Significance
Animals respond to stress in many ways, including initiating cell death to eliminate damaged cells. Protein kinase RNA-activated (PKR) is a protein that senses stress, and it promotes cell death by phosphorylating eIF2α to block protein synthesis in damaged cells. PKR is activated by metabolic stress, such as that associated with obesity, and this activation depends on its RNA-binding domain. Here we investigated whether endogenous RNA triggers PKR activation in response to lipid exposure. Our results indicate that a noncoding RNA, the small nucleolar RNA (snoRNA), binds PKR during cellular metabolic stress, and multiple experiments suggest snoRNAs also activate PKR during metabolic stress. snoRNAs have established roles in RNA modification, and our studies suggest they have additional roles in metabolic stress.
Keywords: PKR, snoRNA, metabolic stress, phosphorylation, RNA-binding protein
Abstract
Protein kinase RNA-activated (PKR) has long been known to be activated by viral double-stranded RNA (dsRNA) as part of the mammalian immune response. However, in mice PKR is also activated by metabolic stress in the absence of viral infection, and this requires a functional kinase domain, as well as a functional dsRNA-binding domain. The endogenous cellular RNA that potentially leads to PKR activation during metabolic stress is unknown. We investigated this question using mouse embryonic fibroblast cells expressing wild-type PKR (PKRWT) or PKR with a point mutation in each dsRNA-binding motif (PKRRM). Using this system, we identified endogenous RNA that interacts with PKR after induction of metabolic stress by palmitic acid (PA) treatment. Specifically, RIP-Seq analyses showed that the majority of enriched RNAs that interacted with WT PKR (≥twofold, false discovery rate ≤ 5%) were small nucleolar RNAs (snoRNAs). Immunoprecipitation of PKR in extracts of UV–cross-linked cells, followed by RT-qPCR, confirmed that snoRNAs were enriched in PKRWT samples after PA treatment, but not in the PKRRM samples. We also demonstrated that a subset of identified snoRNAs bind and activate PKR in vitro; the presence of a 5′-triphosphate enhanced PKR activity compared with the activity with a 5′-monophosphate, for some, but not all, snoRNAs. Finally, we demonstrated PKR activation in cells upon snoRNA transfection, supporting our hypothesis that endogenous snoRNAs can activate PKR. Our results suggest an unprecedented and unexpected model whereby snoRNAs play a role in the activation of PKR under metabolic stress.
Protein kinase RNA-activated (PKR) is a member of a stress-response kinase family that includes PKZ and PEK (1) and is one of four known kinases that can phosphorylate eukaryotic initiation factor 2 (2). It is an IFN-induced kinase found in a latent form in most cells (3) and was initially identified as an antiviral protein involved in innate immunity (4). It is generally accepted that PKR activation is triggered by viral double-stranded RNA (dsRNA) during infection (5). PKR contains two functional domains: tandem dsRNA-binding motifs (dsRBMs) at the N terminus and a catalytic kinase domain at the C terminus (6). The dsRBMs play a key role in the activation and function of PKR. When a single dsRNA molecule binds to both dsRBMs, a conformational change occurs that promotes dimerization and autophosphorylation at multiple serine and threonine residues (7, 8). Point mutations in the dsRBMs of PKR disrupt its interaction with dsRNA and prevent the autophosphorylation required for its kinase activity (9).
Once activated, PKR binds and phosphorylates the α-subunit of eukaryotic initiation factor 2 (eIF2α) on Ser51 (10). Phosphorylation of eIF2α by PKR leads to a general inhibition of protein synthesis (11). In addition to eIF2α, other proteins such as the inflammatory signaling molecules Jun N-terminal protein kinase and IκB kinase can be activated by PKR (12, 13). Interestingly, RNAs derived from the 3′-untranslated regions of certain genes, including tumor necrosis factor α (TNF-α), can activate PKR (14–16), indicating the presence of additional mechanisms linking inflammatory responses with PKR activation.
Recent studies demonstrated that PKR can also respond to metabolic stress in mice and humans (13, 17). For example, obesity and endoplasmic reticulum stress activate PKR, which in turn phosphorylates eIF2α and other targets critical in insulin action and metabolic homeostasis. Genetic loss or chemical inhibition of PKR in obese mice results in improved glucose metabolism (18). The activation of PKR during metabolic stress depends on its dsRBMs, and deletion of either the RNA binding or kinase domains results in similar metabolic outcomes in mice (13, 19), raising the possibility that endogenous RNA is responsible for PKR activation during metabolic stress. Here we explore this question using a combined bioinformatics and experimental approach. Collectively, our data point to a class of small noncoding RNAs, the small nucleolar RNAs (snoRNAs), as potential activators of PKR under metabolic stress.
Results
snoRNAs Are Enriched in PKRWT Immunoprecipitates After PA Treatment.
Mouse embryonic fibroblasts (MEFs) treated with palmitic acid (PA) to induce metabolic stress show an elevated level of PKR autophosphorylation compared with untreated cells (13). An increase in palmitate-induced autophosphorylation is not observed in cells expressing PKR with a point mutation in one of the dsRBMs, raising the possibility that endogenous RNA may be involved in PKR activation during metabolic stress. To identify such endogenous RNAs, we immunoprecipitated PKR from PKR−/− MEFs reconstituted with wild-type PKR (PKRWT) or PKR with a mutation in each dsRBM (PKRRM, K64A, and K154A) after treatment with PA. We then performed RNA immunoprecipitation followed by high-throughput sequencing (RIP-Seq). Our goal was to identify RNAs that were candidates for triggering activation of PKR in the presence of PA in a manner that was dependent on functional dsRBMs.
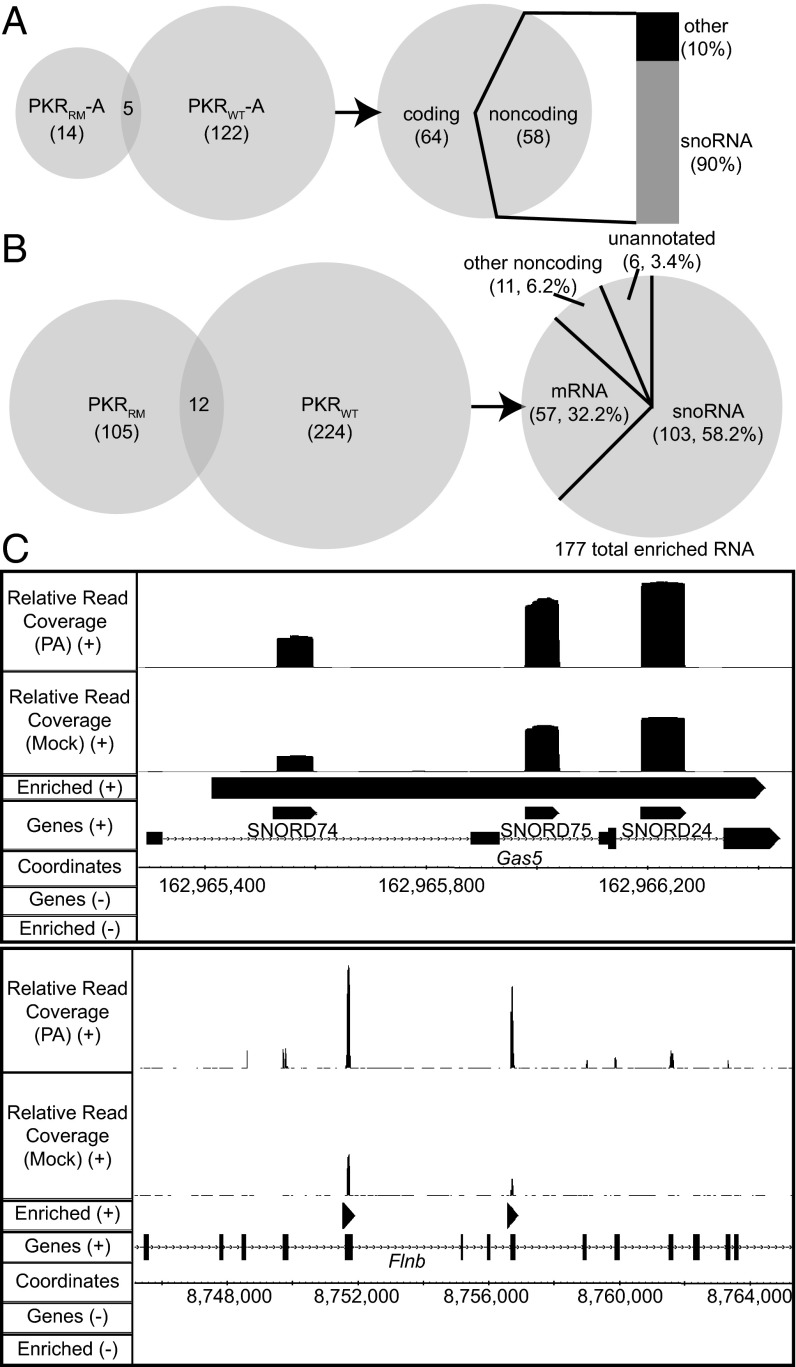
Initially, we sequenced three biological replicates for both PKRWT and PKRRM MEF cells (dataset A, Datasets S1–S4). We used the DefinedRegionDifferentialSeq (DRDS) algorithm from the USeq software package (20) (SI Materials and Methods) to generate lists of annotated genes encoding RNAs enriched in response to PA treatment in PKRWT samples, but not in PKRRM samples (Fig. 1A). Of the 122 genes unique to the PKRWT samples, 58 were noncoding, and surprisingly, 90% of these encoded snoRNAs.
Fig. 1.
snoRNAs are enriched in PKRWT immunoprecipitates after PA treatment. (A) Venn diagram shows the number of annotated genes enriched in PKRWT and PKRRM samples for dataset A [fold increase ≥ 2; false discovery rate (FDR) ≤ 5%]. (Right) Pie chart shows the number of coding and noncoding RNAs enriched only in PKRWT in dataset A (122 RNAs), and column shows the percentage of snoRNAs among these noncoding RNAs. (B) Venn diagram shows number of enriched genomic regions for PKRWT and PKRRM samples, common to datasets A, B and C (fold increase ≥ 2, Storey’s q-value FDR ≤ 5%). Genomic regions encoding RNAs enriched exclusively in PKRWT samples encompass 177 genes, and the pie chart on the right categorizes these regions. Lists of these regions and intersected regions are included in Dataset S2. (C) IGB snapshots of enriched regions (“Enriched”) show an example for one enriched region encompassing three annotated snoRNAs (Upper) encoded in the Gas5 gene, and for two enriched regions within the Flnb gene (Lower). “Relative read coverage” is the number of mapped reads per base/(total number of reads * 106).
To confirm the results of the dataset A analysis, we performed high-throughput sequencing on two additional sets of PKR-immunopurified RNA (datasets B and C), again including samples of PKRWT and PKRRM with or without PA treatment. Dataset B included three additional biological replicates and was analyzed with the DRDS algorithm, whereas dataset C contained a single sample and was analyzed with an algorithm suited to a single sample, the DefinedRegionScanSeqs (DRSS) algorithm, also of the USeq package (20) (Dataset S1). As shown in Fig. S1, and consistent with the analysis of dataset A, for both datasets B and C we observed that over 80% of the noncoding RNAs that were enriched in the PKRWT sample, but not in the PKRRM sample, after PA treatment were snoRNAs. Thus, altogether we sequenced seven biological replicates of both PKRWT and PKRRM in three separate runs. Each sequencing run was analyzed independently, and in each of the three cases, snoRNAs were enriched in response to PA treatment. Pairwise Spearman correlation analyses suggested good agreement between biological replicates within the same dataset (ρ = 0.92–0.99) and between the three datasets (ρ = 0.78–0.89). Fig. S2 shows representative analyses.
Whereas the above analyses considered only reads that aligned to annotated genes, in a second bioinformatics approach, we generated lists of genomic regions, annotated and unannotated, that encoded RNAs enriched in immunoprecipitates of PKRWT, but not PKRRM, after PA treatment. The ScanSeqs algorithm of the USeq package was used with a sliding window of 50 bp (SI Materials and Methods) to analyze each of the three datasets (A, B, and C) individually (Dataset S2). Datasets were then intersected to reveal 224 genomic regions that encoded RNAs enriched in PKRWT immunoprecipitates after PA treatment; these regions were found in all three datasets after excluding regions enriched in PKRRM samples (Fig. 1B). Using the Integrated Genome Browser (IGB) (21), the 224 genomic regions were visualized and manually curated to create a list of corresponding genes. For genomic regions that encompassed multiple annotated snoRNAs, where reads abundantly mapped to each snoRNA but not intervening regions (Fig. 1C, Upper), each snoRNA was included as a distinct gene (103 snoRNA genes; Fig. 1B). Regions that did not include annotated snoRNAs (Fig. 1B) were largely composed of annotated mRNAs (Dataset S2). For this category, when multiple enriched regions were encompassed by a single annotated gene, they were collapsed into a single entry (68 genes for mRNA+other noncoding, Fig. 1 B and C, Lower). Somewhat surprisingly, only six regions were in unannotated regions of the genome. Finally, three enriched regions were discarded as false positives, based on the normalized number of reads aligned to these regions. Our final list totaled 177, and again, the majority were snoRNAs. We compared the predicted thermodynamic stability of enriched snoRNAs and non-snoRNAs with length-matched, random transcribed regions. We found that snoRNAs, but not non-snoRNAs, were predicted to be significantly more stable than the random regions (Fig. S3).
snoRNAs are small noncoding RNAs that function mainly in complex with a conserved set of proteins in the modification of ribosomal RNA (rRNA) and snRNAs (22). The H/ACA class of snoRNA mediates pseudouridylation, and the C/D snoRNAs guide 2′-O-methylation. A small subset of snoRNAs (e.g., U3 snoRNA) guide pre-rRNA cleavage. In addition, there are snoRNAs that do not appear to function as guides for RNA modification, and these are referred to as orphan snoRNAs. Using information for human orthologs (www-snorna.biotoul.fr//), we categorized snoRNAs enriched in PKRWT immunoprecipitates in the presence of PA into guide and orphan snoRNAs. Analyzing the distribution of the two subclasses of snoRNA in all datasets from the two bioinformatics approaches (DRDS/DRSS and ScanSeqs), using ensembl gene IDs (www.ensembl.org), did not show a consistent pattern of snoRNA classes (Fig. S4); some datasets showed a predominance of H/ACA snoRNAs, and some a predominance of C/D snoRNAs. However, the majority of enriched snoRNAs in our datasets function as guides for RNA modification (based on human orthologs, see Datasets S1 and S2).
Validation of snoRNA Enrichment in PKRWT Samples After PA Treatment.
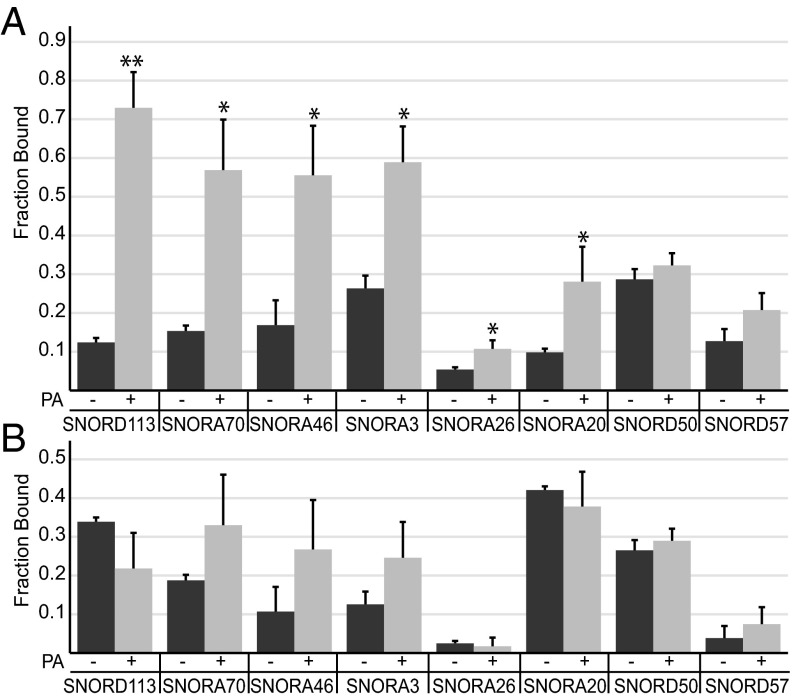
Analysis of multiple biological replicates, with multiple algorithms, indicated that snoRNAs were the most abundant class of RNA in the PKRWT immunoprecipitates after PA treatment. To provide further experimental support for this observation, we exposed cells, with or without prior PA treatment, to 254 nm of UV-C light to cross-link physically interacting molecules. Cells were then lysed, PKR was immunoprecipitated, and immunoprecipitates were washed eight times with high salt to remove non–cross-linked species before RNA extraction. We then used reverse transcription followed by quantitative PCR (RT-qPCR) (Dataset S3) to measure expression levels of eight snoRNAs that showed enrichment in the deep sequencing analysis and found that six of these showed a significant increase in their levels after PA treatment (Fig. 2A). Although the level of some snoRNAs in PKRRM samples showed an increase after PA treatment, these differences were not significant (Student’s t test, P > 0.1) (Fig. 2B).
Fig. 2.
Validation of the enrichment of snoRNAs in PKRWT samples after PA treatment. Graph shows the fraction of specified snoRNA that cross-linked to PKRWT (A) or PKRRM (B) with (+) and without (−) PA treatment. Associated snoRNAs were analyzed by RT-qPCR. “Fraction bound” is the snoRNA level in IP/snoRNA level in input sample. Error bars, mean ± SEM, n = 3. *P < 0.05; **P < 0.005.
UV-C irradiation is a “zero length” cross-linking technique that will only covalently link RNA to proteins that are in direct contact and, further, will not create protein–protein cross-links (23, 24). Thus, these results indicate that PKR is directly interacting with snoRNAs in cells. Furthermore, this association is significantly increased after PA treatment of PKRWT in a manner that is dependent on functional dsRNA-binding motifs. Using purified recombinant PKR and in vitro-transcribed SNORD113 (Box C/D snoRNA), we employed gel shift assays to show that PKR can also interact with snoRNAs in vitro. SNORD113 binds to PKRWT with an estimated Kd of 16.1 nM. Binding was dependent on functional dsRBMs; PKRRM showed only weak affinity for SNORD113 (Fig. S5).
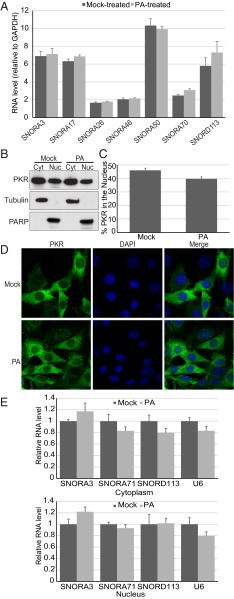
We considered a number of explanations for the observed increase in snoRNAs cross-linked to PKRWT after PA treatment. For example, it seemed possible that the increased association was because PA increased the total levels of snoRNAs. However, using RT-qPCR, we determined that the levels of snoRNAs in total RNA samples did not significantly increase after PA treatment (Fig. 3A). We also considered the possibility that PA induced a change in the localization of either PKR or snoRNAs. However, cell fractionation followed by Western analyses showed that PKR is localized in both cytoplasm and nuclei of MEFs and that there was no significant change in localization after PA treatment (Fig. 3 B and C); immunofluorescence studies confirmed the cell fractionation results (Fig. 3D). We also measured the level of some snoRNAs in nuclear and cytoplasmic fractions using RT-qPCR. We found that the vast majority of snoRNAs were in the nuclear fraction, and we did not detect a change in snoRNA localization after PA treatment (Fig. 3E).
Fig. 3.
PA treatment does not change PKR or snoRNA localization or total snoRNA level. (A) The level of each snoRNA extracted from PKRWT lysate with (PA) and without PA (Mock) treatment was measured by RT-qPCR. RNA levels were normalized to levels of GAPDH mRNA. The mean ± SEM are from four different biological replicates. (B) Western analysis of cytoplasmic (Cyt) and nuclear (Nuc) fractions from MEFs untreated (Mock) or treated (PA) with PA. (C) Quantification of PKR accumulation in nuclear fractions determined as in B. Error bars, mean ± SEM, n = 10. (D) Immunofluorescence of MEFs treated (PA) or untreated (Mock) with PA. Cells were stained using an antibody against PKR, and nuclei were stained with DAPI. (E) The level of each snoRNA extracted from cytoplasmic (Upper) or nuclear fraction (Lower) was measured by RT-qPCR. RNA levels were normalized to 36B4 RNA (25) and plotted as the average ratio of RNA in PA-treated cells relative to mock-treated cells. Error bars, mean ± SEM, n = 3.
snoRNAs Can Activate PKR in Vitro.
To provide an in vitro correlate to our bioinformatics data, we used an in vitro PKR autophosphorylation assay to determine if snoRNAs could directly activate PKR. Representative snoRNAs from class H/ACA and C/D were selected based on the presence in at least one dataset. Titrating RNA against a constant concentration of purified, recombinant PKR showed that snoRNAs can activate PKR in vitro (Fig. 4 A and B). As observed with activation by dsRNA (10), substrate inhibition was observed, and snoRNA-induced PKR activation followed a bell-shaped curve (Fig. S6). Because our RIP-Seq data were derived from MEFs complemented with human PKR, we tested both mouse and human snoRNAs. We found that recombinant human PKR could be efficiently activated by human snoRNAs, with a tendency toward higher activation than mouse snoRNAs for a given RNA concentration (Fig. 4B).
Fig. 4.
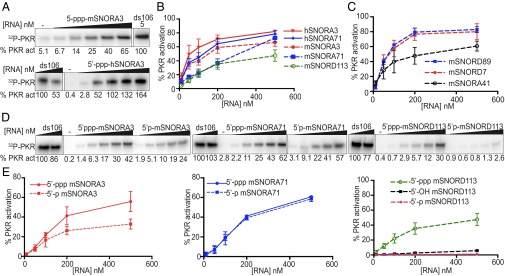
snoRNAs activate PKR in vitro. (A) Autophosphorylation assay with representative snoRNAs. snoRNA concentrations were 10, 50, 100, 200, and 500 nM; perfectly duplexed RNA 106 bp in length (ds106) concentrations were 5 and 10 nM. Phosphorylation activities were normalized with reactions of 5 nM of ds106. Averaged activation data (as assayed in A) for snoRNAs identified (B) or not identified (C) by RIP-Seq. (D) Comparison of autophosphorylation for snoRNAs with different 5′ termini. snoRNA concentrations as in A. (E) Averaged activation data for SNORA3, SNORA71, and SNORD113 with different 5′ termini. mSNORA and hSNORA indicate mouse and human box H/ACA snoRNA, respectively; mSNORD, mouse box C/D snoRNA. Error bars, mean ± SEM, n ≥ 3, except for mSNORD89, mSNORD7 (C), and 5′-OH mSNORD113 (E), which show mean ± SD, n = 2. White spaces indicate where intervening lanes were removed from the figures.
Further experiments were performed to test if the ability of snoRNAs to activate PKR was specific to snoRNAs identified by RIP-Seq or if this was a universal property of snoRNAs. Two box C/D snoRNAs and one box H/ACA snoRNA were selected based on their absence from the enriched annotated genes in PKRWT after PA treatment (Datasets S1 and S2). All three snoRNAs could activate PKR (Fig. 4C).
The snoRNAs that we tested in vitro were transcribed using T7 RNA polymerase and hence have a 5′-triphosphate. Previous work shows that in vitro PKR activation by certain RNAs depends on a 5′-triphosphate (26). Thus, using three different snoRNAs, we compared PKR activation in response to different 5′ termini. For SNORA3 and SNORD113 the presence of a 5′-triphosphate led to a significantly higher level of PKR activity compared with a 5′-monophosphate; for SNORD113, activation by a 5′-monophosphate or 5′-hydroxyl was barely detectable (Fig. 4 D and E). However, PKR activation by SNORA71 was robust and occurred at nearly identical levels for RNAs containing either a 5′-triphosphate or monophosphate. In light of previous studies, the differential requirements for 5′ termini among snoRNAs may reflect different secondary structures (26). Indeed, nuclease mapping of a subset of snoRNAs established that SNORA3 and SNORA71 were more rod-like, with longer helical regions, than SNORD113 (Figs. S7 and S8). Interestingly, of the snoRNAs tested, SNORD113 was most dependent on 5′-triphosphates in autophosphorylation assays (Fig. 4E).
snoRNAs Can Activate PKR in Cells.
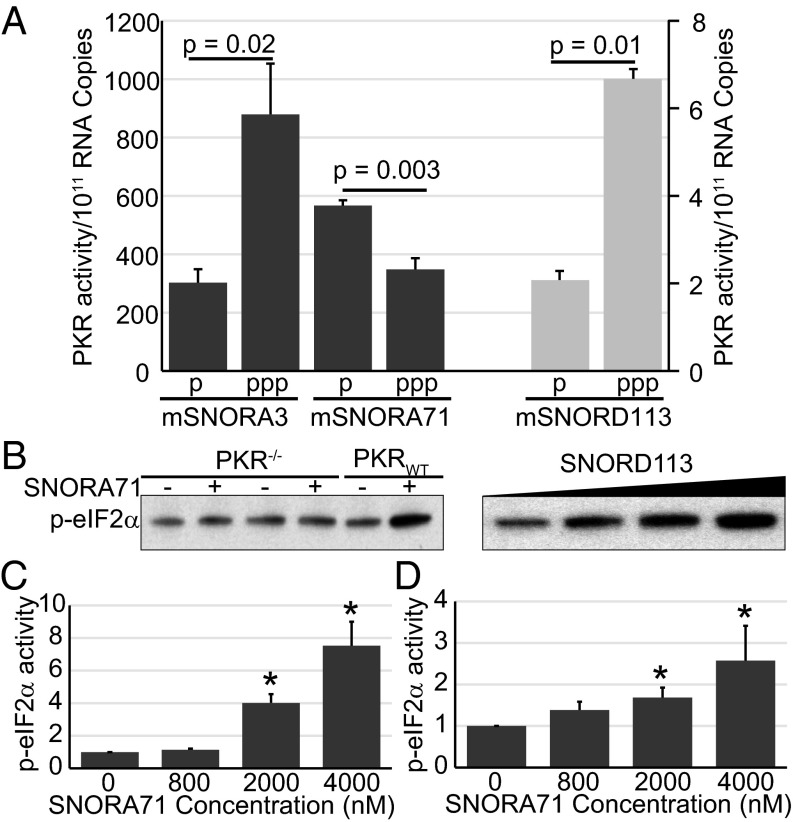
We attempted to knock down snoRNAs in MEFs in hopes of testing effects on PKR activity after PA treatment, but we were unable to attain efficient knockdown. As an alternative, we transfected MEFs with different snoRNAs and assayed PKR activation by Western analysis using an antibody specific for phosphorylated PKR. As shown in Fig. S9, transfecting MEFs with SNORA3, SNORA71, or SNORD113 increased levels of activated PKR compared with mock-treated cells. After normalizing PKR activity to the level of transfected snoRNA, measured by RT-qPCR, we determined that the presence of a triphosphate group at the 5′ end of SNORA3 and SNORD113 led to higher PKR activation compared with a 5′ monophosphorylated snoRNA (Fig. 5A).
Fig. 5.
snoRNAs activate PKR in cells and in lysate. (A) Quantitation of phospho-PKR (p-PKR) activity. PKR activity was calculated per copy number of the transfected snoRNA, as determined from RT-qPCR of transfected cells. PKR activity values per 1011 RNA copy numbers for 5′-monophosphorylated (p) and 5′-triphosphorylated (ppp) snoRNAs are shown. Dark gray bars are graphed relative to the left y axis, and light gray bars are graphed relative to the right y axis. (B) Western blot of p-eIF2α in PKRWT or PKR−/− lysates, Mock-treated (−), 2 μM 5′-ppp-mSNORA71–treated (+) or 5′-ppp-mSNORD113–treated (1.2, 3.2, and 6.4 μM). Quantification of p-eIF2α activity in MEFs (C) and CHO (D) lysates after treating with 5′-ppp-mSNORA71. Fold change in phospho-eIF2α activity is graphed relative to mock-treated lysate. Error bars, mean ± SEM, n = 3. *P < 0.05.
We also found that snoRNAs could induce phosphorylation of eIF2α (a target of PKR) in cell lysates prepared from MEFs. Activation was observed only in extracts prepared from PKR−/− MEFs expressing wild-type PKR (Fig. 5B), indicating that the observed phosphorylation of eIF2α is PKR-dependent. Adding 5′-ppp-mSNORA71 or 5′-ppp-mSNORD113 to the PKRWT MEF lysate increased eIF2α phosphorylation levels (Fig. 5 B and C), and activation was also observed using lysates prepared from CHO cells (Fig. 5D).
Discussion
Here we report studies to identify the endogenous RNA responsible for binding and activating PKR during metabolic stress. Using RIP-Seq protocols, we identified candidate RNAs that bound PKRWT, but not PKRRM, after treatment of cells with PA to mimic metabolic stress. Unexpectedly, the largest category of candidates was snoRNAs. UV–cross-linking experiments confirmed that snoRNAs interact with PKR in MEFs in a PA-dependent manner. In vitro studies with purified, recombinant PKR showed that snoRNAs directly activate PKR, and for some, but not all, snoRNAs this activation was enhanced by the presence of a 5′-triphosphate. Transfection of snoRNAs into cells also activated PKR, and again, for some snoRNAs this activation was enhanced by a 5′-triphosphate. Our studies indicate that snoRNAs bind PKR in cells experiencing metabolic stress, raising the possibility that under certain conditions these noncoding RNAs may activate PKR.
How Does PKR Intersect with Known Roles of snoRNAs?
snoRNAs have well-characterized roles in ribosome biogenesis. They bind to four proteins to form snoRNPs, which guide rRNA base modifications by forming a short duplex with rRNA (27). Our observation that PKR interacts with snoRNAs raises the question of whether PKR is part of these snoRNPs. However, PKR immunoprecipitation from lean and obese liver tissue lysate followed by mass spectrometry did not show association of any guide snoRNP core proteins with PKR (28). This suggests that PKR–snoRNA and snoRNPs may exist as distinct complexes in the cell. At present, we cannot rule out the possibility that snoRNP proteins interact transiently with snoRNA–PKR complexes, but were not captured by immunoprecipitation. Importantly, our RIP-Seq data did not show enrichment of rRNA after PA treatment, suggesting that PKR activation is not mediated by a snoRNA–rRNA duplex.
Where and When Does PKR Interact with snoRNAs?
We find that PKR is in the cytoplasm as well as the nucleus (Fig. 3B), where snoRNPs assemble. Furthermore, our data indicate that PA induces the interaction of PKR and snoRNAs (Fig. 2A); at present, it is entirely unclear how this might occur. A recent study showed that PKR is activated by inverted Alu repeats when these factors mix as the nuclear envelope breaks down during mitosis (29). PA has been shown to induce mitosis (30), and we considered whether mitosis could trigger a snoRNA–PKR interaction. However, our immunofluorescence data (Fig. 3D) did not show MEFs with mitotic morphology after PA treatment. Nevertheless, future studies involving more in-depth analyses will be needed to rule out a cell cycle-specific effect.
Do snoRNAs Activate PKR in Vivo?
We found that snoRNAs activate endogenous PKR in cells and in cell lysates, indicating that snoRNAs could, in theory, activate PKR during metabolic stress. Our in vitro and in-cell data show enhancement of PKR activity by the presence of a 5′-triphosphate for some but not all tested snoRNAs (Figs. 4 and 5). Possibly a 5′-triphosphate increases the binding affinity of some snoRNAs to PKR. Consistent with this hypothesis, PKR contains a potential triphosphate-binding site (31). Most snoRNAs are encoded in introns of host genes. After splicing, exonucleolytic processing of the debranched lariat generates mature snoRNAs with 5′-monophosphates, at least in those snoRNAs tested (32). Other snoRNAs are independent genes that are transcribed by RNA polymerase (pol) II and thus have a 5′-cap (33). According to the current mouse genome annotation from the University of California at Santa Cruz (UCSC) Genome Browser (December 2011, GRCm38/mm10), SNORA3 and SNORA71 are encoded in the introns of RPl27a and of a noncoding RNA, respectively. SNORD113 annotation indicates that it is transcribed from an independent gene. Therefore, SNORA3 and SNORA71 are predicted to have 5′-monophosphates and SNORD113 to have a 5′-cap. In nematodes, yeast, and plants, some snoRNAs are transcribed from independent genes by RNA pol III to generate a 5′-triphosphate on the nascent transcript (33), but to our knowledge a snoRNA transcribed by pol III has not been observed in mammals.
It is interesting to consider the possibility that metabolic stress alters snoRNA processing to create aberrant 5′ termini. For example, blocking trimming and capping might leave a 5′-triphosphate instead of the expected 5′-monophosphate found on mature snoRNAs. However, our RIP-Seq data indicate that the vast majority of snoRNA reads align to the mature, processed form (see Fig. 1C as an example). Alternatively, a report identified a component of the cytosolic capping complex that adds a phosphate to monophosphorylated RNA, generating RNA with a 5′-diphosphate (34). Interestingly, RIG-I, a dsRNA-sensing protein, is activated by RNA with either a 5′-triphosphate or a 5′-diphosphate (35). Although previous studies indicate that PKR is not activated by a diphosphate (26), it seems possible that 5′ termini preferences may vary for different RNAs, and thus, future studies of the effects of 5′-diphosphates are warranted.
In recent years, snoRNAs have been implicated in a wide range of pathways, suggesting that they have functions beyond their roles in RNA modification. snoRNAs maintain chromatin accessibility (36), regulate the coactivator activity of the dyskerin RNP (37), interact with Dicer (38), and are associated with disease (39). Using multiple assays, we show that snoRNAs interact with PKR in a PA-dependent manner. Our studies suggest yet another function for snoRNAs, in particular, as signals of metabolic stress. Here we note that snoRNAs have been previously linked to lipotoxicity (25), and cells deficient in intronic snoRNAs are resistant to cell death induced by PA treatment (40).
Materials and Methods
Details of methodologies that have been previously described are provided in SI Materials and Methods.
RNA Immunoprecipitation.
In all PA treatment experiments, cells were mock-treated or 100 μM PA-treated for 2 h at 37 °C. Immunoprecipitation was performed using MEFs as described (13). RNA was extracted with TRIzol, followed by Turbo DNase treatment (Invitrogen) and ethanol precipitation.
Library Preparation, High-Throughput Sequencing, and Bioinformatics Analysis.
Immunoprecipitated RNA was fragmented by incubating in fragmentation buffer (40 mM Tris⋅acetate, pH 8.1, 30 mM Mg acetate, 100 mM K acetate) for 5 min at 94 °C. cDNA libraries were generated using an Illumina small TruSeq RNA sample preparation kit. The Bioanalyzer DNA 1000 chip was used for evaluating library size. Libraries were subjected to 101 cycles of paired- or single-end sequencing with an Illumina HiSeq-2000 sequencer. Extraction of 101-bp paired-end reads used CASAVA 1.7.0 and CASAVA 1.8.2. Standard FASTAQ files were extracted with the standard Illumina pipeline.
Supplementary Material
Acknowledgments
We thank B. Dalley (University of Utah core facility) for advice and technical support and J. Schaffer (Washington University) for CHO cells and SmD3 primer sequences. Imaging was performed at the Fluorescence Microscopy Core Facility (University of Utah). Microscopy equipment was obtained using National Center for Research Resources Shared Equipment Grant 1S10RR024761-01. This work was supported by funds to B.L.B. from the National Institutes of Aging of the National Institutes of Health under Award 8DP1AG044162 and to G.S.H. in part from Grants DK52539 and HL125753. T.N. was supported by a Scientist Development Grant from the American Heart Association and Precursory Research for Embryonic Science and Technology from the Japan Science and Technology Agency. D.A.N. was supported by National Cancer Institute Cancer Center Support Grant (5P30CA042014-25).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Gene Expression Omnibus database, www.ncbi.nlm.nih.gov/geo (accession no. GSE66540).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1424044112/-/DCSupplemental.
References
- 1.Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2α kinases: Their structures and functions. Cell Mol Life Sci. 2013;70(19):3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140(6):900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog Nucleic Acid Res Mol Biol. 2006;81:369–434. doi: 10.1016/S0079-6603(06)81010-X. [DOI] [PubMed] [Google Scholar]
- 4.Dabo S, Meurs EF. dsRNA-dependent protein kinase PKR and its role in stress, signaling and HCV infection. Viruses. 2012;4(11):2598–2635. doi: 10.3390/v4112598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dauber B, Wolff T. Activation of the antiviral kinase PKR and viral countermeasures. Viruses. 2009;1(3):523–544. doi: 10.3390/v1030523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meurs E, et al. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62(2):379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 7.Taylor DR, et al. Autophosphorylation sites participate in the activation of the double-stranded-RNA-activated protein kinase PKR. Mol Cell Biol. 1996;16(11):6295–6302. doi: 10.1128/mcb.16.11.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romano PR, et al. Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2alpha kinases PKR and GCN2. Mol Cell Biol. 1998;18(4):2282–2297. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMillan NA, et al. Mutational analysis of the double-stranded RNA (dsRNA) binding domain of the dsRNA-activated protein kinase, PKR. J Biol Chem. 1995;270(6):2601–2606. doi: 10.1074/jbc.270.6.2601. [DOI] [PubMed] [Google Scholar]
- 10.Cole JL. Activation of PKR: An open and shut case? Trends Biochem Sci. 2007;32(2):57–62. doi: 10.1016/j.tibs.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6(4):318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 12.Bonnet MC, Weil R, Dam E, Hovanessian AG, Meurs EF. PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex. Mol Cell Biol. 2000;20(13):4532–4542. doi: 10.1128/mcb.20.13.4532-4542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura T, et al. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140(3):338–348. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis S, Watson JC. In vitro activation of the interferon-induced, double-stranded RNA-dependent protein kinase PKR by RNA from the 3′ untranslated regions of human alpha-tropomyosin. Proc Natl Acad Sci USA. 1996;93(1):508–513. doi: 10.1073/pnas.93.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osman F, Jarrous N, Ben-Asouli Y, Kaempfer R. A cis-acting element in the 3′-untranslated region of human TNF-alpha mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev. 1999;13(24):3280–3293. doi: 10.1101/gad.13.24.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nussbaum JM, Gunnery S, Mathews MB. The 3′-untranslated regions of cytoskeletal muscle mRNAs inhibit translation by activating the double-stranded RNA-dependent protein kinase PKR. Nucleic Acids Res. 2002;30(5):1205–1212. doi: 10.1093/nar/30.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carvalho BM, et al. Modulation of double-stranded RNA-activated protein kinase in insulin sensitive tissues of obese humans. Obesity (Silver Spring) 2013;21(12):2452–2457. doi: 10.1002/oby.20410. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura T, Arduini A, Baccaro B, Furuhashi M, Hotamisligil GS. Small-molecule inhibitors of PKR improve glucose homeostasis in obese diabetic mice. Diabetes. 2014;63(2):526–534. doi: 10.2337/db13-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvalho-Filho MA, et al. Double-stranded RNA-activated protein kinase is a key modulator of insulin sensitivity in physiological conditions and in obesity in mice. Endocrinology. 2012;153(11):5261–5274. doi: 10.1210/en.2012-1400. [DOI] [PubMed] [Google Scholar]
- 20.Nix DA, Courdy SJ, Boucher KM. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The Integrated Genome Browser: Free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25(20):2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lafontaine DL, Tollervey D. Birth of the snoRNPs: The evolution of the modification-guide snoRNAs. Trends Biochem Sci. 1998;23(10):383–388. doi: 10.1016/s0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- 23.Pellé R, Murphy NB. In vivo UV-cross-linking hybridization: A powerful technique for isolating RNA binding proteins. Application to trypanosome mini-exon derived RNA. Nucleic Acids Res. 1993;21(10):2453–2458. doi: 10.1093/nar/21.10.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ule J, Jensen K, Mele A, Darnell RB. CLIP: A method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37(4):376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 25.Michel CI, et al. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 2011;14(1):33–44. doi: 10.1016/j.cmet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nallagatla SR, et al. 5′-Triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318(5855):1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 27.Kiss T, Fayet E, Jády BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura T, et al. A critical role for PKR complexes with TRBP in immunometabolic regulation and eIF2a phosphorylation in obesity. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y, et al. PKR is activated by cellular dsRNAs during mitosis and acts as a mitotic regulator. Genes Dev. 2014;28(12):1310–1322. doi: 10.1101/gad.242644.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Yang Y, Wu J. Palmitate impairs cytokinesis associated with RhoA inhibition. Cell Res. 2010;20(4):492–494. doi: 10.1038/cr.2010.33. [DOI] [PubMed] [Google Scholar]
- 31.Toroney R, Hull CM, Sokoloski JE, Bevilacqua PC. Mechanistic characterization of the 5′-triphosphate-dependent activation of PKR: Lack of 5′-end nucleobase specificity, evidence for a distinct triphosphate binding site, and a critical role for the dsRBD. RNA. 2012;18(10):1862–1874. doi: 10.1261/rna.034520.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tycowski KT, Shu MD, Steitz JA. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev. 1993;7(7A):1176–1190. doi: 10.1101/gad.7.7a.1176. [DOI] [PubMed] [Google Scholar]
- 33.Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: A paradigm for gene expression flexibility. Genomics. 2009;94(2):83–88. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 34.Otsuka Y, Kedersha NL, Schoenberg DR. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol. 2009;29(8):2155–2167. doi: 10.1128/MCB.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goubau D, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514(7522):372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubert T, et al. Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin. Mol Cell. 2012;48(3):434–444. doi: 10.1016/j.molcel.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 37.Fong YW, Ho JJ, Inouye C, Tjian R. The dyskerin ribonucleoprotein complex as an OCT4/SOX2 coactivator in embryonic stem cells. eLife. 2014;3:3. doi: 10.7554/eLife.03573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rybak-Wolf A, et al. A variety of dicer substrates in human and C. elegans. Cell. 2014;159(5):1153–1167. doi: 10.1016/j.cell.2014.10.040. [DOI] [PubMed] [Google Scholar]
- 39.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 40.Scruggs BS, Michel CI, Ory DS, Schaffer JE. SmD3 regulates intronic noncoding RNA biogenesis. Mol Cell Biol. 2012;32(20):4092–4103. doi: 10.1128/MCB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.