Significance
The TREX1 enzyme degrades DNA, and mutations in the TREX1 gene cause autoimmune diseases. The TREX1 D18N mutation causes a form of lupus called familial chilblain lupus. We solved the structure of TREX1 D18N bound to dsDNA, showing how the enzyme interacts with dsDNA. We also replaced the TREX1 WT gene in mice with the TREX1 D18N mutated gene and showed how this mutation causes a lupus-like disease. Together, the TREX1 D18N–dsDNA structure and the spontaneous disease exhibited in the TREX1 D18N mouse help to define how TREX1 degrades dsDNA to prevent this molecule from acting as an autoantigen in the mouse and, most likely, in humans to promote autoimmune disease.
Keywords: TREX1, autoimmunity, lupus, structure, DNA
Abstract
The TREX1 gene encodes a potent DNA exonuclease, and mutations in TREX1 cause a spectrum of lupus-like autoimmune diseases. Most lupus patients develop autoantibodies to double-stranded DNA (dsDNA), but the source of DNA antigen is unknown. The TREX1 D18N mutation causes a monogenic, cutaneous form of lupus called familial chilblain lupus, and the TREX1 D18N enzyme exhibits dysfunctional dsDNA-degrading activity, providing a link between dsDNA degradation and nucleic acid-mediated autoimmune disease. We determined the structure of the TREX1 D18N protein in complex with dsDNA, revealing how this exonuclease uses a novel DNA-unwinding mechanism to separate the polynucleotide strands for single-stranded DNA (ssDNA) loading into the active site. The TREX1 D18N dsDNA interactions coupled with catalytic deficiency explain how this mutant nuclease prevents dsDNA degradation. We tested the effects of TREX1 D18N in vivo by replacing the TREX1 WT gene in mice with the TREX1 D18N allele. The TREX1 D18N mice exhibit systemic inflammation, lymphoid hyperplasia, vasculitis, and kidney disease. The observed lupus-like inflammatory disease is associated with immune activation, production of autoantibodies to dsDNA, and deposition of immune complexes in the kidney. Thus, dysfunctional dsDNA degradation by TREX1 D18N induces disease in mice that recapitulates many characteristics of human lupus. Failure to clear DNA has long been linked to lupus in humans, and these data point to dsDNA as a key substrate for TREX1 and a major antigen source in mice with dysfunctional TREX1 enzyme.
The TREX1 gene encodes a powerful DNA exonuclease (1–7). The amino terminal domain of the TREX1 enzyme contains all of the structural elements for full exonuclease activity, and the carboxy terminal region controls cellular trafficking to the perinuclear space (8–10). Mutations in TREX1 cause a spectrum of autoimmune disorders, including Aicardi–Goutieres syndrome, familial chilblain lupus, and retinal vasculopathy with cerebral leukodystrophy and are associated with systemic lupus erythematosus (9, 11–19). The TREX1 disease-causing alleles locate to positions throughout the gene, exhibit dominant and recessive genetics, include inherited and de novo mutations, and cause varied effects on catalytic function and cellular localization. These genetic discoveries have established a causal relationship between TREX1 mutation and nucleic acid-mediated immune activation disease. The spectrum of TREX1-associated disease parallels the diverse effects on enzyme function and localization, indicating multiple mechanisms of TREX1 dysfunction might explain the overlapping clinical symptoms related to failed DNA degradation and immune activation.
The TREX1 enzyme structure reveals the uniquely stable dimer that is relevant to its function and to disease mechanisms. The backbone contacts between the protomer β3-strands generate a stable, central antiparallel β-sheet that stretches the length of the dimer and an extensive hydrogen-bonding network of sidechain–sidechain, sidechain–backbone, and water-bridged contacts that coordinate residues across the TREX1 dimer interface (8, 20). TREX1 catalytic function depends on the dimeric structure, with residues from one protomer contributing to DNA binding and degradation in the opposing protomer (21, 22). Some TREX1 disease-causing mutants exhibit complete loss of catalytic function, whereas others exhibit altered cellular localization (8, 10). A subset of TREX1 catalytic mutants at amino acid positions Asp-18 and Asp-200 exhibit selectively dysfunctional activities on dsDNA. These mutations cause autosomal-dominant disease by retaining DNA-binding proficiency and blocking access to DNA 3′ termini for degradation by TREX1 WT enzyme (21, 23, 24). The TREX1 catalytic sites accommodate four nucleotides of ssDNA, and additional structural elements are positioned adjacent to the active sites for potential DNA polynucleotide interactions.
The connection between failure to degrade DNA by TREX1 and immune activation was first made in the TREX1 null mouse that showed a dramatically reduced survival associated with inflammatory myocarditis (25). However, the origin and nature of the disease-driving DNA polynucleotides resulting from TREX1 deficiency have not been clearly established. One model posits that TREX1 acts in the SET complex to degrade genomic dsDNA during granzyme A-mediated cell death by rapidly degrading DNA from the 3′ ends generated by the NM23-H1 endonuclease (26). Two additional models propose that TREX1 prevents immune activation by degrading ssDNA, but these models differ on the possible source of offending DNA polynucleotide. In TREX1-deficient cells there is an accumulation of ssDNA fragments within the cytoplasm proposed, in one model, to be generated from failed processing of aberrant replication intermediates that result in chronic activation of the DNA damage response pathway (27, 28). Another model proposes the source of accumulating ssDNA in TREX1-deficient cells to be derived from unrestrained endogenous retroelement replication, leading to activation of the cytosolic DNA-sensing cGAS–STING pathway (29–33). This concept is also supported by the participation of TREX1 in degradation of HIV-derived cytosolic DNA (34). Thus, disparate concepts on the DNA polynucleotide-driving immune activation in TREX1 deficiency have been proposed, and it is possible that the robust TREX1 exonuclease participates in multiple DNA degradation pathways. We present here structural and in vivo data supporting the concept that TREX1 degradation of dsDNA is critical to prevent immune activation.
Results and Discussion
The dominant-negative effects of TREX1 D18N in the heterozygous genotype of individuals affected with familial chilblain lupus were revealed in the DNA degradation properties of the hetero- and homodimer forms of TREX1 likely to exist in cells of these individuals. The TREX1 WT homodimers and the WT protomer within heterodimers containing a D18N mutant protomer are fully functional when degrading ssDNA polynucleotides (13). In contrast, TREX1 heterodimers and homodimers containing a D18N mutant protomer are inactive on dsDNA and block the dsDNA degradation activity of TREX1 WT enzyme, providing a genetic and mechanistic explanation linking dysfunctional TREX1 and human disease phenotype (21, 23, 24). The selective catalytic inactivity of TREX1 D18N on dsDNA indicates a significant difference in the interactions of TREX1, with ss- and dsDNA likely linked to DNA unwinding.
TREX1 D18N–dsDNA Structure Reveals a Novel Unwinding Mechanism.
To elucidate the mechanisms of TREX1 D18N-dominant mutant dysfunction, we determined the crystal structure of the TREX1 D18N mutant protein in complex with dsDNA. The structure of the TREX1 D18N–dsDNA complex reveals a novel dsDNA-unwinding mechanism that feeds a single-stranded terminus into the active site and exposes a defect in metal ion binding that contributes to catalytic inactivity. Similar to the DNA repair nucleases that share common elements while exhibiting individually unique mechanisms (35), there is little change in the core TREX1 structure, whereas the flexible regions bind, melt, and reshape the dsDNA to position ssDNA that is specifically required for catalysis into the active site. Together, the tight protein–DNA interactions coupled with a catalytically inactive protein points to the biological dysfunction that connects TREX1-dominant mutants with disease.
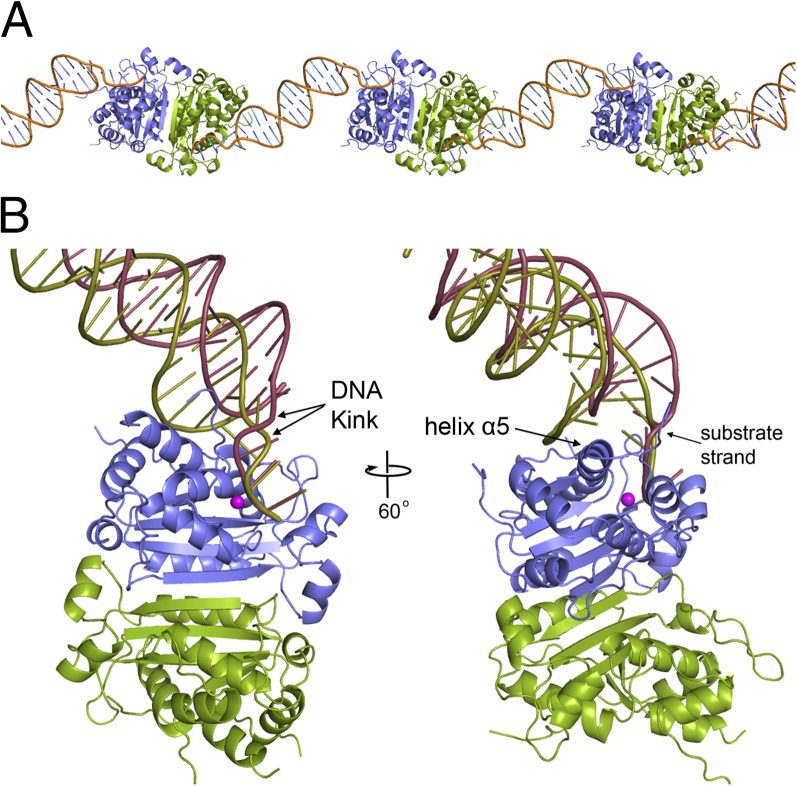
TREX1 uses a nucleic acid-kinking mechanism for unwinding dsDNA to provide ssDNA substrate to the active site. The TREX1–dsDNA structure reveals two distinct DNA-binding steps in its dsDNA-unwinding mechanism. The protein–dsDNA complex was crystallized with two TREX1 dimers and two dsDNA helices in the asymmetric unit. Each dimer has a 3′ end of the DNA bound in one of the active sites with the 3′ end of the complementary strand bound in the active site of an adjacent dimer, creating a “beads on a string” type lattice throughout the crystal (Fig. 1A). A comparison of the four active sites within the TREX1–dsDNA complexes in the asymmetric unit reveals two distinct binding conformations in the ends of the DNA helices (Fig. 1B), representing separate steps in an unwinding process necessary to provide ssDNA for insertion into the active site. Both conformations have the terminal 3′ nucleotide correctly positioned in the active site, and both induce a kink in the substrate strand of the DNA at the point of transition from ds- to ssDNA. There are also marked differences in the conformations of the bound DNA and the associated interactions with each protomer of the TREX1 dimer. A comparison of the TREX1 D18N protein with the WT TREX1 (pdbid 2OA8 (8),) reveals little overall change in the core protein structure, with an overall rmsd between the two of 0.64 Å. Additionally, the positions of the conserved catalytic residues are well maintained.
Fig. 1.
Structure of TREX1–dsDNA complex. (A) The arrangement of TREX1 D18N–dsDNA complex within the crystals has each 3′ end of a dsDNA duplex bound in the active site of adjacent dimers. (B) A superposition of the two protein–DNA complexes in the asymmetric unit reveal dsDNA is bound in two conformations within the active sites. The “tight” conformation (gold DNA) makes extensive interactions to both nucleotide strands and reveals a base-flipping mechanism for dsDNA unwinding. The “loose” conformation (red) makes interactions primarily with the substrate strand.
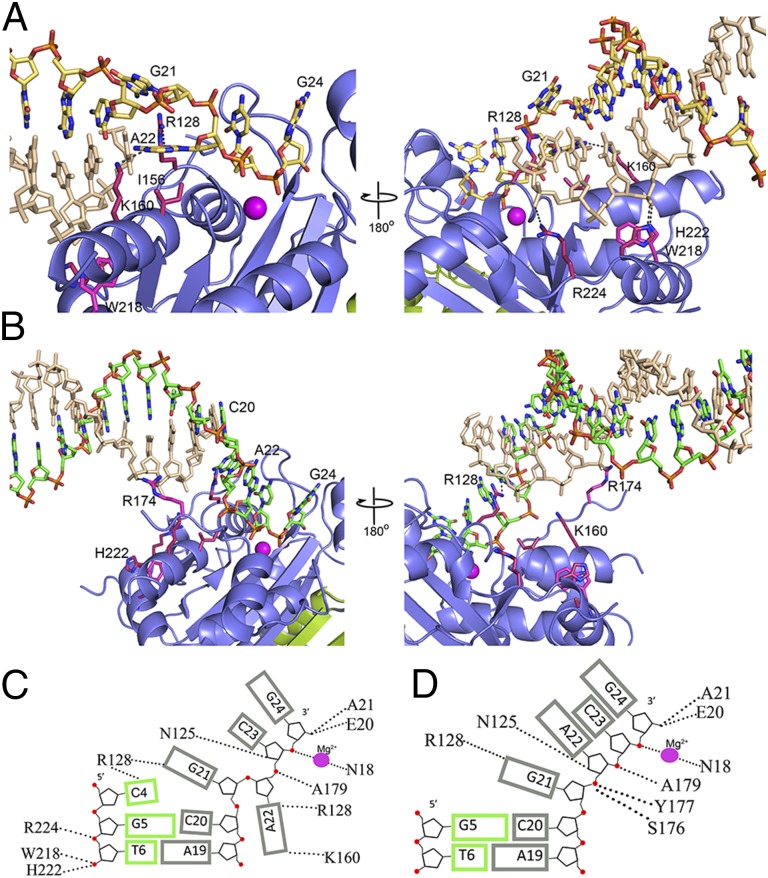
The first conformation, described here as the “tight” conformation, likely represents an early step in DNA unwinding. The TREX1 protein makes contacts with both strands of the DNA. The substrate strand is tethered to the protein through interactions with active site residues, whereas the phosphodiester backbone of the complementary strand straddles helix α5, making contacts with residues W218, H222, and R224 (Fig. 2 A and C). The helix α5 acts as a wedge into the minor groove, inducing a widening of the groove to about 16.5 Å at the point of strand separation. The four nucleotides at the end of the substrate strand are unpaired, with no visible density for the last three on the complementary strand. The last two nucleotides on the substrate strand (C23-G24) are correctly positioned in the active site for hydrolysis of the 3′ terminus. Proximal to this, the DNA backbone is severely distorted, with a ∼95° bend in the phosphodiester backbone between nucleotides G21 and C23. The nucleotide in the middle of this kink (A22) is rotated into the minor groove (Fig. 2 A and C). The flipping of nucleotide A22 by TREX1 is facilitated by residues R128 and K160 directly adjacent to the active site. The hydrophobic face of the flipped base is stabilized by its position directly above the sidechain of I156. R128 also makes cation-pi stacking interactions with the final unpaired nucleotide (C4) of the complementary strand. The importance of residue R128 in the DNA-binding and base-flipping process is consistent with the previously identified TREX1 mutation of R128 to histidine in a patient with neurophychiatric systemic lupus erythematosus (18).
Fig. 2.
TREX1 DNA-unwinding conformations. (A) TREX1–dsDNA “tight” conformation interactions facilitate DNA unwinding through base-flipping of the third nucleotide (A22) on the substrate strand and kinking of strand backbone 5′ to the flipped nucleotide. Base-flipping is facilitated by R128 and K160 on helix α5, which protrudes into and widens the minor groove. The complementary strand (colored beige) is anchored to the protein through interactions with W218, H222, and R224. (B) The TREX1 “loose” conformation makes protein–dsDNA contacts primarily with the substrate strand. The DNA is lifted above the protein, with only residue R174 making contact to the complementary strand (shown in beige). There is no nucleotide flipping, and the four terminal nucleotides in the substrate strand are completely unpaired from the complementary strand. A schematic illustrates the protein–DNA interactions in the (C) tight conformation and (D) loose conformation.
A second, “loose” conformation captures the enzyme after the base-flipping step (Fig. 2 B and D). Here, nucleotide A22 on the substrate strand is rotated back into a base-stacking orientation, with its phosphate group making contact with the amide nitrogen of Y177 and sidechain oxygen of S176. The kink in the DNA backbone is now translated down the strand between nucleotides A22 and C20. Nucleotide C4 on the complementary strand that is visible in the tight conformation is now disordered, indicating an unwinding of the DNA helix by one base pair relative to the tight conformation. Residue R128 makes only a single hydrogen bond interaction with the base of nucleotide G21 of the substrate strand. The complementary strand of DNA is no longer making interactions with W218, H222, and R224, but instead contacts residue R174, allowing the DNA helix to lift above the protein and reducing the widening of the minor groove to about 13 Å (Fig. 2 B and D). This loose DNA-binding conformation may facilitate release of the DNA after hydrolysis, consistent with the nonprocessive nature of the TREX1 enzyme.
The inhibition of dsDNA degradation by the dominant TREX1 D18N mutant can be explained by the enzyme being trapped in an inactive complex, which combines extensive protein–nucleic acid interactions necessary for dsDNA unwinding with a catalytically deficient active site. The TREX1 D18N–dsDNA structure reveals a single magnesium ion in the active site, coordinated by the mutated residue N18, and a phosphate oxygen of the terminal 3′ nucleotide. The second metal ion necessary for catalysis is absent (SI Appendix, Fig. S1). The positions of the two terminal nucleotides of the substrate and the active site residues in both the tight and loose conformations superimpose on each other and also superimpose on a ssDNA substrate bound within a WT TREX1 active site (SI Appendix, Figs. S2 and S3), indicating that the absence of the second divalent metal ion contributes to catalytic inactivity of this mutant. Identification of possible altered dsDNA binding that could contribute to failed catalysis in TREX1 D18N will require comparison with TREX1 WT dsDNA that is not currently available. The selectively dysfunctional degradation of dsDNA by the dominant TREX1 D18N mutant directly points to dsDNA as an important cellular substrate for TREX1. As described in structural studies of the DNA repair enzymes FEN1 (36), MRE11 (37), and RNase T (38) in complexes with DNA substrates, the TREX1 D18N–dsDNA structure reveals a unique protein design that incorporates DNA bending, base flipping, and structural wedges allowing TREX1 to facilitate dsDNA degradation in cells. The TREX1 core is most similar to the RNase T that also requires ssDNA in the active site. However, the TREX1 dimer forms very differently from the RNase T dimer, exposing the necessary structural elements in TREX1 to allow dsDNA unwinding that is not possible in the RNase T.
Spontaneous Lupus-Like Inflammatory Disease in the TREX1 D18N Mouse.
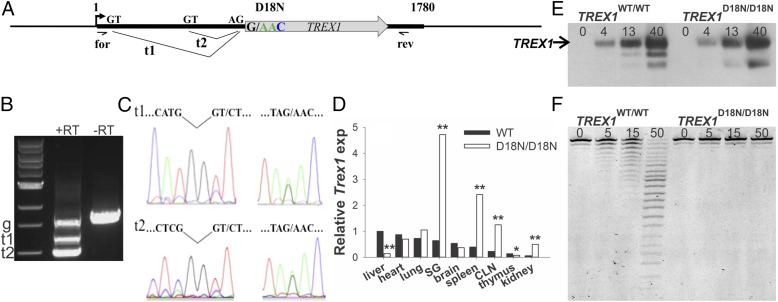
More than 40 TREX1 mutant alleles have been identified that cause a variety of lupus-like autoimmune diseases in humans, but the effects of these TREX1 mutations in mice are not known. To directly determine if dsDNA is a prominent autoantigen when the TREX1 D18N enzyme is present, we tested the effects of the dominant TREX1 D18N mutation in vivo. A genetically precise mouse model that recapitulates the dysfunctional TREX1 pathway was generated using an allelic replacement strategy to express the mouse TREX1 D18N allele from its endogenous promoter that controls the level of expression in the appropriate genomic context on mouse chromosome 9 (SI Appendix, Fig. S4). The TREX1 D18N allele is expressed (Fig. 3A), and the transcript is processed identically to the TREX1 WT allele (Fig. 3 B and C). The levels of TREX1 expression were quantified from selected tissues of animals at 8 wk of age, before the histological detection of significant inflammation. Quantification of TREX1 expression in tissues from TREX1WT/WT and TREX1D18N/D18N mice (Fig. 3D) showed that expression varied in WT tissues and that levels were altered in some of the TREX1D18N/D18N mice tissues as early as 8 wk. Interestingly, increased levels of expression in TREX1D18N/D18N mice were detected in secondary lymphoid organs, which are sites of antigen accumulation, and in some of the tissues found later to be significantly inflamed in older TREX1D18N/D18N mutant mice, including the salivary gland and kidney. Previous work has shown that TREX1 is widely expressed by immune cells and can be induced in macrophages, B cells, and dendritic cells in vitro by proinflammatory stimuli (39). Although it is possible that increased expression in TREX1D18N/D18N mouse tissues reflects populations of macrophages and other inflammatory cells responding to proinflammatory stimuli, immune cell infiltration into tissues of TREX1D18N/D18N mice does not occur to a significant extent at 8 wk, and not all tissues, such as the heart and lung, that exhibit significant inflammation later in life demonstrated increased TREX1 expression. To demonstrate the presence of TREX1 D18N protein in the TREX1D18N/D18N mice, tissues from multiple organs were pooled from TREX1WT/WT and TREX1D18N/D18N mice, TREX1 enzyme was partially purified by ssDNA chromatography, and TREX1 protein was detected by immunoblotting (Fig. 3E) and by DNA exonuclease assay (Fig. 3F) (SI Appendix, Fig. S5). Pooled tissues from TREX1WT/WT and TREX1D18N/D18N mice contained similar levels of TREX1 protein, and exonuclease activity was abolished in TREX1D18N/D18N tissues.
Fig. 3.
The TREX1 D18N allele is expressed, processed, and translated. (A) The TREX1 single exon gene is shown with the two donor (GT) and acceptor (AG) sequences. Transcripts 1 (t1) and 2 (t2) are generated from the two donor sites. (B), TREX1 cDNA products were generated from TREX1WT/D18N mouse liver RNA using forward and reverse primers positioned as shown in A. Reactions containing reverse transcriptase (RT)-generated t1 and t2. The genomic (g) TREX1 sequence was generated in reactions ±RT. (C) Sequencing of t1, t2, and g bands in B demonstrates appropriate t1 and t2 splicing (Left) and expression of TREX1 D18N and WT alleles (Right). (D) TREX1 expression in mouse tissues (six animals of each genotype). SG, salivary gland; CLN, cervical lymph node. (E) Western blot of TREX1 protein (ng) purified from mouse tissues. The position of migration of TREX1 is indicated. The band identified as TREX1 D18N was excised from a gel and confirmed by mass spectroscopic analysis (F). Activity assay of TREX1 protein (pg) purified from mouse tissues. *P < 0.05; **P < 0.001.
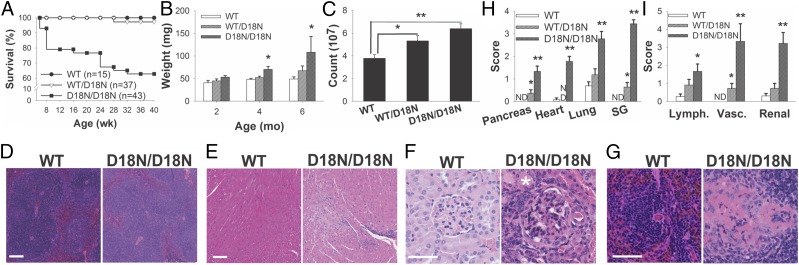
Expression of the mutant TREX1 D18N enzyme in TREX1D18N/D18N mice results in a clinically distinct phenotype from that observed when TREX1 is completely absent, as is the case in the TREX1 knockout mice that do not breed successfully and succumb to cardiomyopathy at a median age of ∼10 wk (25, 31). To determine the clinical phenotype of TREX1D18N/D18N mice we monitored animals from 3 wk to 6 mo of age. TREX1D18N/D18N mice have similar growth characteristics (SI Appendix, Fig. S6) and are typically clinically indistinguishable from WT littermates. TREX1D18N/D18N mice mate successfully up to at least 6 mo of age but have slightly smaller average litters than WT mice (4.6 vs. 5.8, respectively) (SI Appendix, Table S1). However, TREX1D18N/D18N mice have a decreased life span, with losses as early as 6 wk of age (Fig. 4A). The effects of TREX1 D18N expression were similar in males and females and revealed when animals were euthanized for phenotypic examination. The spleens of TREX1D18N/D18N mice were enlarged as early as 4 mo of age (Fig. 4B), which corresponded to increased splenic nucleated cell counts (Fig. 4C). Additionally, by 4–6 mo of age, lymph nodes were enlarged in the majority of TREX1D18N/D18N mice, and the hearts of clinically healthy animals had variously sized regions of pallor and were often mildly to markedly enlarged, with right or biventricular dilation. Of the TREX1D18N/D18N mice that were euthanized due to clinical disease or died before the predetermined euthanasia date, 87.5% had congestive heart failure characterized grossly by bicavitary effusions, chronic passive congestion of the liver, pulmonary atelectasis, and markedly enlarged hearts, with atrial and ventricular dilation and often atrial thromboses. A TREX1D18N/D18N mouse that needed to be euthanized had gross evidence of chronic kidney disease characterized by a shrunken and pitted appearance of the kidneys. Histologic analysis of tissues collected from TREX1D18N/D18N mice revealed four major categories of consistently observed lesions that were the most severe at 6 mo of age, including lymphoid hyperplasia, inflammation, vasculitis, and kidney disease (Fig. 4 D–G). Secondary lymphoid organs of TREX1D18N/D18N mice had expanded lymphoid follicles and increased numbers of germinal centers, the site of B-cell proliferation and maturation. Inflammation of varying severity was present in the heart, lung, salivary gland, pancreas, and occasionally other organs, including the lacrimal gland. The inflammatory infiltrates consisted of lymphocytes and large numbers of antibody-producing plasma cells. Inflammatory cells in the lung and salivary gland were primarily surrounding bronchioles and ducts, respectively, suggesting that inhaled and ingested antigens might contribute to immune stimulation in TREX1D18N/D18N mice. Vasculitis was present in both large- and small-caliber vessels of TREX1D18N/D18N mice and was characterized by influx of inflammatory cells, medial necrosis, and disruption of the vessel wall by protein-rich deposits. Vasculitis is common in lupus patients, and the vascular lesions observed in TREX1D18N/D18N mice mirror those seen in lupus patients. Histopathology of kidney lesions indicated widespread to diffuse, global to segmental membranoproliferative glomerulonephritis and lymphoplasmacytic tubulointerstitial nephritis (Fig. 4 H and I and SI Appendix, Fig. S7 and Tables S2–S4 for histopathology scoring).
Fig. 4.
Trex1D18N/D18N mice have a shortened lifespan and an autoimmune phenotype. (A) Survival curve. (B) Spleen weight (three to eight animals per genotype). (C) Splenocyte cell counts (10 animals per genotype). (D) Spleen H&E. (Scale bar, 100 µm.) (E) Heart H&E. (Scale bar, 100 µm.) (F) Kidney H&E. (Scale bar, 50 µm; * indicates tubular proteinosis.) (G) Splenic vessel H&E. (Bar, 50 µm.) (H) Histology scores for inflammation. SG, salivary gland. (I) Histology scores for splenic lymphoid hyperplasia, vasculitis, and renal disease. ND, not detected. *P < 0.05; **P < 0.001; bars represent the mean, and error bars represent the SEM.
The multiorgan inflammation observed in TREX1D18N/D18N mice is similar to the lupus phenotype in humans where the lung, salivary gland, and heart are often targets and immune-complex glomerulonephritis is a common complication. Increased TREX1 expression in the salivary gland of 8-wk-old TREX1D18N/D18N mice was an unexpected finding, and salivary glands were not significantly enlarged in older TREX1D18N/D18N mice. Although familial chilblain lupus patients have systemic disease (16), there are no reports of clinical involvement of the salivary glands. Skin lesions, including malar rashes and chilblains, are a frequent finding in lupus patients but are not a feature of most spontaneous mouse models of lupus (40–42) and likewise were not observed in TREX1D18N/D18N mice. This is likely because skin lesions in lupus patients are typically induced by environmental conditions, including heat, moisture, and UV light that are not factors in conventionally housed rodents. Disease was most significant in TREX1 D18N homozygous animals, whereas heterozygous animals exhibited normal survival and rather minimal inflammatory differences compared with WT. This is notable because all known cases of TREX1 D18N-mediated disease in humans have been due to the heterozygous genotype. It is possible that environmental stimuli and viral or bacterial infection could exacerbate disease in heterozygous animals.
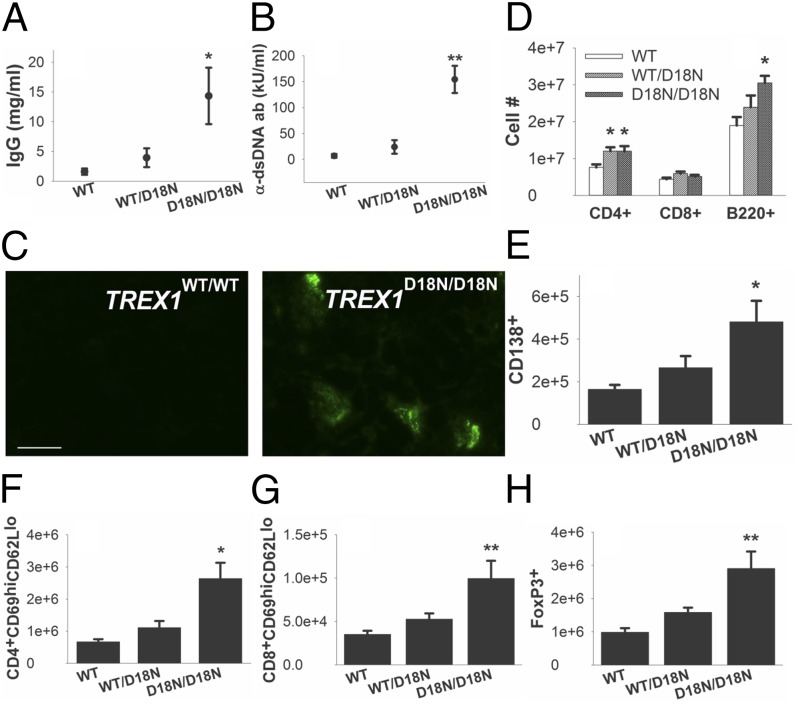
An active inflammatory autoimmune response was present in TREX1D18N/D18N mice that was also detected in TREX1WT/D18N mice, as indicated by the overall plasma cell numbers and productivity. The levels of total serum IgG were significantly increased in TREX1D18N/D18N mice compared with TREX1WT/WT mice (Fig. 5A). To determine if dsDNA was a major antigen we measured serum-total α-dsDNA antibody levels and found significantly increased levels in TREX1D18N/D18N mice compared with TREX1WT/WT mice (Fig. 5B). Furthermore, immunofluorescence of frozen kidney sections revealed immunocomplexes in glomeruli of TREX1D18N/D18N mice containing IgG (Fig. 5C). To characterize the splenic immune cell population, we labeled splenocytes with markers for T and B cells. Consistent with histologic findings, spleens of TREX1D18N/D18N mice contained increased B220+ B cells (Fig. 5D) and CD138+ plasma cells (Fig. 5E). Although the number of splenic CD8+ T cells was similar between WT and mutant mice, the number of CD4+ cells in the spleen of both TREX1WT/D18N and TREX1D18N/D18N mice was increased. Additionally, both CD4+ (Fig. 5F) and CD8+ (Fig. 5G) splenocytes of TREX1D18N/D18N mice had up-regulated expression of the activation marker CD69, demonstrating that cells from mutant mice are more activated and thus more competent at effector functions, such as cytokine production and cell killing. Finally, we found that the number of splenic Treg cells was increased in TREX1D18N/D18N mice (Fig. 5H). The increase in Treg cells could indicate a compensatory response to chronic, uncontrolled inflammation or could indicate that the Treg cells present have decreased function.
Fig. 5.
TREX1D18N/D18N mice produce α-dsDNA antibodies, form glomerular immunocomplexes, and have activated adaptive immune responses. (A) Total IgG ELISA (10–12 animals per genotype). (B) Total α-dsDNA antibody ELISA (10–12 animals per genotype). (C) Immunofluorescence (IF) for IgG in frozen kidney sections. (Bar, 50 µm.) Splenocytes were labeled for (D) CD4, CD8, and B220; (E) CD138; (F) CD4, CD69, and CD62L; (G) CD8, CD69, and CD62L; or (H) CD4, CD25, and FoxP3 and enumerated by flow cytometry. *P < 0.05; **P < 0.001; bars represent the mean, and error bars represent the SEM.
TREX1 Degradation of dsDNA Is Key to Prevent Inappropriate Immune Activation.
The TREX1 D18N–dsDNA structure and the phenotypic characteristics of the TREX1 D18N mouse indicate that TREX1 degrades dsDNA, preventing this polynucleotide from acting as an autoantigen in the mouse, and most likely in humans, to inappropriately activate the immune system. Structure and biochemical analyses of TREX1 disease-causing mutants identified key amino acids positioned adjacent to the active sites, indicating an extended DNA polynucleotide interaction (8, 20–23). The TREX1 D18N–dsDNA structure reveals direct contacts with the DNA duplex on the substrate and nonsubstrate strands. Additionally, important insight into the base-flipping mechanism by which TREX1 separates the DNA strands to efficiently degrade the polynucleotide is learned. Expression of the TREX1 D18N mutant enzyme in mice causes spontaneous autoimmunity, and our findings support the idea that failure to appropriately degrade dsDNA is the cause of disease in TREX1D18N/D18N mice, due to persistent polynucleotide sensing and immune activation. Genetic studies in humans are revealing mutations in key DNA metabolism enzymes, such as DNA polymerase β (43), that cause autoimmune pathology resembling lupus when expressed in mice. To our knowledge, expression of the TREX1 D18N allele represents the first time a monogenic form of lupus in humans has been reproduced by the same genetic change in the mouse. This mutant mouse strain will be a useful tool to further delineate the pathogenic mechanisms of TREX1-mediated autoimmunity specifically, as well as the pathogenesis of nucleic acid-mediated autoimmune disease more broadly.
Materials and Methods
Structure Determination.
The mouse recombinant TREX1 enzyme (amino acids 1–242) was prepared and crystallized with a dsDNA oligonucleotide. The X-ray data were collected and processed (SI Appendix, Table S5), as described in SI Appendix.
Animals.
TREX1 D18N mutant mice were generated on a 129S6/SvEvTac background using an allelic replacement strategy, as shown in SI Appendix, Fig. S4, and described in SI Appendix. Transmission of the TREX1 D18N allele was confirmed by sequencing of tail DNA. Males and females exhibited a similar phenotype, so both sexes were used in these studies. All experiments were performed in accordance with the guidelines set forth by the Institutional Animal Care and Use Committee at Wake Forest Baptist Medical Center.
Immunofluorescence.
Frozen sections were incubated with antibody to mouse IgG (goat α-mouse IgG Alexa Fluor 488, Abcam), washed three times in PBS, then mounted with Fluoroshield Mounting Medium with DAPI (Abcam). See SI Appendix for details.
Total IgG and dsDNA Antibody ELISA.
A total of four to five animals (6 mo old) of each sex and genotype were included in two independent experiments. Total IgG ELISA was performed according to the mouse IgG ELISA Kit (Alpha Diagnostic International) protocol. Total anti-dsDNA antibody ELISA was performed according to the mouse anti-dsDNA Antibodies Total Ig ELISA Kit (Alpha Diagnostics International). Absorbance at 450 nm was obtained using a Tecan Safire 2 spectrophotometer and Tecan Magellan software.
Flow Cytometry.
Single-cell suspensions were prepared from mouse spleen, then labeled with indicated antibody. See SI Appendix for details.
Genotyping, RT-PCR, qPCR, TREX1 Protein Purification, Exonuclease Assay, Western Blot, Histology, and Statistical Analysis.
See SI Appendix.
Supplementary Material
Acknowledgments
We thank Jennifer Schleit and Bradley Preston for providing cloned TREX1 genomic fragments for the targeting vector construct and the Wake Forest Primate Center Histology Laboratory for histology services. This research was supported by funds from the NIH [RO1GM108827 (to T.H.) and R01GM06692 (to F.W.P.)], the American Heart Association [14GRNT20020000 (to T.H.)], and the Alliance for Lupus Research (to F.W.P.). X-ray crystallographic work is supported by the Comprehensive Cancer Center of Wake Forest University National Cancer Institute Cancer Center Support Grant P30CA012197.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates for the D18N TREX–dsDNA complex have been deposited in the Protein Data Bank, www.pdb.org (accession no. 4YNQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423804112/-/DCSupplemental.
References
- 1.Lindahl T, Gally JA, Edelman GM. Properties of deoxyribonuclease 3 from mammalian tissues. J Biol Chem. 1969;244(18):5014–5019. [PubMed] [Google Scholar]
- 2.Perrino FW, Miller H, Ealey KA. Identification of a 3′—>5′-exonuclease that removes cytosine arabinoside monophosphate from 3′ termini of DNA. J Biol Chem. 1994;269(23):16357–16363. [PubMed] [Google Scholar]
- 3.Perrino FW, Mazur DJ, Ward H, Harvey S. Exonucleases and the incorporation of aranucleotides into DNA. Cell Biochem Biophys. 1999;30(3):331–352. doi: 10.1007/BF02738118. [DOI] [PubMed] [Google Scholar]
- 4.Höss M, et al. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 1999;18(13):3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′—>5′ exonucleases. J Biol Chem. 1999;274(28):19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- 6.Mazur DJ, Perrino FW. Structure and expression of the TREX1 and TREX2 3′ —> 5′ exonuclease genes. J Biol Chem. 2001;276(18):14718–14727. doi: 10.1074/jbc.M010051200. [DOI] [PubMed] [Google Scholar]
- 7.Mazur DJ, Perrino FW. Excision of 3′ termini by the Trex1 and TREX2 3′—>5′ exonucleases. Characterization of the recombinant proteins. J Biol Chem. 2001;276(20):17022–17029. doi: 10.1074/jbc.M100623200. [DOI] [PubMed] [Google Scholar]
- 8.de Silva U, et al. The crystal structure of TREX1 explains the 3′ nucleotide specificity and reveals a polyproline II helix for protein partnering. J Biol Chem. 2007;282(14):10537–10543. doi: 10.1074/jbc.M700039200. [DOI] [PubMed] [Google Scholar]
- 9.Richards A, et al. C-terminal truncations in human 3′-5′ DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat Genet. 2007;39(9):1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- 10.Orebaugh CD, et al. The TREX1 C-terminal region controls cellular localization through ubiquitination. J Biol Chem. 2013;288(40):28881–28892. doi: 10.1074/jbc.M113.503391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crow YJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutières syndrome at the AGS1 locus. Nat Genet. 2006;38(8):917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 12.Lee-Kirsch MA, et al. Familial chilblain lupus, a monogenic form of cutaneous lupus erythematosus, maps to chromosome 3p. Am J Hum Genet. 2006;79(4):731–737. doi: 10.1086/507848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee-Kirsch MA, et al. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J Mol Med (Berl) 2007;85(5):531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- 14.Rice G, et al. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. Am J Hum Genet. 2007;81(4):713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crow YJ, Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: Linking nucleic acid metabolism with autoimmunity. Hum Mol Genet. 2009;18(R2):R130–R136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunther C, Berndt N, Wolf C, Lee-Kirsch MA. Familial chilblain lupus due to a novel mutation in the exonuclease III domain of 3′ repair exonuclease 1 (TREX1) JAMA Dermatol. 2014 doi: 10.1001/jamadermatol.2014.3438. in press. [DOI] [PubMed] [Google Scholar]
- 17.Lee-Kirsch MA, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39(9):1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 18.de Vries B, et al. TREX1 gene variant in neuropsychiatric systemic lupus erythematosus. Ann Rheum Dis. 2010;69(10):1886–1887. doi: 10.1136/ard.2009.114157. [DOI] [PubMed] [Google Scholar]
- 19.Namjou B, et al. Evaluation of the TREX1 gene in a large multi-ancestral lupus cohort. Genes Immun. 2011;12(4):270–279. doi: 10.1038/gene.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey SL, Harvey S, Perrino FW, Hollis T. Defects in DNA degradation revealed in crystal structures of TREX1 exonuclease mutations linked to autoimmune disease. DNA Repair (Amst) 2012;11(1):65–73. doi: 10.1016/j.dnarep.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fye JM, Coffin SR, Orebaugh CD, Hollis T, Perrino FW. The Arg-62 residues of the TREX1 exonuclease act across the dimer interface contributing to catalysis in the opposing protomers. J Biol Chem. 2014;289(16):11556–11565. doi: 10.1074/jbc.M114.559252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orebaugh CD, Fye JM, Harvey S, Hollis T, Perrino FW. The TREX1 exonuclease R114H mutation in Aicardi-Goutières syndrome and lupus reveals dimeric structure requirements for DNA degradation activity. J Biol Chem. 2011;286(46):40246–40254. doi: 10.1074/jbc.M111.297903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fye JM, Orebaugh CD, Coffin SR, Hollis T, Perrino FW. Dominant mutation of the TREX1 exonuclease gene in lupus and Aicardi-Goutieres syndrome. J Biol Chem. 2011;286(37):32373–32382. doi: 10.1074/jbc.M111.276287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehtinen DA, Harvey S, Mulcahy MJ, Hollis T, Perrino FW. The TREX1 double-stranded DNA degradation activity is defective in dominant mutations associated with autoimmune disease. J Biol Chem. 2008;283(46):31649–31656. doi: 10.1074/jbc.M806155200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morita M, et al. Gene-targeted mice lacking the Trex1 (DNase III) 3′—>5′ DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24(15):6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhury D, et al. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol Cell. 2006;23(1):133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Lindahl T, Barnes DE, Yang YG, Robins P. Biochemical properties of mammalian TREX1 and its association with DNA replication and inherited inflammatory disease. Biochem Soc Trans. 2009;37(Pt 3):535–538. doi: 10.1042/BST0370535. [DOI] [PubMed] [Google Scholar]
- 28.Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131(5):873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Ablasser A, et al. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J Immunol. 2014;192(12):5993–5997. doi: 10.4049/jimmunol.1400737. [DOI] [PubMed] [Google Scholar]
- 30.Gall A, et al. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity. 2012;36(1):120–131. doi: 10.1016/j.immuni.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134(4):587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volkman HE, Stetson DB. The enemy within: Endogenous retroelements and autoimmune disease. Nat Immunol. 2014;15(5):415–422. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11(11):1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsutakawa SE, Lafrance-Vanasse J, Tainer JA. The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once. DNA Repair (Amst) 2014;19:95–107. doi: 10.1016/j.dnarep.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsutakawa SE, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145(2):198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams RS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135(1):97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsiao YY, Duh Y, Chen YP, Wang YT, Yuan HS. How an exonuclease decides where to stop in trimming of nucleic acids: Crystal structures of RNase T-product complexes. Nucleic Acids Res. 2012;40(16):8144–8154. doi: 10.1093/nar/gks548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pereira-Lopes S, et al. The exonuclease Trex1 restrains macrophage proinflammatory activation. J Immunol. 2013;191(12):6128–6135. doi: 10.4049/jimmunol.1301603. [DOI] [PubMed] [Google Scholar]
- 40.Pathak S, Mohan C. Cellular and molecular pathogenesis of systemic lupus erythematosus: Lessons from animal models. Arthritis Res Ther. 2011;13(5):241–249. doi: 10.1186/ar3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perry D, Sang A, Yin Y, Zheng YY, Morel L. Murine models of systemic lupus erythematosus. J Biomed Biotechnol. 2011;2011(271694):1–19. doi: 10.1155/2011/271694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rottman JB, Willis CR. Mouse models of systemic lupus erythematosus reveal a complex pathogenesis. Vet Pathol. 2010;47(4):664–676. doi: 10.1177/0300985810370005. [DOI] [PubMed] [Google Scholar]
- 43.Senejani AG, et al. Mutation of POLB causes lupus in mice. Cell Reports. 2014;6(1):1–8. doi: 10.1016/j.celrep.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.