In a recent study, Xiang et al. (1) analyze ancient chicken mtDNA from northern China, together with published modern chicken data, and they proposed that the chicken was domesticated in northern China as early as 10,000 y ago. However, a reanalysis of their data suggested that the data are overinterpreted.
Xiang et al. (1) use two rounds of PCR amplification and direct Sanger sequencing to obtain a mtDNA control region fragment of 326 bp. The primers were designed in terms of NC_001323, which was suggested to contain errors (2). When aligning the singleplex PCR primers (compare their table S2) and ancient mtDNA sequences with the redefined reference NC_007235 (2), we found that the sequences KC456215–KC456222 [nucleotide positions (np) 233–558] reported by Xiang et al. (1) contained the primer sequences of CR1-F (np 233–253) and CR2-R (np 538–558). This generates errors of artificial recombination (3): it is impossible to see the scored mutation 246 in NC_001323 in all eight ancient DNA sequences belonging to different haplogroups, and mutation 243 of nonhaplogroup ABZ (Fig. 1) was missing in KC456215–KC456217 (Table 1). Due to these errors, the haplogroup assignments and the haplotype-sharing status for those sequences should be treated with caution.
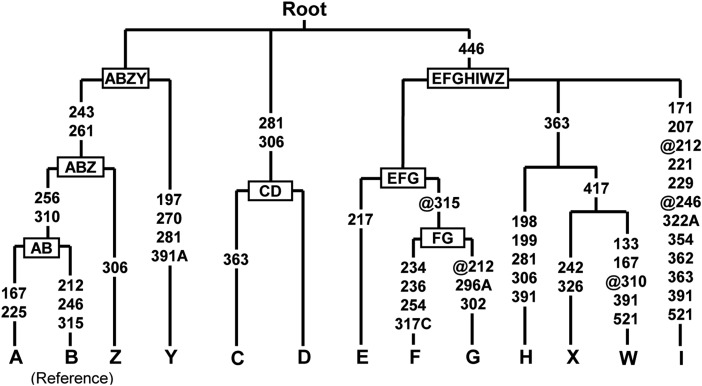
Fig. 1.
Classification tree of the mtDNA haplogroups in chickens. The tree based on control region information is taken from the cited paper (2). The diagnostic mutations considered relative to the reference NC_007235 belonging to haplogroup B are indicated on the branches. Nucleotide changes are specified for transversions by suffixes. Mutations toward a base identical-by state to the reference sequences are indicated with the prefix @.
Table 1.
Phylogenetic analyses of eight ancient chicken mtDNA sequences
| Sequences | Samples | Scored variants (np 233–558) | Haplogroup |
| CR1-F | 246 | ||
| NC_001323 | 246 | A | |
| KC456222 | WY2 | 246-446 | AB? Or B? |
| KC456221 | WY1 | 246-256-261-310-315 | ABZY? Or ABZ? |
| KC456220 | NZT3 | 246-315-362-446 | AB? Or A? |
| KC456219 | NZT2 | 246-272G-446 | AB? Or B? |
| KC456218 | NZT1 | 246-315 | AB? Or A? |
| KC456217 | JLD2 | @243-246-256-261-281-306-310-315-415 | CD |
| KC456216 | JLD1 | @243-246-256-261-281-306-310-315 | CD |
| KC456215 | CS1 | @243-246-256-261-310-315-446 | EFGHIWZ |
The np in the sequences (GenBank accession nos. KC456215–KC456222) are scored relative to the reference sequence NC_007235. These sequences are located in np 233–558 relative to NC_007235. The primers CR1-F, CR1-R, CR2-F, and CR2-R are located in np 233–253, 395–418, 385–405, and 538–559 relative to NC_007235, respectively. The haplogroup nomenclature is taken from the published chicken mtDNA haplogroup tree (2). Suspected mutations due to errors are in bold. Mutations toward a base identical-by state to the reference sequences are indicated with the prefix @.
Xiang et al. (1) claim that the ancient DNA provided evidence of chicken domestication in northern China around the early Holocene. However, the short sequence would offer limited information for a firm conclusion. When we removed the primer sequences from these mtDNA fragments (as a result, 285 bp left, np 254–538; Table 1), sequences KC456218–KC456220 from the Nanzhuangtou site could be tentatively assigned as basal branches within macrohaplogroup AB or haplogroups A or B (Fig. 1). Sequence KC456215 from the Cishan site could be allocated into macrohaplogroup EFGHIWZ (Fig. 1). Its mutation motif (256-261-310-315-446) was found in 894 chickens belonging to haplogroup E with global distribution and in 5 red junglefowls (2). Therefore, it is hard to make a conclusion of whether the early Holocene samples were from wild junglefowls or early domesticated chickens.
Finally, deciphering short fragments of the ancient mtDNA sequence is not easy. A reliable phylogeny of chicken mtDNA lineages, together with more information from the coding region or even complete mtDNA sequences, will justify haplogroup assignments (4). For instance, according to mutation motif 256-261-281-306-310-315 in KC456216, sample JLD1 from the Jiuliandun Chu Tombs could be assigned to macrohaplogroup CD (Table 1). With mutation 7155 in the COI sequence (KC456203), sample JLD1 can now be classified into haplogroup D2, which occurs in Turpan (Tulufan) chickens from northwest China (2). Evidently, by reference to the available phylogeny (2), we can further refine the mtDNA haplogrouping, which will be helpful in related phylogeographic analyses. Moreover, archaeological approaches, such as stable isotope biochemistry of ancient bones, should be considered to add to the story depicted by ancient DNA in an attempt to distinguish the relics from the domestic and wild animals (5).
Acknowledgments
This work was supported by the Breakthrough Project of Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB13020600. The Youth Innovation Promotion Association, Chinese Academy of Sciences, provided support to M.-S.P.
Footnotes
The authors declare no conflict of interest.
References
- 1.Xiang H, et al. Early Holocene chicken domestication in northern China. Proc Natl Acad Sci USA. 2014;111(49):17564–17569. doi: 10.1073/pnas.1411882111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miao YW, et al. Chicken domestication: An updated perspective based on mitochondrial genomes. Heredity (Edinb) 2013;110(3):277–282. doi: 10.1038/hdy.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi NN, Fan L, Yao YG, Peng MS, Zhang YP. Mitochondrial genomes of domestic animals need scrutiny. Mol Ecol. 2014;23(22):5393–5397. doi: 10.1111/mec.12955. [DOI] [PubMed] [Google Scholar]
- 4.Yao YG, Kong QP, Man XY, Bandelt HJ, Zhang YP. Reconstructing the evolutionary history of China: A caveat about inferences drawn from ancient DNA. Mol Biol Evol. 2003;20(2):214–219. doi: 10.1093/molbev/msg026. [DOI] [PubMed] [Google Scholar]
- 5.Barton L, et al. Agricultural origins and the isotopic identity of domestication in northern China. Proc Natl Acad Sci USA. 2009;106(14):5523–5528. doi: 10.1073/pnas.0809960106. [DOI] [PMC free article] [PubMed] [Google Scholar]