Significance
How membrane proteins are guided into the membrane is a fundamental question of cell biology. Translocons are known to create a polypeptide-conducting transmembrane (TM) channel having a lateral gate to allow lipid-phase partitioning of the substrate. Here, we show that YidC guides a class of membrane proteins in a channel-independent fashion. Our experiments using intact Bacillus subtilis cells show that SpoIIIJ, a YidC homolog, forms a water-accessible cavity in the cell membrane and that the cavity’s overall hydrophilicity, as well as the presence of an Arg residue at one of several alternative places on the cavity, is functionally important. The extracellular part of substrate is probably first attracted to the YidC cavity before establishment of a TM configuration through hydrophobic partitioning.
Keywords: membrane protein insertion, MifM, SpoIIIJ, YidC, Oxa1
Abstract
The recently solved crystal structure of YidC protein suggests that it mediates membrane protein insertion by means of an intramembrane cavity rather than a transmembrane (TM) pore. This concept of protein translocation prompted us to characterize the native, membrane-integrated state of YidC with respect to the hydropathic nature of its TM region. Here, we show that the cavity-forming region of the stage III sporulation protein J (SpoIIIJ), a YidC homolog, is indeed open to the aqueous milieu of the Bacillus subtilis cells and that the overall hydrophilicity of the cavity, along with the presence of an Arg residue on several alternative sites of the cavity surface, is functionally important. We propose that YidC functions as a proteinaceous amphiphile that interacts with newly synthesized membrane proteins and reduces energetic costs of their membrane traversal.
Biogenesis of membrane proteins, a fundamental cellular process essential for all living organisms, includes insertion of a newly synthesized membrane protein into the membrane followed by its folding and assembly with other cellular components. In the Sec-dependent pathway in bacteria, membrane insertion is mediated by the protein-conducting channel SecYEG in the plasma (cytoplasmic) membrane (1–3), whereas acquisition of the native conformation is facilitated by the conserved YidC/Oxa1/Alb3 family of membrane proteins (4–7). In a Sec-independent pathway, YidC protein facilitates insertion of a class of membrane proteins independent of SecYEG. Thus, YidC is a dual-function protein that serves as a chaperone or an insertase in membrane protein biogenesis (4–7).
Bacillus subtilis possesses two YidC homologs, SpoIIIJ (YidC1) and YidC2 (YqjG). These proteins, in combination with their substrate MifM, have provided us a unique in vivo experimental system with which to study YidC. Although SpoIIIJ and YidC2 share growth-essential functions, as indicated from the synthetic lethal phenotype of their deletion (8, 9), SpoIIIJ is constitutively expressed and YidC2 is induced upon dysfunction of SpoIIIJ (10, 11) in a manner that is repressible autogenously (12). MifM is encoded from the upstream ORF of yidC2 and plays an essential role in this cross-feedback and autogenous regulation by undergoing regulated elongation arrest in its translation (12–14). The ribosome stalling at mifM leads to exposure of the Shine–Dalgarno sequence of yidC2 to enhance its translation. Importantly, elongation arrest of mifM is released upon the YidC-dependent membrane insertion of the nascent MifM polypeptide, enabling the yidC2 translation to be up-regulated when cellular YidC activity declines. In this manner, MifM enables the cell to maintain the capacity of the YidC pathways of membrane protein biogenesis under changing intracellular and extracellular conditions (11, 12). This regulatory system also enables us to monitor the in vivo activities of YidC proteins; expression of a yidC2′-lacZ translational fusion gene, and hence the β-gal activity, will increase in response to a decrease in the SpoIIIJ activity (11).
Although both SecYEG and YidC could facilitate membrane protein insertion, their modes of actions are fundamentally different. For instance, whereas SecYEG can mediate membrane insertion of proteins with multiple transmembrane (TM) segments, as well as those proteins having large extracytoplasmic (periplasmic) domains, YidC, as an insertase, is specialized in insertion of small membrane proteins that possess a single or two TM segment(s) and short extracytoplasmic region(s) (7). Crystal structures of archaeal and bacterial SecYEβ and SecYE(G) complexes reveal an hourglass-shaped TM pore formed by the TM segments of SecY. The pore can also open laterally to the lipid phase of the membrane, allowing release of a TM segment of substrates out of the translocon pore to establish membrane protein integration (15–17). Although earlier electron microscopic studies of Escherichia coli YidC and Saccharomyces cerevisiae Oxa1 led to a proposal that YidC forms a homodimer, which creates a channel-like structure at the subunit interface (18), more recent evidence suggests that a monomer of YidC interacts with the ribosome that is translating a membrane protein (19, 20). The crystal structures of YidC from Bacillus halodurans at a resolution up to 2.4 Å (21) revealed that the five TM segments of YidC form a cavity presumably in the lipid bilayer. This cavity appears to be open to the lipidic phase and the cytoplasm but not to the extracytoplasmic environment (Fig. 1 A and B), arguing against the dimeric insertion pore model. Strikingly, the concave surface of the cavity is enriched in hydrophilic amino acid residues, including a conserved Arg. Genetic analyses of a B. subtilis YidC homolog, SpoIIIJ, and its substrate membrane protein, MifM, revealed that the positive charge of the conserved Arg (Arg73 in SpoIIIJ) (Fig. S1) and the negatively charged residues in the extracytoplasmic and TM regions of MifM are essential for insertion of MifM into the membrane (21). From these results, we proposed that SpoIIIJ mediates insertion of a class of membrane proteins such as MifM by a channel-independent mechanism, in which electrostatic attraction between the SpoIIIJ cavity and the substrate initiates the reaction (21). The other B. subtilis YidC homolog, YidC2 (YqjG), also functions by means of a similar mechanism for insertion of MifM (12). The importance of the cavity was also supported by photo–cross-linking experiments showing that the inner surface of the cavity of SpoIIIJ interacts with substrate protein in vivo (21). Together with the crystal structure of E. coli YidC (22), it is suggested that having a hydrophilic and positively charged cavity is a feature shared by the YidC family members.
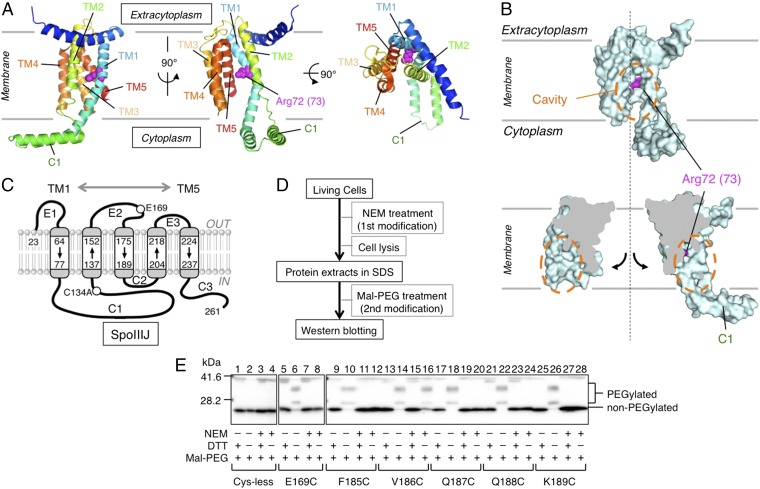
Fig. 1.
Experimental design of NEM reactivity assay to assess water accessibility to the SpoIIIJ cavity. (A) Ribbon diagram representations of the crystal structure of B. halodurans YidC2 (Protein Data Bank ID code 3WO6). Shown are the side views (Left and Center) and a top view (Right). Arg72 (corresponding to Arg73 in B. subtilis SpoIIIJ; Fig. S1) is shown by magenta spheres. TM and C1 indicate the TM segments (with numbers) and the first cytoplasmic region, respectively. (B) Surface model (Upper) and cut-away molecular surface representation (Lower) of B. halodurans YidC2. Orange dashed-line circles encircle the intramembrane cavity. Arg72 (Arg73 in SpoIIIJ) is shown in magenta. (C) Schematic representation of the membrane integration topology of B. subtilis SpoIIIJ and the sites where a unique Cys was introduced for NEM reactivity assay. TM1–TM5, C1–C3, and E1–E3 show the TM, the cytoplasmic, and the extracytoplasmic regions, respectively. (D) Work flow of the assay. Intact cells were treated with NEM. Proteins were then extracted with SDS and subjected to PEGylation of the remaining thiols under denaturing conditions. Finally, SpoIIIJ species were visualized after SDS/PAGE. (E) Electrophoretic separation of the PEG-modified and unmodified SpoIIIJ species. Positions of Cys introduced into SpoIIIJ are shown at the bottom. Each sample received four different treatments, as indicated by + and −. DTT in excess was included in alternate samples at the PEGylation step to give unmodified controls. PEGylated SpoIIIJ forms multiple slow-migrating bands due to heterogeneity of the Mal-PEG preparation. The bands near the 42-kDa position are nonspecific.
Because the unprecedented hydropathic arrangement of YidC is of crucial importance in our understanding of membrane protein biogenesis, its occurrence in the native membrane must be verified using intact living cells. Here, we explored the hydropathic nature and the functional requirements of the cavity-forming TM region of SpoIIIJ in intact cells. The YidC cavity indeed proved to be accessible by water, and its hydrophilicity, including an Arg residue somewhere in the cavity, proved to be important functionally. That YidC creates an aqueous microenvironment in the membrane gives strong support to the channel-independent mode of its action.
Results
Water Accessibility of the SpoIIIJ Intramembrane Cavity.
YidC forms an intramembrane cavity that is open laterally, presumably toward the lipid phase of the membrane and the cytoplasm, whereas it is inaccessible from the extracytoplasm (Fig. 1 A and B). The inner surface of the cavity contains several hydrophilic amino acid residues, including the essential Arg, raising a possibility that YidC forms a hydrophilic local environment in the otherwise hydrophobic lipid bilayer. To verify this unusual hydropathic arrangement experimentally, we examined the water accessibility of YidC TM residues using intact living cells and the N-ethylmaleimide (NEM) reactivity assay. NEM is membrane-permeable and alkylates the thiol group of a Cys residue of protein in a water-dependent reaction (23–25), enabling us to assess the water availability of a specific site of the target protein in intact cells by strategically placing a Cys residue.
We first constructed the Cys-less SpoIIIJ (SpoIIIJ-C134A), which proved to be functional as shown by the low β-gal activity of the yidC2′-lacZ reporter (Fig. S2), whose expression level inversely correlates with the efficiency of membrane insertion of MifM, a YidC substrate (11). We then introduced a Cys residue into selected single positions of the Cys-less SpoIIIJ. Mutant proteins were expressed from the native chromosomal locus under the control of the native spoIIIJ promoter. Intact cells were then treated with NEM directly without any cell disruption (Fig. 1D).
Proteins extracted from NEM-treated cells are solubilized, denatured in SDS, and then subjected to countermodification with maleimide-PEG (Mal-PEG), an alkylating reagent of ∼5 kDa (Fig. 1D). An NEM-unmodified fraction of the target protein, still having free thiol, is now modified by Mal-PEG and mobility-shifted, whereas the NEM-modified fraction of the protein resists the countermodification and does not show any appreciable mobility shift. The efficiency of NEM modification was assessed by the extent of countermodification with Mal-PEG (26).
To characterize this assay system, we first replaced Glu169 in the second extracytoplasmic loop with Cys. SpoIIIJ-C134A/E169C thus constructed was expected to have a fully water-accessible Cys (Fig. 1C). When NEM was omitted from the first reaction, the protein was efficiently modified with Mal-PEG with concomitant disappearance of the unmodified species (Fig. 1E, lane 6). The Cys-less SpoIIIJ did not show this mobility shift, and the intensity of the unmodified band remained unchanged (Fig. 1E, lanes 1–4). By contrast, NEM treatment of the E169C-expressing cells almost completely blocked the Mal-PEG modification even after denaturation (Fig. 1E, lane 8), indicating that the Cys at this position was fully accessible by NEM and water as expected.
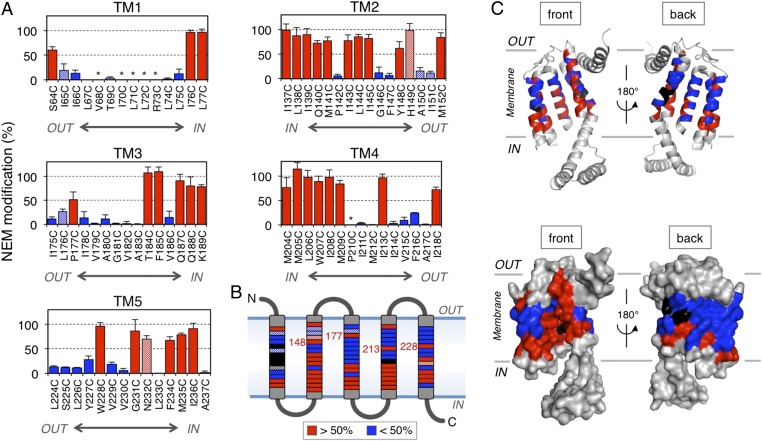
We examined a total of 74 single-Cys mutant derivatives of SpoIIIJ to cover the TM regions of SpoIIIJ (Fig. 1C). Images of immunoblotting in the NEM reactivity assay are shown in Fig. 1E (lanes 9–28) for selected target positions and in Fig. S3 for all of the positions examined. Cys at positions 185, 187, 188, and 189 was fully reactive with NEM, as judged from the lack of countermodification with Mal-PEG (Fig. 1E, lane 10 vs. lane 12, lane 18 vs. lane 20, lane 22 vs. lane 24, and lane 26 vs. lane 28, respectively). By contrast, Cys at position 186 was not markedly reactive with NEM, as judged from the evident countermodification (Fig. 1E, lane 14 vs. lane 16). These results suggest that water molecules can access residues 185, 187, 188, and 189 but cannot effectively access residue 186. Average NEM modification efficiencies in at least three independent experiments, calculated as described in Experimental Procedures, are shown in Fig. 2A. The summarized water accessibility features are depicted in Fig. 2 B and C (efficiently modified residues are shown in red). We note that Cys at positions 68, 70, 71, 72, 73, and 210 (shown by asterisks in Fig. 2A and shown in black in Fig. 2 B and C and Fig. S3) was not efficiently modified with Mal-PEG, even without the first NEM modification. These positions may have been buried in a manner inaccessible by the alkylating agents and/or water, even after denaturation with SDS.
Fig. 2.
Water accessibility profiles of the TM regions of SpoIIIJ as assessed by NEM reactivity. (A) NEM modification efficiencies (mean ± SD, n ≥ 3) of Cys at the indicated positions. Red columns show efficiencies of >50%, and blue columns show lower (<50%) modification efficiencies. Asterisks indicate SpoIIIJ derivatives that could not be assessed because of the lack of PEGylation even without NEM treatment. Striped columns represent nonfunctional SpoIIIJ mutants. (B) Schematic representation of the sites of Cys and their NEM modification efficiencies. The sites of higher and lower modification, as well as nonreactive sites, are color-coded as in A. The four numbered positions are located in the extracytoplasmic half of the membrane and are NEM-modified efficiently. The sites of nonfunctional Cys substitutions are striped. (C) Ribbon (Upper) and surface (Lower) representations of the front and back views of the B. halodurans YidC2 structure with the color-coded water accessibility.
The results presented above show, strikingly, that numerous positions of the TM segments of SpoIIIJ are facing an aqueous environment. We note a tendency of the residues located closer to the cytoplasm to exhibit higher modification efficiencies than those residues located on the distal side, with some exceptions (Fig. 2 A–C). It is remarkable that the eight positions (S64, Y148, H149, M152, P177, I213, I218, and W228) are still highly accessible by water, even though they are located in the extracytoplasmic half of the bilayer membrane. Among these residues, S64, M152, and I218 are likely to be exposed to the extracytoplasmic environment, as shown from the crystal structure, explaining their high reactivity. By contrast, the other five NEM-modifiable residues in the distal half are likely embedded in the lipid bilayer, among which Y148, P177, I213, and W228 project their side chains toward the interior of the concave cavity.
In summary, our systematic in vivo NEM modification assay suggests that the SpoIIIJ cavity creates an aqueous environment in the living cell membrane. Functional assays showed that most of the mutant SpoIIIJ derivatives were functional, although some others were less functional (Fig. S2). Although we included the nonfunctional SpoIIIJ mutants (Fig. S2 and those mutants shown in striped colors in Fig. 2 A and B) in our analysis, omitting them does not essentially affect our conclusion.
Functional Importance of General Hydrophilicity of the YidC Cavity.
We next addressed whether the hydrophilicity of the cavity is important for the YidC function. In our previous genetic studies, single Ala substitutions for the conserved hydrophilic residues in the cavity did not deteriorate SpoIIIJ functions, except for Arg73 (21). We reason that single Ala substitutions may be insufficient to reduce the overall hydrophilicity of the cavity. We therefore selected six hydrophilic residues in the cavity (Gln140, Thr184, Gln187, Gln188, Gly231, and Asn232) that were efficiently modified by NEM (Fig. 2) for their simultaneous replacement with either Ala (SpoIIIJ-6A) or Leu (SpoIIIJ-6L) to make the cavity more hydrophobic. As a control, we constructed a mutant in which the six residues were replaced either by hydrophilic Asn or Gln (SpoIIIJ-5N1Q, having mutations Q140N, T184N, Q187N, Q188N, G231N, and N232Q). NEM reactivity of Cys introduced at either the 213th or the 228th position was significantly lowered by the spoIIIJ-6A and the spoIIIJ-6L mutations (Fig. 3B). By contrast, the NEM reactivity remained unaffected at the high level by the spoIIIJ-5N1Q mutation.
Fig. 3.
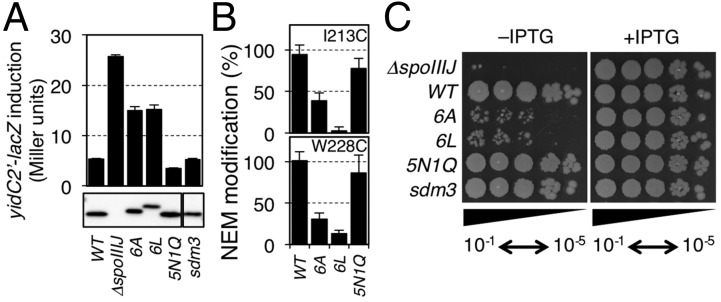
Functional importance of hydrophilicity of the SpoIIIJ cavity. (A) Efficiencies of MifM insertion into the membrane by the SpoIIIJ variants. (Upper) β-Gal activities (mean ± SD, n = 3) of the spoIIIJ mutant strains harboring the yidC2′-lacZ reporter gene, which inversely correlate with the efficiencies of MifM insertion. (Lower) Cellular accumulation of SpoIIIJ derivatives determined by anti-SpoIIIJ immunoblotting. (B) NEM modification efficiencies of Cys introduced either at the 213rd or the 228th position of the WT and SpoIIIJ cavity mutants indicated at the bottom. (C) Growth-supporting abilities of the SpoIIIJ cavity mutants. A complementation assay of B. subtilis was carried out using strains lacking the yidC2 gene and having a rescue plasmid encoding IPTG-inducible spoIIIJ-FLAG. The chromosome contained the indicated spoIIIJ alleles shown on the left. Cultures were serially diluted (from 10−1 to 10−5) and spotted onto LB agar plates containing 0 mM (Left) or 1 mM (Right) IPTG, which were then incubated for 17.5 h at 37 °C.
We then assessed the insertase activity of the SpoIIIJ mutants using the yidC2′-lacZ reporter. Whereas cells expressing WT spoIIIJ had β-gal activity of 5.3 units (Fig. 3A, column 1), the spoIIIJ-deletion strain (ΔspoIIIJ) had 25.7 units of β-gal activity (Fig. 3A, column 2). β-Gal activity of cells expressing spoIIIJ-6A was 15.0 units, and β-gal activity of spoIIIJ-6L–expressing cells was 15.2 units, showing defects in SpoIIIJ function. By contrast, cells expressing spoIIIJ-5N1Q had only 3.5 units of β-gal activity, showing full functionality of SpoIIIJ. Immunoblotting showed that cellular abundance was similar for the SpoIIIJ variants examined, except for SpoIIIJ-6L, which was at a slightly lower level (Fig. 3A, Lower). However, this slight decrease in the accumulation level does not explain the lower activity of SpoIIIJ-6L, because cells expressing WT SpoIIIJ at a similarly decreased abundance due to a mutation in the Shine–Dalgarno sequence (sdm3-spoIIIJ) showed normal reporter expression (Fig. 3A; sdm3). Thus, the spoIIIJ-6A and the spoIIIJ-6L mutations impair the activity of SpoIIIJ to insert MifM into the membrane.
Deletion of yidC2 makes spoIIIJ essential for cell viability (8, 9), allowing us to examine the functionality of the SpoIIIJ mutant derivatives in supporting the growth of B. subtilis. We used plasmid expressing spoIIIJ-FLAG under the isopropyl-β-d-thiogalactopyranoside (IPTG)–inducible promoter to assess the growth phenotypes of spoIIIJ mutations on the chromosome that was also deleted for yidC2; in the absence of IPTG, the chromosomal spoIIIJ (with a mutation to be tested) was the sole source of YidC. We observed severe growth defects for strains having the spoIIIJ-6A or the spoIIIJ-6L mutation in the absence of IPTG. By contrast, the spoIIIJ-5N1Q and the sdm3-spoIIIJ cells grew normally even in the absence of IPTG. These results show that the hydrophilicity of the cavity is required for the growth-supporting function of SpoIIIJ. Taken together with the results obtained from the lacZ reporter assay, the SpoIIIJ cavity must be hydrophilic to function normally.
Flexible Positional Requirements for the Essential Positive Charge Within the YidC Cavity.
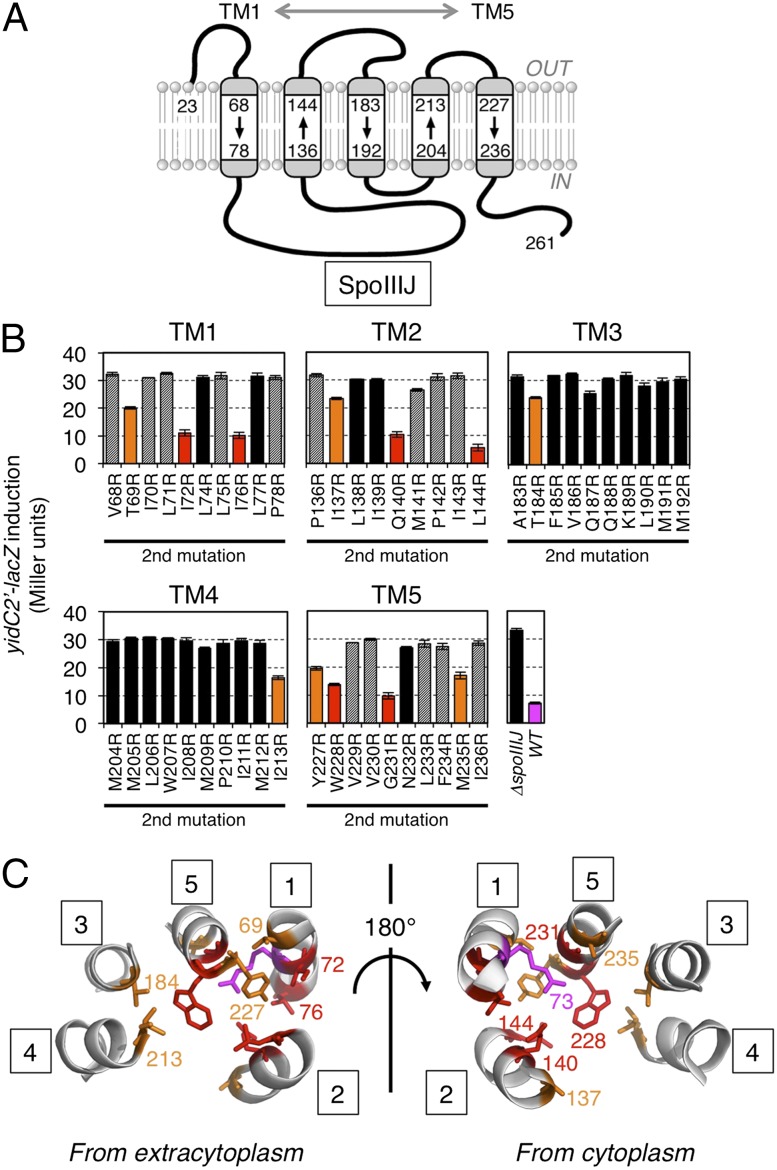
The SpoIIIJ cavity contains an Arg residue that is functionally essential, leading us to propose a charge attraction model for the initiation of translocation of MifM-like substrates (21). In this case, substrate recognition may not be based on strict structural complementarity and electrostatic interaction may allow certain positional flexibility. We addressed whether the Arg residue can be relocated to different positions on the cavity by constructing a series of SpoIIIJ mutants with the original Arg73 replaced with Ala and having a unique Arg at various positions within the TM segments of SpoIIIJ. Western blotting experiments showed that the Arg-relocating mutations sometimes destabilized the SpoIIIJ protein (Fig. S4). Most of the unstable protein had an Arg residue outside the cavity (shown in blue in Fig. S4), which may have caused severe hydrophobic mismatches.
We used the yidC2′-lacZ reporter assay and the growth complementation assay to assess the functionality of the mutant forms of SpoIIIJ. Although many Arg-relocated mutants gave elevated β-gal activity, comparable to the activity observed with the spoIIIJ-deleted cells (Fig. 4B, mutants are shown in black columns), as well as with cells carrying the spoIIIJ-R73A mutation (21), several mutants expressed β-gal at levels significantly lower than the above-mentioned class of mutants. The latter SpoIIIJ variants can still support MifM insertion even though they have lost the crucial Arg at the original position and instead contain a relocated Arg at a different position (Fig. 4B). Six of them (termed class I, which includes I72R, I76R, Q140R, L144R, W228R, and G231R; shown in red in Fig. 4 B and C) had β-gal activities of lower than 15 units, indicative of nearly full functionality in inserting MifM into the membrane. The remaining five mutants (termed class II, which includes T69R, I137R, T184R, I213R, and M235R) had β-gal activities ranging from 15 to 25 units, indicative of partial functionality. The class I mutations were found only in TM1, TM2, and TM5, whereas the class II mutations were found in all five of the TM segments. Locations of these residues on the crystal structure of B. halodurans YidC2 revealed that, with the exception of I72, they project their side chains toward the inside of the cavity (Fig. 4C). The side chain of I72 projects toward TM2 but still seems to be accessible from the cavity interior. TM2 and TM5 are both geometrically close to TM1, where Arg73 originally resided, possibly explaining why the class I mutations occurred only in TM1, TM2, and TM5.
Fig. 4.
Consequences of relocation of the essential Arg in SpoIIIJ. (A) Target boundary of Arg relocation experiments shown in the SpoIIIJ topology model. (B) Efficiencies of MifM insertion into the membrane by the SpoIIIJ variants. Reported are β-gal activities (mean ± SD, n = 3) of the spoIIIJ mutant strains harboring the yidC2′-lacZ reporter gene. All strains, except ΔspoIIIJ and WT, had the Arg73Ala mutation and an additional second site mutation indicated at the bottom in SpoIIIJ. Red, orange, and black columns represent SpoIIIJ mutants with low (<15 units), intermediate (15–25 units), and high (>25 units) activities of β-gal, indicative of high, intermediate, and low efficiencies of MifM insertion, respectively. Mutant proteins that did not significantly accumulate in the cell are striped. β-Gal activity of the WT strain is shown in magenta. (C) Ribbon representations of the TM regions (numbered in squares) of B. halodurans YidC2 with the sites of functional Arg relocation (in SpoIIIJ) highlighted by side chains colored in red (high activity) and orange (intermediate activity). The B. subtilis numbering is used. The original Arg73 is shown in magenta.
A growth complementation assay showed that all of the class I and the class II R73A/I213R mutations fully supported cell growth in the absence of YidC2. The class II mutations other than R73A/I213R resulted in poor growth (Fig. S5). Thus, the abilities of the SpoIIIJ variants to support cell growth correlated well with their insertase activities. These systematic analyses strongly support the idea that the concave surface of the cavity must be positively charged to maintain the SpoIIIJ activity but that some flexibility is allowed about exact positions of the positive charge, being consistent with the charge attraction model.
Discussion
Translocation of hydrophilic regions of a newly synthesized polypeptide across the hydrophobic lipid bilayer is an energetically challenging process in the membrane protein insertion pathways. Although the SecYEG translocon overcomes this difficulty by forming a polypeptide-conducting channel that sequesters a translocating polypeptide from the lipidic environment (15–17), several lines of evidence (19, 20, 27), most notably the crystal structure of B. halodurans YidC2 (21), suggest that YidC uses a channel-independent mechanism.
The results of our systematic NEM probing analysis of the monocysteine derivatives of SpoIIIJ indicate that the SpoIIIJ cavity provides an aqueous environment within the membrane of living cells. Although Cys substitution at certain positions, such as in the midst of consecutive hydrophobic residues, could itself have altered the local disposition of the polypeptide, we envisage that such cases are rare except for the nonfunctional mutations. The overall conclusion obtained from our in vivo analysis agrees well with the crystal structures of YidC, as well as with the results of molecular dynamics simulation of YidC, showing the presence of water molecules in the cavity (21). The hydrophilic residues on the concave surface of the cavity should contribute to maintaining the local aqueous environment, because simultaneous substitution of nonpolar Ala or Leu for the six selected hydrophilic residues on the cavity significantly reduced the efficiencies of NEM modification of a Cys introduced into the cavity. Importantly, SpoIIIJ’s activities to insert MifM, as well as to support cell growth, are compromised significantly by the spoIIIJ-6A and the spoIIIJ-6L mutations, corroborating the physiological importance of the cavity hydrophilicity.
A role of the YidC cavity may be to provide a hydrophilic environment in the otherwise hydrophobic lipid bilayer, thereby reducing the energetic cost required for insertion of hydrophilic regions of substrate into the membrane en route to the trans side. It is also conceivable that the hydrophobic mismatch at the protein–lipid interface could elicit local structural rearrangements of the lipid bilayer (28, 29) so as to affect substrate–membrane interactions, and thereby facilitate membrane insertion. We speculate that the YidC cavity is designed not simply as a hydrophilic platform but to allow for the unusual arrangement of intramembrane aqueous space to be compatible with the thermodynamic principle. Although it is unknown how this compatibility is accomplished, the notion is consistent with the observations that placement of an Arg residue is possible within the cavity, but not in its outer regions without severely destabilizing the protein (Fig. S4).
The functional Arg does not strictly require unique positioning in the cavity, because we were able to relocate it from the original 73rd position to several other positions within the cavity without loss of function. Such positional flexibility appears to be consistent with the electrostatic attracting force serving as a primary driving force for insertion of substrate. As discussed previously, the cytosolic C1 region with hairpin-like helices may provide a substrate entry point (21). Therefore, Arg that is closer to the C1 region may have higher functionality as an insertase element, although such a positioning should also be compatible with the subsequent step of translocation completion.
The hydrophilic surface within the membrane interior might be also important for the chaperone functions of YidC in the Sec-dependent insertion pathway. For instance, TM regions of membrane proteins may contain functionally important polar residues, which might be unstable in the lipidic environment upon release from translocons until assembling with a partner TM polypeptide also containing complementary polar residues (30, 31). It is tempting to speculate that the hydrophilic cavity of YidC provides a transient docking surface that binds a newly inserted TM segment before it finds a partner of assembly. Such a function is analogous to the one provided by regular aqueous-phase chaperones, although they interact with hydrophobic, instead of hydrophilic, surfaces of substrates.
The YidC family contains divergent members in different organisms, which differ in their modes of cooperation with other factors, including the signal recognition particle and the ribosome (32–35). Moreover, each homolog can have multiple functions and reaction mechanisms (7, 36–38). We envision that the peculiarity of having a hydrophilic cavity in the membrane may be a common feature conserved in many of the family members. Still, it is possible that the hydrophilic local environment is used differently in different YidC homologs. For instance, the cavity Arg in the E. coli YidC was reported to be dispensable for the insertase activity of the Pf3 coat protein, which requires the Arg when handled by Streptococcus mutans YidC2 (39). Further studies on this interesting membrane protein will advance our understanding of how living organisms manage to solve problems associated with the movement of macromolecules across hydropathic borders.
Experimental Procedures
Bacterial Strains and Plasmids.
The B. subtilis strains and plasmids used in this study are listed in Tables S1 and S2, respectively. Construction procedures of B. subtilis strains are summarized in Tables S3 and S4 presented in SI Experimental Procedures.
Media and Conditions for Growth of B. subtilis.
B. subtilis cells were cultured at 37 °C in LB containing appropriate antibiotic(s). Samples were withdrawn from 3-mL cultures at an A600 (OD600) of 0.5–1.0 for NEM reactivity assay, β-gal assay, or Western blotting. Growth conditions for the growth complementation assay are described in SI Experimental Procedures.
NEM Reactivity Assay, β-Gal Assay, and Western Blotting.
NEM and Mal-PEG modifications were carried out as described in SI Experimental Procedures. Efficiency of NEM modification of SpoIIIJ was calculated by the equation NEM modification (%) = 100 × (a − b)/a (26), where a is the PEGylation efficiency obtained without the NEM treatment of the cell and b is the PEGylation efficiency obtained from NEM-treated cells. The PEGylation efficiency (%) was calculated by the formula 100 × (i0 − i)/i0, in which i0 and i represent the intensity of SpoIIIJ at the non-PEGylated position, the former before PEGylation and the latter after PEGylation. We used the decrease in the band intensity of non-PEGylated species after Mal-PEG treatment (without taking the band intensity of the PEGylated species into account) because the PEGylated proteins were heterogeneous in size and low in transfer efficiency upon blotting. β-Gal activity assays (10) and Western blotting (12) using anti-SpoIIIJ antiserum (21) were performed as described previously.
Supplementary Material
Acknowledgments
We thank Chika Tsutsumi, Tomoe Takino, and Sayuri Shikata for technical support and Akiko Nakashima for secretarial assistance. This work was supported by the Grants-in-Aid for Scientific Research provided by MEXT (Ministry of Education, Culture, Sports, Science and Technology) and by Japan Society for the Promotion of Science [Grants 26116008, 25291006, and 24657095 (to S.C.); Grant 20247020 (to K.I.); Grants 26119007 and 26291023 (to T.T.); and Grant 24227004 (to O.N.)], as well as by the Private University Strategic Research Foundation Support Program from the MEXT [Grant S1219 (to K.I.)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423817112/-/DCSupplemental.
References
- 1.Pohlschröder M, Hartmann E, Hand NJ, Dilks K, Haddad A. Diversity and evolution of protein translocation. Annu Rev Microbiol. 2005;59:91–111. doi: 10.1146/annurev.micro.59.030804.121353. [DOI] [PubMed] [Google Scholar]
- 2.Park E, Rapoport TA. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biophys. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- 3.du Plessis DJ, Nouwen N, Driessen AJ. The Sec translocase. Biochim Biophys Acta. 2011;1808(3):851–865. doi: 10.1016/j.bbamem.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 4.Dalbey RE, Wang P, Kuhn A. Assembly of bacterial inner membrane proteins. Annu Rev Biochem. 2011;80:161–187. doi: 10.1146/annurev-biochem-060409-092524. [DOI] [PubMed] [Google Scholar]
- 5.Wang P, Dalbey RE. Inserting membrane proteins: The YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim Biophys Acta. 2011;1808(3):866–875. doi: 10.1016/j.bbamem.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Xie K, Dalbey RE. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat Rev Microbiol. 2008;6(3):234–244. doi: 10.1038/nrmicro3595. [DOI] [PubMed] [Google Scholar]
- 7.Dalbey RE, Kuhn A, Zhu L, Kiefer D. The membrane insertase YidC. Biochim Biophys Acta. 2014;1843(8):1489–1496. doi: 10.1016/j.bbamcr.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 8.Murakami T, Haga K, Takeuchi M, Sato T. Analysis of the Bacillus subtilis spoIIIJ gene and its Paralogue gene, yqjG. J Bacteriol. 2002;184(7):1998–2004. doi: 10.1128/JB.184.7.1998-2004.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tjalsma H, Bron S, van Dijl JM. Complementary impact of paralogous Oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J Biol Chem. 2003;278(18):15622–15632. doi: 10.1074/jbc.M301205200. [DOI] [PubMed] [Google Scholar]
- 10.Rubio A, Jiang X, Pogliano K. Localization of translocation complex components in Bacillus subtilis: Enrichment of the signal recognition particle receptor at early sporulation septa. J Bacteriol. 2005;187(14):5000–5002. doi: 10.1128/JB.187.14.5000-5002.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiba S, Lamsa A, Pogliano K. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J. 2009;28(22):3461–3475. doi: 10.1038/emboj.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiba S, Ito K. MifM monitors total YidC activities of Bacillus subtilis, including that of YidC2, the target of regulation. J Bacteriol. 2015;197(1):99–107. doi: 10.1128/JB.02074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiba S, Ito K. Multisite ribosomal stalling: A unique mode of regulatory nascent chain action revealed for MifM. Mol Cell. 2012;47(6):863–872. doi: 10.1016/j.molcel.2012.06.034. [DOI] [PubMed] [Google Scholar]
- 14.Chiba S, et al. Recruitment of a species-specific translational arrest module to monitor different cellular processes. Proc Natl Acad Sci USA. 2011;108(15):6073–6078. doi: 10.1073/pnas.1018343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427(6969):36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 16.Tsukazaki T, et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455(7215):988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455(7215):936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler R, et al. YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Mol Cell. 2009;34(3):344–353. doi: 10.1016/j.molcel.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 19.Seitl I, Wickles S, Beckmann R, Kuhn A, Kiefer D. The C-terminal regions of YidC from Rhodopirellula baltica and Oceanicaulis alexandrii bind to ribosomes and partially substitute for SRP receptor function in Escherichia coli. Mol Microbiol. 2014;91(2):408–421. doi: 10.1111/mmi.12465. [DOI] [PubMed] [Google Scholar]
- 20.Wickles S, et al. A structural model of the active ribosome-bound membrane protein insertase YidC. eLife. 2014;3:e03035. doi: 10.7554/eLife.03035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumazaki K, et al. Structural basis of Sec-independent membrane protein insertion by YidC. Nature. 2014;509(7501):516–520. doi: 10.1038/nature13167. [DOI] [PubMed] [Google Scholar]
- 22.Kumazaki K, et al. Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci Rep. 2014;4:7299. doi: 10.1038/srep07299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura-Someya T, Iwaki S, Yamaguchi A. Site-directed chemical modification of cysteine-scanning mutants as to transmembrane segment II and its flanking regions of the Tn10-encoded metal-tetracycline/H+ antiporter reveals a transmembrane water-filled channel. J Biol Chem. 1998;273(49):32806–32811. doi: 10.1074/jbc.273.49.32806. [DOI] [PubMed] [Google Scholar]
- 24.Kaback HR, et al. Site-directed alkylation and the alternating access model for LacY. Proc Natl Acad Sci USA. 2007;104(2):491–494. doi: 10.1073/pnas.0609968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogdanov M, Zhang W, Xie J, Dowhan W. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM(TM)): Application to lipid-specific membrane protein topogenesis. Methods. 2005;36(2):148–171. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maegawa S, Koide K, Ito K, Akiyama Y. The intramembrane active site of GlpG, an E. coli rhomboid protease, is accessible to water and hydrolyses an extramembrane peptide bond of substrates. Mol Microbiol. 2007;64(2):435–447. doi: 10.1111/j.1365-2958.2007.05679.x. [DOI] [PubMed] [Google Scholar]
- 27.Kedrov A, et al. Elucidating the native architecture of the YidC: ribosome complex. J Mol Biol. 2013;425(22):4112–4124. doi: 10.1016/j.jmb.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 28.White SH. Membrane protein insertion: The biology-physics nexus. J Gen Physiol. 2007;129(5):363–369. doi: 10.1085/jgp.200709741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Contreras FX, Ernst AM, Wieland F, Brügger B. 2011. Specificity of intramembrane protein-lipid interactions. Cold Spring Harb Perspect Biol 3(6). pii: a004705.
- 30.Saint-Georges Y, Hamel P, Lemaire C, Dujardin G. Role of positively charged transmembrane segments in the insertion and assembly of mitochondrial inner-membrane proteins. Proc Natl Acad Sci USA. 2001;98(24):13814–13819. doi: 10.1073/pnas.251503098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price CE, Driessen AJ. Conserved negative charges in the transmembrane segments of subunit K of the NADH:ubiquinone oxidoreductase determine its dependence on YidC for membrane insertion. J Biol Chem. 2010;285(6):3575–3581. doi: 10.1074/jbc.M109.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Funes S, et al. Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc Natl Acad Sci USA. 2009;106(16):6656–6661. doi: 10.1073/pnas.0809951106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia L, et al. Yeast Oxa1 interacts with mitochondrial ribosomes: The importance of the C-terminal region of Oxa1. EMBO J. 2003;22(24):6438–6447. doi: 10.1093/emboj/cdg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 2003;22(24):6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis NE, et al. A dynamic cpSRP43-Albino3 interaction mediates translocase regulation of chloroplast signal recognition particle (cpSRP)-targeting components. J Biol Chem. 2010;285(44):34220–34230. doi: 10.1074/jbc.M110.160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Errington J, et al. Structure and function of the spoIIIJ gene of Bacillus subtilis: A vegetatively expressed gene that is essential for sigma G activity at an intermediate stage of sporulation. J Gen Microbiol. 1992;138(12):2609–2618. doi: 10.1099/00221287-138-12-2609. [DOI] [PubMed] [Google Scholar]
- 37.Saller MJ, et al. Bacillus subtilis YqjG is required for genetic competence development. Proteomics. 2011;11(2):270–282. doi: 10.1002/pmic.201000435. [DOI] [PubMed] [Google Scholar]
- 38.Gray AN, et al. 2011. Unbalanced charge distribution as a determinant for dependence of a subset of Escherichia coli membrane proteins on the membrane insertase YidC. MBio 2(6). pii: e00238-e11.
- 39.Chen Y, Soman R, Shanmugam SK, Kuhn A, Dalbey RE. The role of the strictly conserved positively charged residue differs among the Gram-positive, Gram-negative, and chloroplast YidC homologs. J Biol Chem. 2014;289(51):35656–35667. doi: 10.1074/jbc.M114.595082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.