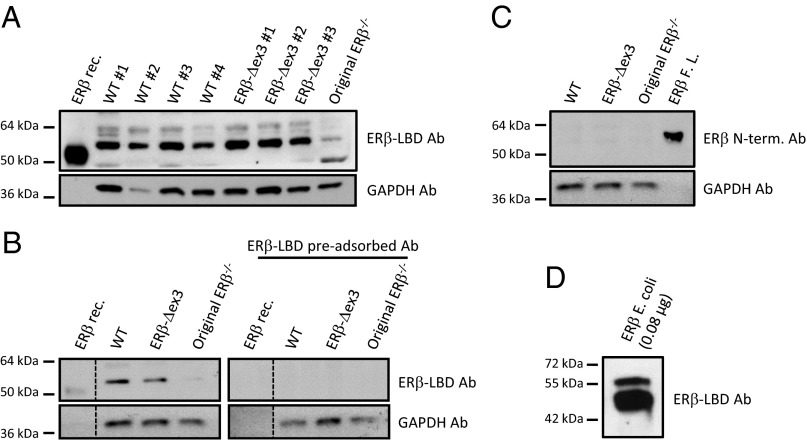
Although the ERβ transcript produced by the recombination had two in-frame stop codons, the mouse expressed an ERβ protein including an in-frame LBD, but lacking the first zinc finger. Indeed, on Western blots with cellular extracts from the ventral prostate, ERβ protein of molecular weight 55 kDa was detected (Fig. 3 A and B). The antibody used was raised against the ERβ LBD. As a positive control for ERβ detection, we used a commercial ERβ recombinant protein whose predicted molecular weight is 53.4 kDa due to the deletion of part of the N-terminal domain of the protein. Increasing concentrations of ERβ recombinant protein loaded onto the gel (1–30 pmol) led to a concentration-dependent increase in the ERβ signal detected by the ERβ–LBD antibody (Fig. S3). In ventral prostate extracts from the original ERβ−/− mouse, no ERβ protein was detectable (Fig. 3 A and B). Fig. 3B also shows loss of detection of ERβ protein when the ERβ–LBD antibody was preabsorbed with the ERβ protein, thereby establishing the specificity of the antibody. Therefore, not only was there an ERβ protein migrating on SDS/PAGE as a 55-kDa band but ERβ was also detected in the nuclei of prostate epithelium and lungs by immunohistochemistry. Three different anti-ERβ antibodies whose epitopes target the LBD of ERβ1, the N-terminal domain of ERβ, and the C-terminal peptide of ERβ showed that in the ERβ-Δex3 mouse there is an in-frame LBD and C terminus, with absence of N terminus (Fig. S4).
Fig. 3.
ERβ protein expression in the WT and ERβ-Δex3 mouse ventral prostate. Western blot using ERβ–LBD antibody shows that bands of 55 kDa were detected in both WT as well as ERβ-Δex3 mouse VP (A). The specificity of the ERβ–LBD antibody was assessed by preabsorbing the antibody with ERβ protein (B). ERβ protein expression was not detectable in the original ERβ−/− mouse VP (B). An antibody raised against the N-terminal part of ERβ did not detect the receptor in mouse protein extracts (C). As ERβ protein expressed in E. coli frequently undergoes in vitro protein degradation, it displays full-length and N-terminally truncated isoforms of the receptor at the same time (D). Ab, antibody; ERβ E. coli, ERβ protein expressed in E. coli; ERβ FL, full-length ERβ recombinant protein; ERβ rec, ERβ recombinant protein.
In vitro transcription/translation of ERβ mRNA extracted from ERβ-Δex3 mouse ovaries produced a truncated protein whose apparent molecular weight is about 20 kDa (Fig. S5). Thus, in vivo, but not in vitro, the ERβ-Δex3 mRNA produces an ERβ protein that seems to retain the normal translational reading frame. With the deletion of exon 3, the molecular weight of the ERβ-Δex3 protein should be lower than that in the WT mouse. However, the protein in the WT mouse prostate extracts migrated on SDS gels with a similar molecular weight as the ERβ-Δex3 protein. To explain the apparent comigration of ERβ in WT and ERβ-Δex3 mice on Western blots, we used an anti-ERβ N-terminal antibody to determine whether there was an intact N terminus in the WT mouse protein. With this antibody raised against amino acid 1–153 in ERβ1, no band was detected in extracts from the prostates of WT mice (Fig. 3C) but we detected a 59-kDa band in purified full-length commercial ERβ (Fig. 3C).
Thus, it appears that there is degradation of ERβ with removal of the N terminus during preparation of the tissue extracts resulting in a 55-kDa protein. ERβ expressed in Escherichia coli purified to homogeneity and stored at −80 °C in the presence of protease inhibitor mixture also degrades with time and is converted from a single band of 59 kDa to a 50-kDa band (Fig. 3D). The cause of the degradation remains unclear but may be related to the unstructured nature of the N terminus, which may make it more sensitive to proteolytic cleavage. We conclude that the 55-kDa band in the WT is due to a degradation of ERβ1, which occurs during handling of tissue.
Morphological Study of the WT and ERβ-Δex3 Mouse Ventral Prostate.
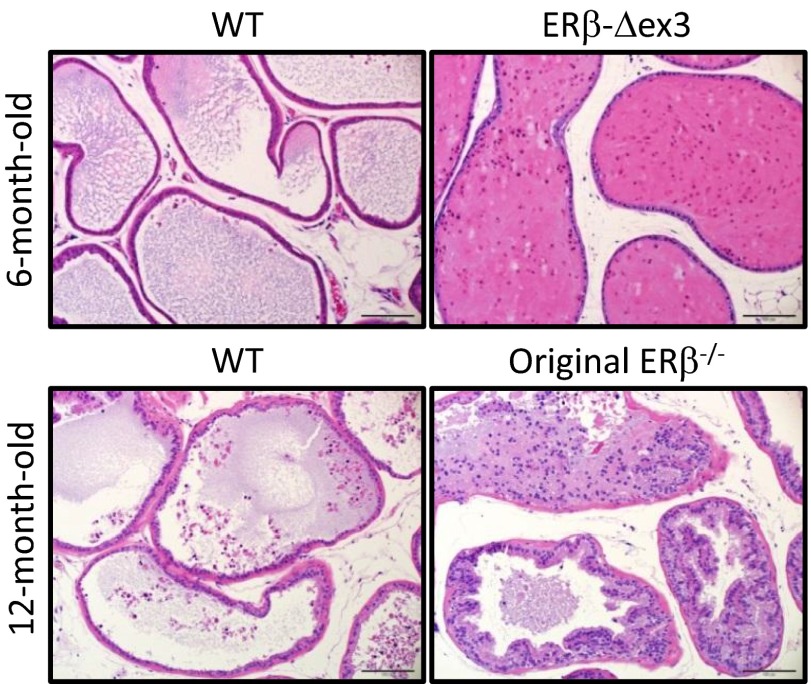
The ducts of the ventral prostates of 6- and 12-mo-old WT mice are organized in a single layer of columnar or cuboidal epithelium surrounded by thin layers of stroma (Fig. 4). After H&E staining, the ventral prostates of ERβ-Δex3 mice appeared histologically normal and morphologically indistinguishable from those of age-matched WT littermates. In 6-mo-old mice, proliferation in the ERβ-Δex3 mouse VP was similar compared with the WT (Fig. S6). In the ventral prostate of 12-mo-old original ERβ−/− mice, there are multiple foci of hyperplasia characterized by proliferation of epithelial cells, as previously observed by Imamov et al. and Weihua et al. (8, 22). In the epithelial cell layer, there were multiple infoldings and piling up, with accumulation of cells inside the lumen of the prostatic ducts (Fig. S7). Taken together, these results indicate that deletion of exon 3 of ERβ does not affect the morphological structure of the VP.
Fig. 4.
Histological structure of WT and ERβ-Δex3 mouse VP. Representative sections of the ventral prostates collected from WT, ERβ-Δex3 mice, or original ERβ−/− mice were stained with hematoxylin and eosin (H&E). H&E-stained ventral prostate tissues displayed no differences in histology between aged-matched WT and ERβ-Δex3 mouse tissues. In the original mutant mouse ventral prostate, the prostatic epithelium was disorganized and the ducts were filled with multiple layers of proliferative epithelial cells. (Scale bar, 100 µm.)
Lack of Changes in Gene Expression in ERβ-Δex3 Mouse Ventral Prostate.
Immunostaining for androgen receptor and Dachshund-1 (DACH-1) revealed that in both WT and ERβ-Δex3 mice, there was a clear nuclear staining of these proteins in the prostatic epithelium (Fig. 5 and Fig. S8A). Androgen receptor was also expressed in immune cells, as indicated by black arrows. DACH-1 expression was particularly strong and specific for the epithelial cells. There was no significant difference in androgen receptor or DACH-1 expression between the WT and ERβ-Δex3 ventral prostate. In contrast, in the original ERβ−/− ventral prostate, protein expression of androgen receptor as well as DACH-1 was higher than that in WT mice (Fig. 5 and Fig. S8 A and B).
Fig. 5.
Androgen receptor immunostaining in the WT and ERβ-Δex3 mouse ventral prostates. In the ventral prostates of 6- or 12-mo-old WT, ERβ-Δex3, or original ERβ−/− mice strong nuclear expression of androgen receptor was observed in the epithelial cells with some stromal cells also positive. Androgen receptor-positive immune cells are indicated by black arrows. No detectable changes in the intensity or the number of positive cells were observed in the ERβ-Δex3 mouse ventral prostate compared with the WT littermate. In contrast, in the original ERβ−/− ventral prostate, expression of androgen receptor was increased. (Scale bar, 50 µm.)
In the ventral prostate, the scaffolding protein caveolin-1 is specifically expressed at the plasma membranes of epithelial and endothelial cells, as well as in the stroma surrounding the epithelial ducts. By immunostaining, we demonstrated that caveolin-1 was expressed in the epithelium and stromal layer of the prostatic ducts in WT and ERβ-Δex3 mice. Expression was significantly reduced in the epithelium and stroma of original ERβ−/− ventral prostate ducts (Fig. S9).
ERβ and AP-1 DNA Binding Activity in WT and ERβ-Δex3 Mice.
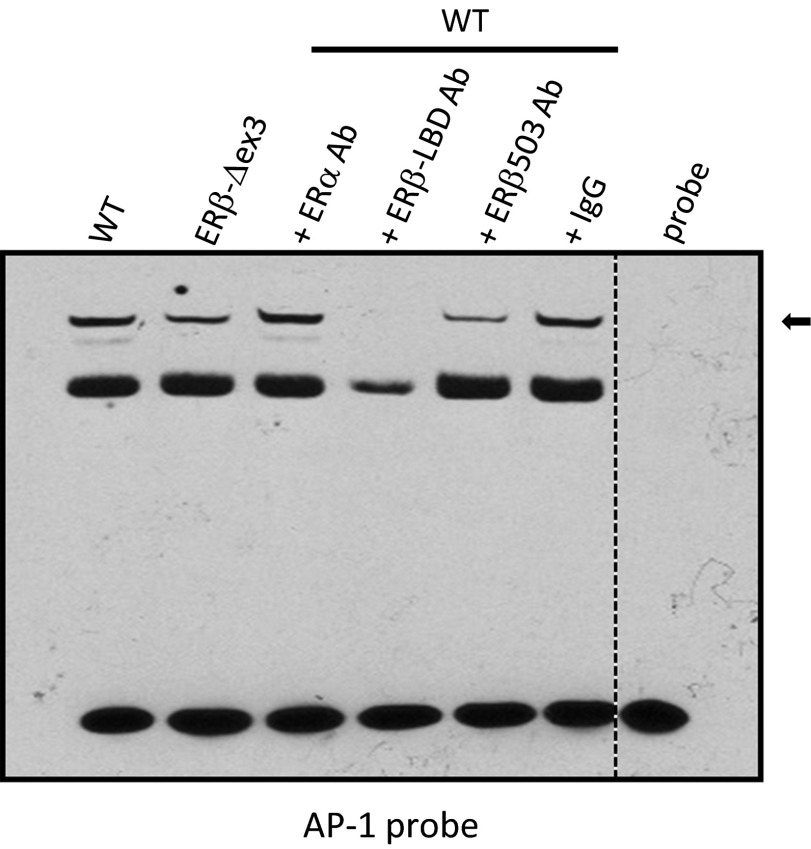
Both WT and ERβ-Δex3 ventral prostate nuclear extracts formed complexes with an AP-1 response element and the addition of an anti-ERβ antibody prevented and/or strongly decreased the binding of the AP-1 complex to its responsive element (Fig. 6). In contrast, addition of an anti-ERα antibody did not alter complex formation with the AP-1 probe. These results indicate that the ERβ protein produced by the ERβ-Δex3 mouse is able to form AP-1/ERβ protein complexes able to bind AP-1 responsive elements.
Fig. 6.
ERβ and AP-1 DNA binding activity in WT and mutant mouse ventral prostate. EMSAs using the 5′-biotinylated double-stranded AP-1 probes with nuclear extracts from WT or ERβ-Δex3 mouse ventral prostate revealed that the ERβ protein produced in the ERβ-Δex3 mice binds to the AP-1 complex at the AP-1 responsive elements in the ventral prostate. Formation of the DNA–protein complexes was strongly decreased by the addition of ERβ–LBD or ERβ 503 antibodies. Negative controls lacking nuclear extracts are shown (probe). ERβ and AP-1 protein–DNA complexes are indicated by an arrow.
Discussion
Since the report of its discovery in 1996 (23), the physiological role of ERβ has often been debated. Its absence from the uterus and pituitary caused some endocrinologists to question whether it could be a mediator of the actions of 17β-estradiol. However, ERβ signaling actually does mediate many of the nonreproductive functions of estradiol and some of those which were called “indirect” because of the absence of ERα in estrogen-responsive tissues.
In addition to the classical mode of ER transcriptional activity, ER can signal through tethering mechanisms, in which ER acts as a coactivator or corepressor of other transcription factors, without the involvement of EREs (14, 24). The predominant role of this nonclassical pathway in ERβ signaling was revealed by genomic landscaping (19). In several systems, the capacity of ERβ to activate ERE-regulated reporter genes in transactivation assays was much weaker than that of ERα (21). Despite this finding, the DBD continued to be the target for generating ERβ knockout (KO) mice. It is this strategy which has caused the recent conclusion that ERβ is of little physiological relevance (12). This misunderstanding has, unfortunately, reduced the enthusiasm for development of ERβ agonists, which would otherwise be actively pursued as pharmaceuticals for treatment of diseases of the prostate and central nervous and immune systems.
In contrast to the ERβ-Δex3 mice, in ERα-Δex3 mice generated with a comparable Cre/loxP-mediated recombination, ERα function was completely lost, indicating the dominance of ERE binding for the function of ERα (25). The reason why a similar recombination leads to divergent results while targeting two closely related genes remains to be elucidated. Ribosomal subunits do not efficiently dissociate from the translated mRNA when stalling at a premature stop codon if it is not closely followed by a poly(A) signal. Indeed, proper translation termination depends on a stimulating signal from the poly(A) binding protein and therefore only occurs when the stop codon is in spatial proximity of the poly(A) tail region (26–28). For that reason, we speculate that the transcription machinery can read through the two stop codons (TAA followed by TGA) in the case of the manipulated ERβ gene. In addition, the read-through efficiency was found to associate with the identity of the premature stop codon and its sequence context. In our study, the ERβ-Δex3 cDNA sequence determination showed that the stop codon TGA is followed by a cytosine residue. Interestingly, the highest read-through efficiency is of the TGA stop codon, followed by TAG and, to a lesser extent, TAA (29). The sequences upstream and downstream of the stop codon also have an important role in determining its susceptibility to read through. For example, a cytosine residue after either the stop codon TGA or TAA (position +4) is correlated with high levels of read through (29). In addition, removal of exon 3 should have mediated a frame shift in the coding sequence. As ERβ expressed in ERβ-Δex3 mouse is being detected by several anti-ERβ antibodies directed against the LBD and C terminus part of the receptor, we can also hypothesize that ribosomes undergo a frame shift while reading the mRNA sequence and that the resulting ERβ protein retains the regular reading frame.
Because the ERβ-Δex3 mouse was normal except for the inability of females to ovulate, we have concluded that most of the functions of ERβ were preserved in ERβ-Δex3 mice. In contrast, removal of the exon 3 from the ERα gene was sufficient to interrupt ERα signaling (26), indicating that most of ERα signaling occurs via binding of ERα to EREs. Indeed, no ERα protein was detected in these mice and ERα−/− female mice develop obesity with hyperglycemia and display hemorrhagic polycystic ovaries and atrophic uteri (25). A study by Price et al. (30) showed that, in the presence of ER agonists, an exon 3-deleted splice variant of ERβ missing the second zinc finger in the DBD could transactivate luciferase reporter constructs containing an AP-1 site, but not an ERE. More recently, re-ChIP studies performed on a genome-wide scale showed co-occupancy of ERβ and AP-1 on chromatin, with a decrease in ERβ recruitment to chromatin when siRNAs targeting c-Fos or c-Jun were used (19). Our results show that the addition of an anti-ERβ antibody prevented and/or strongly decreased the binding of the AP-1 protein complex to its responsive elements. Taken together, our data confirm the major role of AP-1 in mediating estrogen signaling in the mouse ventral prostate.
ERβ reduces or limits the growth stimulating effects of androgen receptor in the prostatic epithelium (31, 32). In accordance with our previous studies (22), our results show that the content of androgen receptor is higher in the original ERβ−/− ventral prostate than in those of WT littermates. We also found that Dachshund-1 protein is an ERβ-regulated gene, which is coexpressed with ERβ in the epithelium of the ventral prostate (Fig. S8C). DACH-1 physically associates with androgen receptor and inhibits the transcriptional activity of androgen-dependent androgen receptor functions like cellular growth, DNA synthesis, and proliferation (33). Therefore, the loss of DACH-1 expression may lead to enhanced androgen receptor activity. This finding helps to explain one mechanism through which ERβ may repress androgen receptor activity. DACH-1 is expressed in normal prostatic epithelial cells but its expression is strongly reduced in prostate cancer (33) and this loss correlates with tumor progression and invasiveness. In the original ERβ−/− mouse, expression of androgen receptor as well as DACH-1 in the nuclei of the ventral prostate was higher than in WT littermates but there were no detectable changes in androgen receptor or DACH-1 expression in the ventral prostate epithelium of the ERβ-Δex3 mouse. These data indicate that control of prostatic growth is probably mediated mostly through ERE-independent pathways like the AP-1 pathway. As DACH-1 was also shown to repress a variety of AP-1 responsive genes, and to physically interact with c-Jun and repress its function (34), it is possible that a protein complex involving ERβ and DACH-1 could influence the transcriptional activity of the androgen receptor gene in the prostate.
In humans, levels of caveolin-1 in tumor epithelial cells increase during prostate cancer progression. Conversely, current evidence indicates that caveolin-1 loss in prostate cancer-associated stroma contributes to the metastatic behavior of tumor cells in advanced and metastatic prostate cancer (35) and loss of caveolin-1 expression in the prostatic tumor-associated stroma is associated with high Gleason score (36). In clinical studies, significant down-regulation of the protein in prostate stroma has been shown to mediate progression to the castration-resistant phase of prostate cancer through diverse pathways (37). Previously, our laboratory had shown that caveolin-1 expression was lower in original ERβ−/− mouse gastrocnemius muscle than in WT littermates (38). In the present study, we found strong caveolin-1 staining in the stroma of the WT and ERβ-Δex3 mouse VP. However, there was very little expression of caveolin-1 in the stroma of original ERβ−/− ventral prostate. Low levels of caveolin-1 in the stroma of the original ERβ−/− ventral prostate might contribute to the development of prostate hyperplasia and prostatic intraepithelial neoplasia in aging original ERβ−/− male mice.
The inability of ERβ-Δex3 mice to ovulate is one dysfunction common to both mutant ERβ strains of mice and suggests involvement of EREs in the ovulation process. Estrogen signaling to gonadotropin releasing hormone (GnRH) neurons is critical for coordinating the preovulatory surge of luteinizing hormone (LH) with follicular maturation. The GnRH gene is directly down-regulated by estrogen in the hypothalamus, and, at the time of the preovulatory surge, there is a paradoxical increase in GnRH secretion that triggers ovulation (39). More precisely, the human GnRH gene has been shown to be directly inducible by estrogens via an ERE located between −547 and −516 bp of its promoter (40). Although some studies have reported ERα immunoreactivity in GnRH neurons (41), others have claimed that ERβ is the predominant receptor (42). Even though we present evidence that very few functions of ERβ require its direct interaction with EREs, one exception may be the estrogen-dependent regulation of GnRH secretion. The individual role of ERα and ERβ in feedback regulation of GnRH is still unresolved. Loss of ERα from the arcuate nucleus does not inhibit acute feedback of estrogen on LH release from the pituitary (43). Neuronal inactivation of ERβ results in failure of estradiol to suppress LH secretion. However, this does not appear to be due to effects on GnRH synthesis or secretion because knockout of ERβ in GnRH neurons did not affect cyclicity or feedback regulation of estradiol on LH secretion (44). ERβ is not expressed in the adult pituitary nor is it expressed in the arcuate nucleus. So the cells in which ERβ regulates LH secretion remain to be identified.
In conclusion, the main finding in this study is that most of the physiological functions of ERβ do not involve ERE binding. More specifically, our results suggest that ERβ control of prostatic growth is probably mediated mostly through non-ERE mechanisms like the AP-1 pathway and suggest novel pathways as targets for treating abnormal prostatic growth.
Materials and Methods
Western Blotting.
The cell extract-associated proteins (20 µg) were resolved on a 10% polyacrylamide gel, using 1% SDS/Tris glycine buffer. The proteins were then electrotransferred onto polyvinylidene difluoride (PVDF) membrane (Biorad) and the free protein-binding sites of the PVDF membranes were blocked for 1 h in Tris buffer saline (TBST, 20 mM Tris, 137 mM NaCl, 0.1% Tween 20) containing 5% (wt/vol) nonfat dry milk. The membranes were incubated with a rabbit anti-ERβ LBD antibody, a mouse anti-ERβ N-terminal antibody, or a rabbit anti-GAPDH (FL-335-HRP, Santa Cruz Biotechnology) antibody in TBST supplemented with 5% (wt/vol) nonfat dry milk. After overnight incubation at 4 °C, the membranes incubated with the anti-ERβ antibody were rinsed in TBST buffer and incubated for 1 h with the appropriate secondary antibody at 1:6,000 dilution, whereas the membranes incubated with the anti-GAPDH antibody were washed and directly processed for development. Protein signals were revealed using an Amersham ECL Plus Western Blotting Detection Reagent (GE Healthcare). The ERβ signal was normalized to that of GAPDH. The ERβ human recombinant protein (ERβ rec) used as a positive control of ERβ detection was purchased from Invitrogen (reference P2466). The estimated molecular weight of this N-terminal truncated protein, calculated by the vendor, is 53.4 kDa. As a second positive control for ERβ detection, 0.08 µg of ERβ protein expressed in E. coli (ERβ E. coli) was directly subjected to the gel used for resolution of the cellular extracts. This ERβ protein was produced through a bacterial expression system [BL21 (DE3) cells] and purified by heparin affinity chromatography columns. A full-length human ERβ1 recombinant protein (ERβ FL) was a generous gift from Christophoros Thomas, Center for Nuclear Receptors and Cell Signaling, Department of Biology and Biochemistry, University of Houston, Houston, and was initially purchased from Pan Vera.
Immunohistochemistry.
Five-micrometer paraffin-embedded sections were dewaxed in xylene, rehydrated, and processed for antigen retrieval with 10 mM citrate buffer (pH 6.0) in a Lab Vision PT module (Thermo Scientific). The cooled sections were incubated in a buffer composed of 50% (vol/vol) methanol and 3% (vol/vol) H2O2 for 30 min to quench endogenous peroxidase, and then unspecific binding was blocked by incubating the slides in 3% (wt/vol) BSA with 0.1% Nonidet P-40 in PBS for 1 h. Sections were then immunostained with anti-ERβ 503 (anti-ERβ antibody mapping the C-terminus part of the receptor), antiandrogen receptor, or anti-Ki67 antibodies in 1% BSA with 0.1% Nonidet P-40 in PBS overnight at 4 °C. The 1% BSA with 0.1% Nonidet P-40 in PBS replaced primary antibodies in negative controls. After washing, sections stained with the anti-ERβ antibody were incubated with a biotinylated goat anti-chicken secondary antibody (1:200 dilution) for 1 h at room temperature and then Vectastain ABC kit (Vector Laboratories) was used for the avidin–biotin complex method according to the manufacturer's instructions. Rabbit-on-Rodent HRP-Polymer reagent (Biocare Medical) was used for the antiandrogen receptor and anti-Ki67 antibodies. After sections were washed in PBS, peroxidase activity was visualized with 3,3′-diaminobenzidine (DAKO or Thermo Scientific). The sections were lightly counterstained with Mayer’s hematoxylin (Sigma-Aldrich), dehydrated through an ethanol series to xylene, and mounted with Permount (Fisher Scientific).
EMSAs.
DNA–protein binding assays were carried out with 5 µg of prostate nuclear extracts from WT or ERβ-Δex3 mice. Synthetic 5′-biotinylated complementary oligonucleotides were purchased from IDT and annealed for 5 min at 95 °C in Tris-EDTA buffer (10 mM Tris, 1 mM EDTA). The forward sequence of the double-stranded oligonucleotides used is 5′-CGCTTGATGACTCAGCCGGAA-3′ for the AP-1 probe. The reactions were carried out for 10 min at room temperature followed by 10 min on ice in the presence of 1× binding buffer composed of 50 ng/μL poly (dI-dC), 20 mM Tris pH 7.9, 1 mM EDTA, 2 mM DTT, 100 mM NaCl, 1 mM Na3Vo4, and 0.02% BSA, using 20 fmoles of biotin-end-labeled target. The 2 μL of anti-ERβ 503, anti-ERβ-LBD, anti-ERα antibody or corresponding control IgG were added per 20 μL of binding reaction where indicated. Assays were loaded onto native 5% polyacrylamide gels (Biorad) preelectrophoresed for 40 min in 0.5× Tris borate/EDTA (TBE), and electrophoresed for 50 min at 100 V before being transferred onto a positively charged nylon membrane (Biodyne B, Pierce) in 0.5× TBE at 100 V for 45 min. Transferred protein–DNA complexes were cross-linked to the membrane on a transilluminator equipped with 312-nm bulbs for 15 min and detected using HRP-conjugated streptavidin (LightShift Chemiluminescent EMSA kit, Pierce) according to the manufacturer's instructions.
Animal Experiments.
The animal studies were approved by the Stockholm South ethical review board and the local Animal Experimentation Ethics Committee for animal experimentation (University of Houston animal protocol 09-036).
Supplementary Material
Acknowledgments
We thank Bilqees Bhatti, Dr. Kaberi Das, and Christopher Brooks for excellent technical assistance and Dr. Xiaohua Lou for providing the ERβ protein expressed in E. coli. This study was supported by the Swedish Cancer Society, the Cancer Prevention and Research Institute of Texas (Grant RP110444-P1), the Texas Emerging Technology Fund under Agreement 18 No. 300-9-1958, and the Robert A. Welch Foundation (E-0004).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504944112/-/DCSupplemental.
References
- 1.Krege JH, et al. Generation and reproductive phenotypes of mice lacking estrogen receptor beta. Proc Natl Acad Sci USA. 1998;95(26):15677–15682. doi: 10.1073/pnas.95.26.15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L, Andersson S, Warner M, Gustafsson JA. Estrogen receptor (ER)beta knockout mice reveal a role for ERbeta in migration of cortical neurons in the developing brain. Proc Natl Acad Sci USA. 2003;100(2):703–708. doi: 10.1073/pnas.242735799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Andersson S, Warner M, Gustafsson JA. Morphological abnormalities in the brains of estrogen receptor beta knockout mice. Proc Natl Acad Sci USA. 2001;98(5):2792–2796. doi: 10.1073/pnas.041617498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shim GJ, et al. Disruption of the estrogen receptor beta gene in mice causes myeloproliferative disease resembling chronic myeloid leukemia with lymphoid blast crisis. Proc Natl Acad Sci USA. 2003;100(11):6694–6699. doi: 10.1073/pnas.0731830100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patrone C, et al. Regulation of postnatal lung development and homeostasis by estrogen receptor beta. Mol Cell Biol. 2003;23(23):8542–8552. doi: 10.1128/MCB.23.23.8542-8552.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morani A, et al. Lung dysfunction causes systemic hypoxia in estrogen receptor beta knockout (ERbeta-/-) mice. Proc Natl Acad Sci USA. 2006;103(18):7165–7169. doi: 10.1073/pnas.0602194103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wada-Hiraike O, et al. Role of estrogen receptor beta in colonic epithelium. Proc Natl Acad Sci USA. 2006;103(8):2959–2964. doi: 10.1073/pnas.0511271103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imamov O, et al. Estrogen receptor beta regulates epithelial cellular differentiation in the mouse ventral prostate. Proc Natl Acad Sci USA. 2004;101(25):9375–9380. doi: 10.1073/pnas.0403041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muthusamy S, et al. Estrogen receptor β and 17β-hydroxysteroid dehydrogenase type 6, a growth regulatory pathway that is lost in prostate cancer. Proc Natl Acad Sci USA. 2011;108(50):20090–20094. doi: 10.1073/pnas.1117772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath LG, et al. Frequent loss of estrogen receptor-beta expression in prostate cancer. Cancer Res. 2001;61(14):5331–5335. [PubMed] [Google Scholar]
- 11.Latil A, et al. Evaluation of androgen, estrogen (ER alpha and ER beta), and progesterone receptor expression in human prostate cancer by real-time quantitative reverse transcription-polymerase chain reaction assays. Cancer Res. 2001;61(5):1919–1926. [PubMed] [Google Scholar]
- 12.Antal MC, Krust A, Chambon P, Mark M. Sterility and absence of histopathological defects in nonreproductive organs of a mouse ERbeta-null mutant. Proc Natl Acad Sci USA. 2008;105(7):2433–2438. doi: 10.1073/pnas.0712029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Lone R, Frith MC, Karlsson EK, Hansen U. Genomic targets of nuclear estrogen receptors. Mol Endocrinol. 2004;18(8):1859–1875. doi: 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]
- 14.Webb P, et al. The estrogen receptor enhances AP-1 activity by two distinct mechanisms with different requirements for receptor transactivation functions. Mol Endocrinol. 1999;13(10):1672–1685. doi: 10.1210/mend.13.10.0357. [DOI] [PubMed] [Google Scholar]
- 15.Leung YK, Gao Y, Lau KM, Zhang X, Ho SM. ICI 182,780-regulated gene expression in DU145 prostate cancer cells is mediated by estrogen receptor-beta/NFkappaB crosstalk. Neoplasia. 2006;8(4):242–249. doi: 10.1593/neo.05853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing D, et al. Estrogen modulates NFκB signaling by enhancing IκBα levels and blocking p65 binding at the promoters of inflammatory genes via estrogen receptor-β. PLoS ONE. 2012;7(6):e36890. doi: 10.1371/journal.pone.0036890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vivar OI, et al. Estrogen receptor beta binds to and regulates three distinct classes of target genes. J Biol Chem. 2010;285(29):22059–22066. doi: 10.1074/jbc.M110.114116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saville B, et al. Ligand-, cell-, and estrogen receptor subtype (alpha/beta)-dependent activation at GC-rich (Sp1) promoter elements. J Biol Chem. 2000;275(8):5379–5387. doi: 10.1074/jbc.275.8.5379. [DOI] [PubMed] [Google Scholar]
- 19.Zhao C, et al. Genome-wide mapping of estrogen receptor-beta-binding regions reveals extensive cross-talk with transcription factor activator protein-1. Cancer Res. 2010;70(12):5174–5183. doi: 10.1158/0008-5472.CAN-09-4407. [DOI] [PubMed] [Google Scholar]
- 20.Kuiper GG, et al. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinology. 1997;138(3):863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 21.McInerney EM, Weis KE, Sun J, Mosselman S, Katzenellenbogen BS. Transcription activation by the human estrogen receptor subtype beta (ER beta) studied with ER beta and ER alpha receptor chimeras. Endocrinology. 1998;139(11):4513–4522. doi: 10.1210/endo.139.11.6298. [DOI] [PubMed] [Google Scholar]
- 22.Weihua Z, et al. A role for estrogen receptor beta in the regulation of growth of the ventral prostate. Proc Natl Acad Sci USA. 2001;98(11):6330–6335. doi: 10.1073/pnas.111150898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson JA. Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci USA. 1996;93(12):5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paech K, et al. Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science. 1997;277(5331):1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- 25.Antonson P, Omoto Y, Humire P, Gustafsson JA. Generation of ERα-floxed and knockout mice using the Cre/LoxP system. Biochem Biophys Res Commun. 2012;424(4):710–716. doi: 10.1016/j.bbrc.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 26.Bordeira-Carriço R, Pêgo AP, Santos M, Oliveira C. Cancer syndromes and therapy by stop-codon readthrough. Trends Mol Med. 2012;18(11):667–678. doi: 10.1016/j.molmed.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 27.Mühlemann O, Eberle AB, Stalder L, Zamudio Orozco R. Recognition and elimination of nonsense mRNA. Biochim Biophys Acta. 2008;1779(9):538–549. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 28.Silva AL, Ribeiro P, Inácio A, Liebhaber SA, Romão L. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA. 2008;14(3):563–576. doi: 10.1261/rna.815108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manuvakhova M, Keeling K, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA. 2000;6(7):1044–1055. doi: 10.1017/s1355838200000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price RH, Jr, et al. A splice variant of estrogen receptor beta missing exon 3 displays altered subnuclear localization and capacity for transcriptional activation. Endocrinology. 2001;142(5):2039–2049. doi: 10.1210/endo.142.5.8130. [DOI] [PubMed] [Google Scholar]
- 31.McPherson SJ, et al. Estrogen receptor-beta activated apoptosis in benign hyperplasia and cancer of the prostate is androgen independent and TNFalpha mediated. Proc Natl Acad Sci USA. 2010;107(7):3123–3128. doi: 10.1073/pnas.0905524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonkhoff H, Berges R. The evolving role of oestrogens and their receptors in the development and progression of prostate cancer. Eur Urol. 2009;55(3):533–542. doi: 10.1016/j.eururo.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 33.Wu K, et al. The cell fate determination factor dachshund inhibits androgen receptor signaling and prostate cancer cellular growth. Cancer Res. 2009;69(8):3347–3355. doi: 10.1158/0008-5472.CAN-08-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu K, et al. DACH1 is a cell fate determination factor that inhibits cyclin D1 and breast tumor growth. Mol Cell Biol. 2006;26(19):7116–7129. doi: 10.1128/MCB.00268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ayala G, et al. Loss of caveolin-1 in prostate cancer stroma correlates with reduced relapse-free survival and is functionally relevant to tumour progression. J Pathol. 2013;231(1):77–87. doi: 10.1002/path.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giatromanolaki A, Koukourakis MI, Koutsopoulos A, Mendrinos S, Sivridis E. The metabolic interactions between tumor cells and tumor-associated stroma (TAS) in prostatic cancer. Cancer Biol Ther. 2012;13(13):1284–1289. doi: 10.4161/cbt.21785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freeman MR, Yang W, Di Vizio D. Caveolin-1 and prostate cancer progression. Adv Exp Med Biol. 2012;729:95–110. doi: 10.1007/978-1-4614-1222-9_7. [DOI] [PubMed] [Google Scholar]
- 38.Barros RP, Machado UF, Warner M, Gustafsson JA. Muscle GLUT4 regulation by estrogen receptors ERbeta and ERalpha. Proc Natl Acad Sci USA. 2006;103(5):1605–1608. doi: 10.1073/pnas.0510391103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sisk CL, Richardson HN, Chappell PE, Levine JE. In vivo gonadotropin-releasing hormone secretion in female rats during peripubertal development and on proestrus. Endocrinology. 2001;142(7):2929–2936. doi: 10.1210/endo.142.7.8239. [DOI] [PubMed] [Google Scholar]
- 40.Radovick S, et al. Evidence for direct estrogen regulation of the human gonadotropin-releasing hormone gene. J Clin Invest. 1991;88(5):1649–1655. doi: 10.1172/JCI115479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butler JA, Sjöberg M, Coen CW. Evidence for oestrogen receptor alpha-immunoreactivity in gonadotrophin-releasing hormone-expressing neurones. J Neuroendocrinol. 1999;11(5):331–335. doi: 10.1046/j.1365-2826.1999.00347.x. [DOI] [PubMed] [Google Scholar]
- 42.Wolfe A, Wu S. Estrogen receptor-β in the gonadotropin-releasing hormone neuron. Semin Reprod Med. 2012;30(1):23–31. doi: 10.1055/s-0031-1299594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeo SH, Herbison AE. Estrogen-negative feedback and estrous cyclicity are critically dependent upon estrogen receptor-α expression in the arcuate nucleus of adult female mice. Endocrinology. 2014;155(8):2986–2995. doi: 10.1210/en.2014-1128. [DOI] [PubMed] [Google Scholar]
- 44.Cheong RY, Porteous R, Chambon P, Abrahám I, Herbison AE. Effects of neuron-specific estrogen receptor (ER) α and ERβ deletion on the acute estrogen negative feedback mechanism in adult female mice. Endocrinology. 2014;155(4):1418–1427. doi: 10.1210/en.2013-1943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.