Significance
There is an urgent need for new antibiotics active against resistant bacterial pathogens like Pseudomonas aeruginosa. Target-directed drug development provides a potential path to such drugs, and essential gene products represent potential targets. Accordingly, the work reported here defines a highly verified set of such functions for P. aeruginosa required for growth under a variety of different conditions.
Keywords: Tn-seq, ESKAPE, cystic fibrosis, antibiotic target, sputum
Abstract
The essential functions of a bacterial pathogen reflect the most basic processes required for its viability and growth, and represent potential therapeutic targets. Most screens for essential genes have assayed a single condition—growth in a rich undefined medium—and thus have not distinguished genes that are generally essential from those that are specific to this particular condition. To help define these classes for Pseudomonas aeruginosa, we identified genes required for growth on six different media, including a medium made from cystic fibrosis patient sputum. The analysis used the Tn-seq circle method to achieve high genome coverage and analyzed more than 1,000,000 unique insertion positions (an average of one insertion every 6.0 bp). We identified 352 general and 199 condition-specific essential genes. A subset of assignments was verified in individual strains with regulated expression alleles. The profile of essential genes revealed that, compared with Escherichia coli, P. aeruginosa is highly vulnerable to mutations disrupting central carbon-energy metabolism and reactive oxygen defenses. These vulnerabilities may arise from the stripped-down architecture of the organism’s carbohydrate utilization pathways and its reliance on respiration for energy generation. The essential function profile thus provides fundamental insights into P. aeruginosa physiology as well as identifying candidate targets for new antibacterial agents.
There is a pressing need for new drugs to treat bacterial infections, particularly those caused by pathogens frequently resistant to multiple antibiotics (1, 2). One of these organisms, the Gram-negative bacterium Pseudomonas aeruginosa, has emerged as a central threat in healthcare settings and can cause a variety of infections, including: acute pneumonia, bacteremia and wound, urinary tract, intra-abdominal, and chronic airway infections (3). Mortality rates for some of these infections can exceed 60%. Accordingly, P. aeruginosa is one of six pathogens for which new antibacterial agents are most desperately needed (1).
One approach to identify new drugs active against a pathogen like P. aeruginosa is to target processes required for its growth and survival (4). Such processes depend on the organism’s essential gene set. Both general and growth condition-specific essential genes can be distinguished. General essential genes are required under virtually all growth conditions, whereas condition-specific essential genes are required under a subset of conditions. General essential functions can be further subdivided into those that are required in all organisms for fundamental processes like DNA replication, and those more limited in phylogenetic distribution, which are more likely to be important for the particular lifestyle of an organism. General essential functions represent potential drug targets, although essentiality is only one of several features that characterize good targets (5–7).
Essential genes have been identified at genome scale using several approaches (8). Targeted deletion mutagenesis was used to define a group of about 300 Escherichia coli genes essential for growth on nutrient medium (9). This group has been refined and validated, and serves as a high quality reference set (10). An alternative method, sequencing of saturation-level pools of transposon mutants (“Tn-seq”), has become popular for essential gene screening because of its technical ease (11–15). However, most Tn-seq studies have included only limited confirmation of findings using individual mutants (8).
Three previous studies have identified P. aeruginosa essential genes. Two projects creating ordered transposon mutant libraries helped define such genes by exclusion, and the results were combined to specify candidate essential genes conserved in the strains analyzed (16, 17). More recent work identified genes essential for growth of strain PA14 at high genome coverage using Tn-seq. (18). All three studies examined bacteria grown only on Luria-Bertani (LB) nutrient agar and did not include follow-up verifications.
In the study reported here, we defined P. aeruginosa genes essential for growth under multiple conditions. The analysis used Tn-seq and achieved greater genome coverage than reached previously for any species. The present effort identified 352 general essential genes and 199 conditionally essential genes. These results provide insights into central elements of P. aeruginosa physiology and reveal vulnerabilities that could potentially be exploited therapeutically.
Results and Discussion
Identifying Essential Genes.
We sought to identify both general and growth medium-specific essential genes of P. aeruginosa. To do this, we generated 13 fully independent, saturation-level transposon mutant pools on six different media and characterized them using Tn-seq. (15). To achieve high insertion density, we used a transposon with low insertion specificity derived from Tn5, and to limit false assignments caused by polarity, the transposon carried an outward facing promoter designed to express downstream genes (16, 19). Essential genes were defined as those with few or no insertions (SI Materials and Methods).
We analyzed three media in depth: LB nutrient agar, MOPS-pyruvate agar, and media made from cystic fibrosis sputa. We chose LB because it is a standard rich medium promoting rapid growth, and because it enables a direct comparison with the previous essentiality studies. We expected LB to be the most permissive medium, but this turned out not to be true. MOPS-pyruvate was used as a minimal medium to provide maximum nutritional contrast with LB. Sputum medium was used because it corresponds to the bacterial growth medium in vivo in cystic fibrosis infections, in which bacteria reach high cell densities. The makeup of mutant pools selected on each medium was defined using the Tn-seq circle method (15), with 49,000–253,000 unique insertion sites identified per pool (Table 1). Technical and biological replicate assays gave reproducible results (Fig. S1). Unique insertions at 1,043,276 sites were detected in the analysis, a genome average of one unique insertion per 6.0 bp.
Table 1.
Tn-seq assays
| Mutant pool | Tn-seq run | Colonies pooled | Mapped reads | Unique insertion sites | Essential coding genes |
| LB-1 | 1 | 139,100 | 2,303,025 | 94114 | 468 |
| 2 | 139,100 | 13,436,777 | 88202 | 485 | |
| 3 | 139,100 | 8,997,565 | 63546 | 561 | |
| LB-2 | 1 | 106,400 | 14,904,269 | 81677 | 429 |
| 2 | 106,400 | 18,640,542 | 81550 | 495 | |
| LB-3 | 1 | 172,800 | 24,322,071 | 109951 | 467 |
| Minimal-1 | 1 | 240,650 | 28,214,377 | 193993 | 421 |
| Minimal-2 | 1 | 312,000 | 13,427,239 | 205614 | 406 |
| Minimal-3 | 1 | 285,800 | 5,160,085 | 252948 | 450 |
| Sputum-1 | 1 | 76,400 | 5,661,970 | 79723 | 446 |
| Sputum-2 | 1 | 144,000 | 7,630,737 | 181477 | 378 |
| Sputum-3 | 1 | 46,800 | 14,522,500 | 48635 | 564 |
| Sputum-4 | 1 | 96,600 | 2,992,340 | 91843 | 469 |
| 0.1X LB | 1 | 180,800 | 8,736,912 | 121357 | 423 |
| Human Serum | 1 | 155,400 | 754,804 | 127933 | 488 |
| BHI | 1 | 216,000 | 4,439,772 | 174631 | 483 |
| Total | 2,172,750 | 1,043,276* |
The Tn-seq results for 16 experiments corresponding to 13 independent mutant pools are summarized. The essential coding genes for each run were combined to identify consensus essentials for each media type.
Corresponds to one insertion every 6.0 bp.
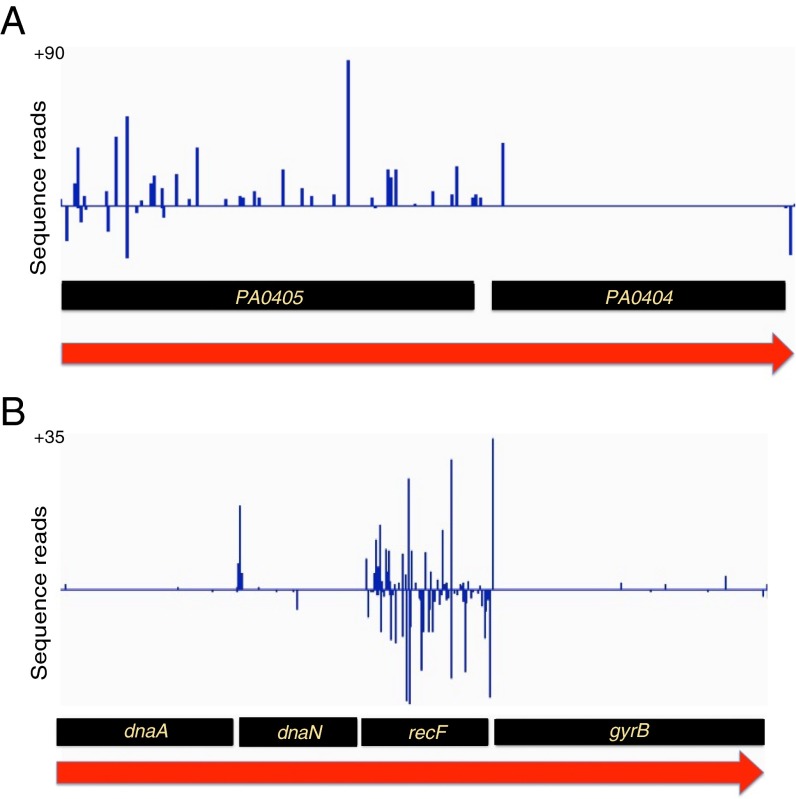
Two representative transposon insertion patterns help illustrate how essential genes were identified (Fig. 1). Fig. 1A shows an operon with two genes (20). The downstream gene on the right (PA0404) lacks insertions except at the very 5′ end and is assumed to be essential. The upstream gene on the left (PA0405) has a high density of insertions and is considered nonessential. Note that the orientation of the insertions in PA0405 is highly biased (bars above the line) such that the transposon outward-facing promoter should promote expression of PA0404. The forward-oriented insertions in PA0405 apparently circumvent polarity on PA0404 expression through the activity of the promoter. Fig. 1B shows an operon with three essential genes (dnaA, dnaN, and gyrB) and one nonessential gene (recF). In this case, insertions in recF were not polar on gyrB, regardless of orientation. This result suggests that a previously unrecognized secondary promoter exists for the gyrB gene. Even long operons of essential genes tolerated insertions in the appropriate orientation between genes (Fig. S2), indicating that polar effects were not a significant source of false essential assignments in this study.
Fig. 1.
Transposon insertions in two regions of the genome. The distributions of insertions in two predicted operons are shown. The height of each bar shows the Tn-seq read number at the corresponding position, with the placement above or below the line corresponding to the transposon orientation. (A) PA0405-PA0404 operon; (B) dnaA-dnaN-recF-gyrB operon.
Note that although the four essential genes shown in Fig. 1 were nearly entirely lacking insertions, they were not totally devoid of them. We assume that the exceptional insertions were not inactivating: for example, because of insertion in one copy of a transiently duplicated gene or to the presence of a suppressor mutation in the target cell (21).
Histograms of the number of insertion sequencing reads per annotated protein-coding gene identified well-separated groups corresponding to genes without, and with, insertions for each Tn-seq run (Fig. S3). Candidate essential genes for a given run were defined as those falling significantly below the main peak on the right (SI Materials and Methods). Genes falling in this region for more than half of the independent mutant pools prepared per condition were considered to be confirmed essentials.
To include additional genes with essential domains only, or essential genes that were absent from the genome annotation (20), we searched independently of the annotation for genome regions lacking insertions (SI Materials and Methods and Dataset S1) (13). We found 22 protein-coding genes with essential domains and one gene that had not previously been recognized in the annotation (PA0728.1) (see below). In addition, the analysis identified 18 candidate essential regions that overlap possible small regulatory RNA coding sequences (22) (Dataset S1).
General and Condition-Specific Essential Genes.
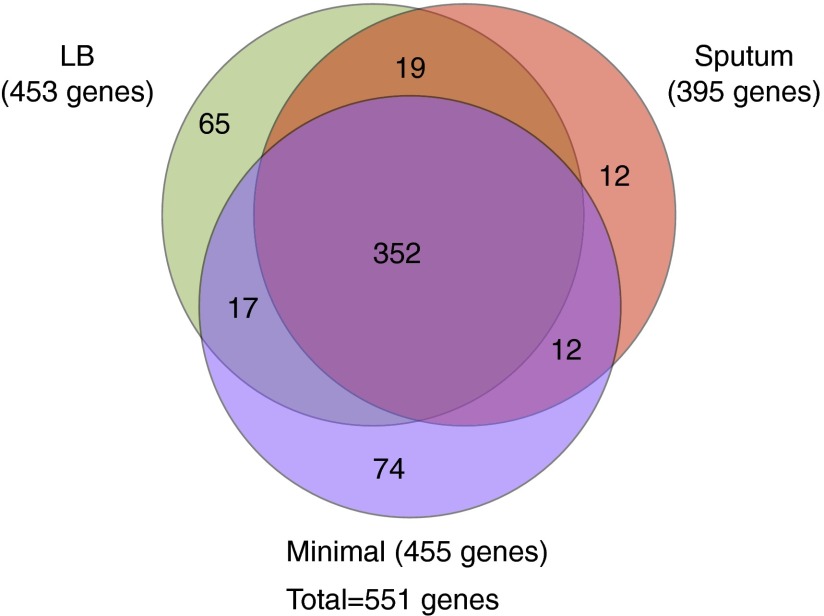
For the three primary growth conditions analyzed, we identified consensus sets of confirmed essential genes (Dataset S1). We found that 352 genes were required on all three media, and refer to these as “general” essential genes (Fig. 2). An additional 199 genes were essential under a subset of the conditions and are called “condition-specific” essentials. Depending on the medium, 11–23% of essential genes were condition-specific.
Fig. 2.
Overlap among genes essential under different growth conditions. The distribution of 551 genes found to be essential under at least one of the three growth conditions examined in depth is shown.
The distribution of the general essential genes in functional categories was typical of that found in previous studies (8), with “translation, post-translational modification, and degradation” being most enriched (Table S1). Among the LB-specific essentials, there was an unexpected enrichment of genes in the “cell wall/LPS/capsule” category, which traced primarily to genes needed for LPS O-antigen synthesis (see below). Genes of unknown function were common in both general and condition-specific classes of essentials.
Nearly all of the general and condition-specific essential genes (99% and 90%, respectively) belong to the P. aeruginosa core genome (23) (Dataset S1). Two of the exceptional general essential genes absent from the core genome (imm2 and PA0728.1) encode functions that block the expression or action of toxic proteins that are encoded by the corresponding strains (discussed below).
Comparison with previous studies.
Three previous reports identified candidate essential genes for P. aeruginosa growth on LB medium (16–18). Comparison with our general essentials identifies 141 genes found in all four studies (Dataset S1). This overlap group represents a highly confirmed set of genes, but appears to be a severe underestimate based on its small number (8), and because it lacks many genes generally assumed to be essential, like the replication gene dnaB, the RNA polymerase gene rpoC, and the secretion apparatus gene secE.
We also compared the P. aeruginosa essential genes to the highly verified E. coli set (10). There are 270 P. aeruginosa orthologs of the 304 E. coli genes essential for growth on nutrient agar (24). Nearly all (87%) of the P. aeruginosa orthologs were essential for growth on LB (Table 2). This group corresponds to about half (48%) of all of the P. aeruginosa genes essential on LB. We assume that the remainder of the genes in the set consists of P. aeruginosa-specific essentials, unrecognized homologs, and any false assignments that may have been included. The general essential set of P. aeruginosa genes also provided good coverage of the orthologs of the E. coli essentials (85%), with fewer additional genes (35% of the total). Two previous lists of P. aeruginosa essential genes provided somewhat lower coverage with higher fractions of additional genes. The results imply that orthologs of most E. coli essential genes are also essential in P. aeruginosa, and that such genes are nearly all generally essential.
Table 2.
Comparison of P. aeruginosa and E. coli essential genes
| Essential genes (present study) | Essential genes (previous studies) | |||
| Category | LB | General | LB (17) | LB (18) |
| Strain | PAO1 | PAO1 | PA14/ PAO1* | PA14 |
| Candidate essential genes | 453 | 352 | 335 | 634† |
| Coverage of essential E.coli orthologs | 0.87 (236/270) | 0.85 (229/270) | 0.52 (139/269) | 0.58 (156/269) |
| Additional PA genes (fraction of total) | 217 (0.48) | 123 (0.35) | 196 (0.59) | 478 (0.75) |
Additional media.
Genes classified as general essentials based on the three primary media should be required for growth on most or all additional media. As a test of this, we carried out Tn-seq analyses of individual mutant pools generated on three additional media: another high nutrient medium (brain-heart infusion, BHI), a dilute nutrient medium with supplements (0.1 × LB), and medium made from human serum (SI Materials and Methods). We found that 97–100% of the general essential genes were also required on these media (Dataset S1), strongly supporting their assignments.
Mutants with regulated expression of individual genes.
A potential limitation of Tn-seq screening for essential genes is that because mutants grown in competition are analyzed, slow-growing as well as nongrowing mutants may be lost from the population and their genes counted as essential (15). As a test of the extent to which this is true, we constructed strains with regulated versions of a sample of general essential genes. The strains were constructed by chromosomal integration of plasmids carrying inducible promoters with different expression ranges (E. coli araBAD and trp-lac) (SI Materials and Methods). Of 46 operons examined (corresponding to 62 essential genes), 43 led to inducer-stimulated growth and 25 showed nearly complete inducer-dependence (Table S2). We assume that some of the strains may show only partial inducer-dependence because of residual expression in the absence of inducer. The results indicate that most of the general essential genes are required for optimal growth of individual colonies, and that at least half are strongly required.
Phylogenetically widespread essential functions.
The 551 essential genes we identified underlie diverse biological processes. About two-thirds of the P. aeruginosa general essential genes correspond to genes essential in E. coli and most other bacteria (8). These genes underlie the most fundamental cellular processes: DNA replication, transcription and translation, protein folding and export, RNA degradation, peptidoglycan synthesis, and cofactor, nucleotide, and lipid synthesis (Table S1). The functions represent potential broad host range antibiotic targets.
Less-widespread functions.
A number of essential genes of P. aeruginosa are not essential in E. coli. Two processes in which such genes are conspicuous are central carbon-energy metabolism and protection from reactive oxygen damage.
Central carbon-energy metabolism.
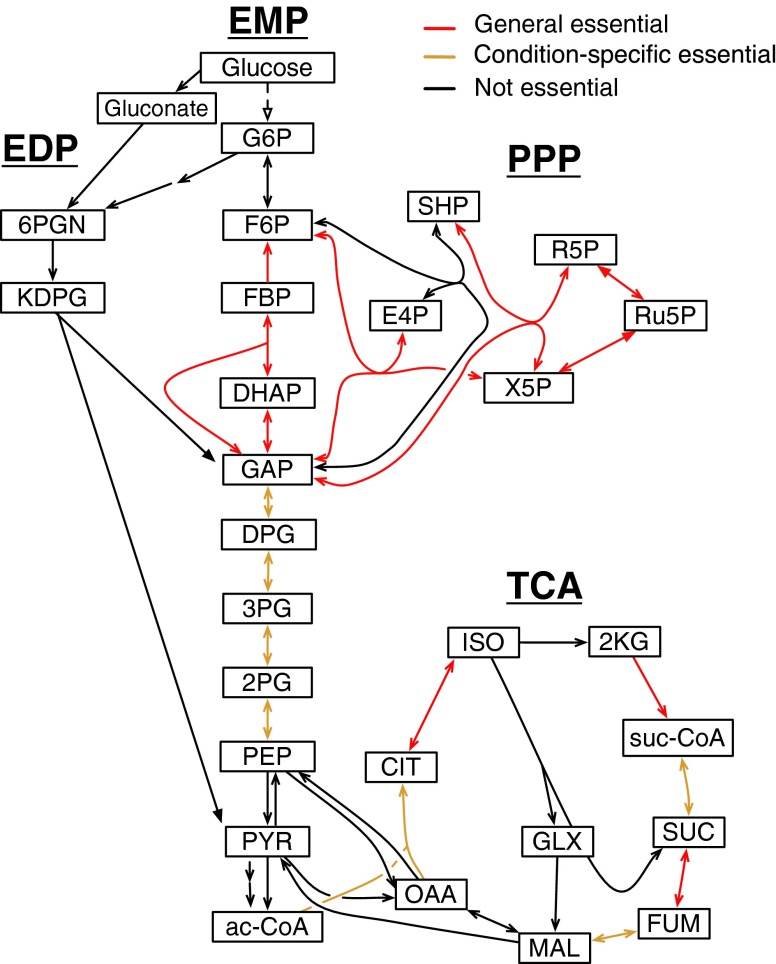
Sixteen steps of central carbon metabolism in P. aeruginosa are essential under multiple growth conditions, with eight essential under all of the conditions examined (Table S3). The essential functions are found in glycolysis/gluconeogenesis (Embden–Meyerhof pathway, EMP), the pentose phosphate shunt (pentose phosphate pathway, PPP) and the tricarboxylic acid cycle (TCA) (Fig. 3). Only four of the genes in these pathways are essential in E. coli for growth on LB. The difference between the two species reflects their remarkably different central carbon metabolisms.
Fig. 3.
Essential functions of central carbon metabolism. The essentialities of different steps of the EDP, glycolysis (EMP), PPP, and TCA are indicated. Some biochemical steps may be essential but not detected because of genetic redundancy: pyruvate kinase (pyrA, pyrK), malate oxidoreductase (mqoA, mqoB, PA1252), pyruvate carboxylase (PA1400, PA5435-PA5436), and isocitrate dehydrogenase (icd, idh).
E. coli metabolizes glucose and other sugars by three pathways: glycolysis (EMP), the PPP, and the Entner–Doudoroff pathway (EDP) (25). In P. aeruginosa, glycolysis and the upper (oxidative) branch of the PPP are not available because of the absence of key enzymes (phosphofructokinase and phosphogluconate dehydrogenase) (26, 27). In addition, P. aeruginosa has a highly active glucose oxidation pathway that limits direct glucose phosphorylation by glucokinase under aerobic growth conditions. As a result of these differences, sugars are metabolized in P. aeruginosa primarily by the EDP (26, 27). Accordingly, essential intermediates of the EMP and PPP, such as fructose-6-phosphate and ribose-5-phosphate, must be made by gluconeogenesis and the lower (nonoxidative) branch of the PPP. Thus, in P. aeruginosa, in contrast to E. coli, there are only single—rather than double—routes of biosynthesis available for such intermediates. The constraints introduced by this architecture apparently make upper EMP and PPP pathway functions, such as fructose bisphosphate phosphatase and transketolase, which are dispensable for E. coli, essential in P. aeruginosa.
P. aeruginosa relies mainly on respiration for energy, with very limited fermentation capability (28). This reliance helps explain the essentiality of most steps of the TCA, because the TCA is the primary source of reducing equivalents for electron transport. The finding that not all TCA steps are essential may be rationalized by known metabolic redundancies (Fig. 3). In addition, the essentiality of aconitase (AcnB) on all primary media, but citrate synthase only on MOPS-pyruvate, seems paradoxical but may be because of secondary activity of 2-methylcitrate synthase (29).
Its orientation to respiration for ATP production probably also explains why, unlike E. coli, the P. aeruginosa proton ATPase genes (PA5554–PA5560) are general essentials. E. coli ATPase mutants are viable because they can grow on substrates supporting fermentative ATP production, bypassing the need for oxidative phosphorylation (30). Cytochrome oxidase genes are not essential for aerobic growth of P. aeruginosa, presumably because of the multiplicity of such functions (31). Remarkably, however, three general essential genes (PA2951– PA2953) were identified whose (mitochondrial) homologs function in respiration to transfer electrons from dehydrogenases to the ubiquinone pool (32, 33). In addition, a putative cytochrome c1 (PA4429) is essential for growth on LB.
Reactive oxygen defense.
Several P. aeruginosa functions that protect cells from superoxide and hydrogen peroxide are essential. One of two cytoplasmic superoxide dismutases (SodB) is a general essential under our conditions, consistent with the slow-growth phenotype of mutants characterized previously (34). Other essential functions may protect cells from reactive oxygen species (ROS) through repair of damaged Fe-S clusters (PA0759, YfgZ), replenishing NADPH pools depleted by superoxide (ferredoxin-NADPH reductase, Fpr) and controlling free iron levels in the cytoplasm (ferric uptake regulator, Fur) (35–37). E. coli mutations in the corresponding genes increase sensitivity to ROS, but are not lethal under unstressed growth conditions (although inactivation of both cytoplasmic superoxide dismutases is lethal) (38).
Two factors that may contribute to the high sensitivity of P. aeruginosa to loss of ROS defenses are the organism’s production of redox active phenazines and the essentiality of iron-containing proteins sensitive to inactivation by ROS. Phenazines are toxic to organisms ranging from other bacteria to metazoans, and it may be that an important role of superoxide defenses is to protect P. aeruginosa from its own poison. Fe-S–dependent dehydratases and mononuclear iron enzymes are key targets of superoxide and hydrogen peroxide (38). Several such enzymes of central carbon metabolism are essential in P. aeruginosa: aconitase (AcnB, PA1787) and fumarase (PA4333) of the TCA, and ribulose-5-phosphate 3-epimerase of the PPP (39). An additional target of reactive oxygen is the Isc Fe-S cluster synthesis system (40), which is essential in P. aeruginosa. The corresponding system in E. coli is not essential because it has a backup system (Suf); inactivation of both systems is lethal (41). Other targets of reactive oxygen that are essential, such as peptide deformylase (PA0019) and Fur (PA4764), presumably also contribute to the P. aeruginosa dependence on reactive oxygen defense functions.
Other Essential Processes.
Lipopolysaccharide synthesis.
As in E. coli and other Gram-negative bacteria, genes required for production of lipid A and KDO (3-deoy-d-manno-octulosonic acid) are essential in P. aeruginosa. However, inner-core heptose and outer-core galactosamine are also generally essential in P. aeruginosa (42).
LPS outer-core rhamnose and O-antigen were essential on LB nutrient agar. Previous studies have found that loss of O-antigen can reduce PAO1 growth because it leads to sensitivity to a potent self-produced bacteriocin (pyocin R2) (43). To test whether this mechanism explained our finding, we constructed three mutants deleted of O-antigen biosynthetic genes in pyocin-producing and nonproducing strains. In the pyocin+ genetic background, all of the mutations severely inhibited growth on LB agar, confirming the Tn-seq findings (Fig. S4). However, in a strain background deleted of pyocin R2 genes, the O-antigen mutations had little effect on growth. Thus, it appears that O-antigen was indeed essential in our screens because it protects cells from the pyocin. Because pyocin R2 orthologs are found in many P. aeruginosa strains, this mechanism of outer core and O-antigen essentiality may also be widespread (44).
Antitoxin and pyocin repression genes.
Our analysis identified a general essential gene absent from the PAO1 genome annotation that appears to encode the antitoxin member of a ParDE-like toxin-antitoxin pair. The new gene (PA0728.1) is situated in the phage Pf1 region and presumably contributes to maintenance of the phage genome. Two additional essential genes (PA0125 and PA4674) also encode putative antitoxins of RelBE- and HigBA-like toxin-antitoxin pairs (45), and two others inhibit pyocin expression (prtR) or activity (imm2). In the absence of any of these inhibitory functions, toxin or pyocin activity presumably kills producer cells (43, 46, 47).
Asparagine-tRNAAsn formation.
The GatABC complex generates Asn-tRNAAsn by transamidation of Asp-tRNAAsn, an essential reaction that compensates for the absence of an asparginyl-tRNA synthetase in P. aeruginosa (48). The process is not essential in mammals (49) and represents a potential drug target.
Medium-specific essentials.
A number of processes were essential on a subset of the media examined and help identify differences in growth physiology on them.
Growth on nutrient media (LB and BHI).
Mutants lost from nutrient media may be sensitive to components of the media. For example, both a sodium-proton antiporter (PA1054-1059) and a membrane protease (FtsH), essential only on LB and BHI, are probably necessary for bacteria to cope with the NaCl in the media (50, 51). Glycerol-3-phosphate dehydrogenase (GlpD) may be essential because toxic glycerol-3-phosphate made from glycerol in the nutrient media accumulates in its absence (52).
Other genes selectively essential on nutrient media may be needed for rapid growth and division. For example, the Rep and PriA helicases may be essential during rapid growth to resolve replication-transcription conflicts (53). A 16S rRNA methylase (RsmH) that acts at the decoding center may be needed for translational fidelity at high growth rates (54), and a highly conserved GTPase (ObgE) may be essential to modulate the stringent response to a level commensurate with rapid growth (55).
Growth on minimal medium (MOPS-pyruvate).
Nearly all of the genes specifically required for growth on MOPS-pyruvate medium are annotated as amino acid, nucleotide, and cofactor biosynthetic functions, and our findings help confirm these assignments. An additional function strongly essential on minimal medium was RpoN, an RNA polymerase σ-factor involved in nitrogen assimilation and other processes (56).
Growth on sputum.
Several functions associated with outer-membrane integrity and synthesis were specifically essential for sputum growth. These functions included the abundant outer-membrane OprI lipoprotein covalently attached to murein that helps stabilize the cell envelope (57) and the function attaching it to murein (PA2854). An ortholog of outer-membrane chaperone protein Skp (PA3647) was also specifically essential on sputum medium (58). The global translational regulator RsmA (59) was required for growth on sputum and LB, but not on minimal medium.
Conversely, most genes for amino acid and nucleotide biosynthesis were not essential on sputum. The result fits well with studies showing that cystic fibrosis patient sputum is rich in free amino acids (60). Genes needed for unsaturated fatty acid biosynthesis (fabA, fabB, PA5174, and fabV/PA2950) were essential on minimal and LB but not sputum medium, suggesting that sputum contains unsaturated fatty acids. Unsaturated fatty acid synthesis has been proposed as an antimicrobial drug target, but this finding suggests that its inhibition in P. aeruginosa may not be effective for treating cystic fibrosis pulmonary infections (61).
Concluding Remarks.
The principle goal of this work was to specify gene functions that are essential in different growth media, and may thus have the greatest potential as antibiotic targets in diverse conditions. To identify such functions, we screened for genes required for P. aeruginosa growth using Tn-seq and then identified the subset required on all of the media. This group of genes is likely to represent the most fundamental processes needed for P. aeruginosa viability and growth.
The principle advantage of using Tn-seq to identify essential genes is that it makes it possible to carry out a genomic-level analysis in a single experiment. The analysis of multiple independent mutant pools grown under a given condition provides statistically robust essential gene assignments. In addition, the analysis is not necessarily tied to a genome annotation and can identify new genes, as was done here. Tn-seq also has two significant limitations. First, Tn-seq has a limited capacity to distinguish mutants growing very slowly from those not growing at all. Thus, confirmatory experiments with individual mutants are crucial to verify Tn-seq findings. Second, because Tn-seq assays the fitness of mutants in pools, mutants whose phenotypes are “complemented” by secreted factors from other bacteria may be missed.
The identification of essential genes provides an important starting point for further tests to distinguish the most promising functions for further development as targets. Examples of useful tests are examining whether partial loss of a function inhibits growth and whether a function is essential during infection (6, 7). Strains with conditional expression of different essential genes should allow such attributes to be systematically evaluated.
Materials and Methods
Strains and Plasmid.
The P. aeruginosa parent strain was MPAO1 (16). The donor strain for transposon mutagenesis was E. coli SM10λpir/pIT2. Plasmid pIT2 is a suicide plasmid that carries Tn5 derivative ISlacZhah-Tc (16).
Transposon Mutagenesis.
Transposon mutagenesis was carried out by conjugation of P. aeruginosa with E. coli SM10λpir/pIT2, followed by selection for P. aeruginosa derivatives carrying the transposon tetracycline-resistance determinant (16). Donor E. coli cells (SM10λpir/pIT2) in midexponential growth (OD600 0.8–1.0) were mixed with overnight cultures of MPAO1 grown without agitation at 42 °C (OD600 1.1–1.4). Cultures were mixed 1:1 and pelleted, then resuspended in 10 mM MgSO4, spotted onto 0.45-µm Millipore filters, placed on LB agar, and incubated for 90 min at 37 °C. Mating mixtures were then suspended in either LB or MOPS-pyruvate medium, DMSO was added to 5% (vol/vol) final concentration and cell suspensions flash-frozen in a dry ice-ethanol bath.
Tn-seq.
The makeup of transposon mutant pools was profiled using the Tn-seq circle method (15). DNA from mutant pools was isolated using DNeasy Blood & Tissue Kit, (Qiagen No. 69506), and 8–18 µg of DNA per pool was sheared to a target size of 300 bp (Covaris E210 focused-ultrasonicator, duty cycle 10%, intensity 5, 100 cycles per burst, duration of 250 s per tube), end-repaired (New England Biolabs Next End Repair Module), and dA-tailed (New England Biolabs Klenow), followed by Illumina adaptor attachment (New England Biolabs Quick Ligation Kit). DNA was purified following the end-repair, A-tailing and adaptor ligation steps using AMPure XP beads (Agencourt, No. A63881), with the beads carried through and recharged for the final two purifications by addition of PEG 8,000 + 2.5 M NaCl. Restriction enzyme digestion, size selection, circularization, and exonuclease treatment were carried out as described previously (15). The transposon–genome junctions were amplified from the circularized DNA fragments by real-time PCR (Bio-Rad Mini Opticon) in 50-µL reactions containing 3-µL template DNA, 25 µL 2× KAPA HiFi HotStart Readymix, 0.125 µL 100× SYBR Green dye (Life Technologies), and 300 nM each of primers T8_SLXA_PAIR_AmpF_1 and SLXA_PAIR_REV_AMP. Thermocycling was 95 °C for 3 min, then 33 cycles of 95 °C for 20 s, 66 °C for 15 s, and 72 °C for 30 s. Samples were removed at 50% maximum amplification and purified (QIAgen MinElute Kit), then sequenced on Illumina Genome Analyzer II or MiSeq platforms using sequencing primers T8_SEQ_G or T8_INDEX_1. Sequence data analysis (SI Materials and Methods) included gap size analysis (Fig. S5).
Nucleotide Accession Numbers.
The Tn-seq sequence reads have been deposited in the Sequence Read Archive (SRA) (www.ncbi.nlm.nih.gov/sra) under accession number SRP052838.
Supplementary Material
Acknowledgments
We thank Mike Vasil, Paul Phibbs, and Pat Secor for helpful discussions, and Elizabeth Ramage for experimental contributions. This work was supported by grants from the Cystic Fibrosis Foundation (MANOIL08GO) and the National Institutes of Health (NIH) (1 R21 AI105898). P.K.S. was supported by grants from the NIH (R01HL110879, R01AI101307, and K24 HL102246), European Commission (Seventh Framework Programme Project 603038 CFMatters), and the Burroughs Wellcome Fund. B.J.S. was supported by the Cystic Fibrosis Foundation LeRoy Matthews Physician Scientist award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Tn-seq sequence reads have been deposited in the Sequence Read Archive (SRA) (www.ncbi.nlm.nih.gov/sra) under accession number SRP052838.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422186112/-/DCSupplemental.
References
- 1.Rice LB. Progress and challenges in implementing the research on ESKAPE pathogens. Infect Control Hosp Epidemiol. 2010;31(Suppl 1):S7–S10. doi: 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- 2.Moir DT, Opperman TJ, Butler MM, Bowlin TL. New classes of antibiotics. Curr Opin Pharmacol. 2012;12(5):535–544. doi: 10.1016/j.coph.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Page MG, Heim J. Prospects for the next anti-Pseudomonas drug. Curr Opin Pharmacol. 2009;9(5):558–565. doi: 10.1016/j.coph.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Juhas M, Eberl L, Church GM. Essential genes as antimicrobial targets and cornerstones of synthetic biology. Trends Biotechnol. 2012;30(11):601–607. doi: 10.1016/j.tibtech.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Gwynn MN, Portnoy A, Rittenhouse SF, Payne DJ. Challenges of antibacterial discovery revisited. Ann N Y Acad Sci. 2010;1213:5–19. doi: 10.1111/j.1749-6632.2010.05828.x. [DOI] [PubMed] [Google Scholar]
- 6.Murima P, McKinney JD, Pethe K. Targeting bacterial central metabolism for drug development. Chem Biol. 2014;21(11):1423–1432. doi: 10.1016/j.chembiol.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 7.Projan SJ. Whither antibacterial drug discovery? Drug Discov Today. 2008;13(7-8):279–280. doi: 10.1016/j.drudis.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Luo H, Lin Y, Gao F, Zhang CT, Zhang R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014;42(Database issue):D574–D580. doi: 10.1093/nar/gkt1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Rudd KE. EcoGene 3.0. Nucleic Acids Res. 2013;41(Database issue):D613–D624. doi: 10.1093/nar/gks1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barquist L, Boinett CJ, Cain AK. Approaches to querying bacterial genomes with transposon-insertion sequencing. RNA Biol. 2013;10(7):1161–1169. doi: 10.4161/rna.24765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Opijnen T, Camilli A. Transposon insertion sequencing: A new tool for systems-level analysis of microorganisms. Nat Rev Microbiol. 2013;11(7):435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christen B, et al. The essential genome of a bacterium. Mol Syst Biol. 2011;7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barquist L, et al. A comparison of dense transposon insertion libraries in the Salmonella serovars Typhi and Typhimurium. Nucleic Acids Res. 2013;41(8):4549–4564. doi: 10.1093/nar/gkt148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher LA, Shendure J, Manoil C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. MBio. 2011;2(1):e00315–e10. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs MA, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2003;100(24):14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liberati NT, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA. 2006;103(8):2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skurnik D, et al. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog. 2013;9(9):e1003582. doi: 10.1371/journal.ppat.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oberhardt MA, Puchałka J, Fryer KE, Martins dos Santos VA, Papin JA. Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. J Bacteriol. 2008;190(8):2790–2803. doi: 10.1128/JB.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winsor GL, et al. Pseudomonas Genome Database: Improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2011;39(Database issue):D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reams AB, Kofoid E, Savageau M, Roth JR. Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics. 2010;184(4):1077–1094. doi: 10.1534/genetics.109.111963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gómez-Lozano M, Marvig RL, Molin S, Long KS. Genome-wide identification of novel small RNAs in Pseudomonas aeruginosa. Environ Microbiol. 2012;14(8):2006–2016. doi: 10.1111/j.1462-2920.2012.02759.x. [DOI] [PubMed] [Google Scholar]
- 23.Ozer EA, Allen JP, Hauser AR. Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genomics. 2014;15:737. doi: 10.1186/1471-2164-15-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whiteside MD, Winsor GL, Laird MR, Brinkman FS. OrtholugeDB: A bacterial and archaeal orthology resource for improved comparative genomic analysis. Nucleic Acids Res. 2013;41(Database issue):D366–D376. doi: 10.1093/nar/gks1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fraenkel DG. Mutants in glucose metabolism. Annu Rev Biochem. 1986;55:317–337. doi: 10.1146/annurev.bi.55.070186.001533. [DOI] [PubMed] [Google Scholar]
- 26.Lessie TG, Phibbs PV., Jr Alternative pathways of carbohydrate utilization in pseudomonads. Annu Rev Microbiol. 1984;38:359–388. doi: 10.1146/annurev.mi.38.100184.002043. [DOI] [PubMed] [Google Scholar]
- 27.Temple LM, Sage AE, Schweizer HP, Phibbs PV. Carbohydrate catabolism in Pseudomonas aeruginosa. In: Montie T, editor. Pseudomonas. Plenum; New York: 1998. pp. 35–72. [Google Scholar]
- 28.Vander Wauven C, Piérard A, Kley-Raymann M, Haas D. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: Evidence for a four-gene cluster encoding the arginine deiminase pathway. J Bacteriol. 1984;160(3):928–934. doi: 10.1128/jb.160.3.928-934.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerike U, Hough DW, Russell NJ, Dyall-Smith ML, Danson MJ. Citrate synthase and 2-methylcitrate synthase: Structural, functional and evolutionary relationships. Microbiology. 1998;144(Pt 4):929–935. doi: 10.1099/00221287-144-4-929. [DOI] [PubMed] [Google Scholar]
- 30.Futai M, Kanazawa H. Structure and function of proton-translocating adenosine triphosphatase (F0F1): Biochemical and molecular biological approaches. Microbiol Rev. 1983;47(3):285–312. doi: 10.1128/mr.47.3.285-312.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arai H, et al. Enzymatic characterization and in vivo function of five terminal oxidases in Pseudomonas aeruginosa. J Bacteriol. 2014;196(24):4206–4215. doi: 10.1128/JB.02176-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai MH, Saier MH., Jr Phylogenetic characterization of the ubiquitous electron transfer flavoprotein families ETF-alpha and ETF-beta. Res Microbiol. 1995;146(5):397–404. doi: 10.1016/0923-2508(96)80285-3. [DOI] [PubMed] [Google Scholar]
- 33.Watmough NJ, Frerman FE. The electron transfer flavoprotein: ubiquinone oxidoreductases. Biochim Biophys Acta. 2010;1797(12):1910–1916. doi: 10.1016/j.bbabio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Hassett DJ, Schweizer HP, Ohman DE. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177(22):6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waller JC, et al. Evidence that the folate-dependent proteins YgfZ and MnmEG have opposing effects on growth and on activity of the iron-sulfur enzyme MiaB. J Bacteriol. 2012;194(2):362–367. doi: 10.1128/JB.06226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giró M, Carrillo N, Krapp AR. Glucose-6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli. Microbiology. 2006;152(Pt 4):1119–1128. doi: 10.1099/mic.0.28612-0. [DOI] [PubMed] [Google Scholar]
- 37.Barton HA, Johnson Z, Cox CD, Vasil AI, Vasil ML. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol Microbiol. 1996;21(5):1001–1017. doi: 10.1046/j.1365-2958.1996.381426.x. [DOI] [PubMed] [Google Scholar]
- 38.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat Rev Microbiol. 2013;11(7):443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobota JM, Imlay JA. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc Natl Acad Sci USA. 2011;108(13):5402–5407. doi: 10.1073/pnas.1100410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nachin L, Loiseau L, Expert D, Barras F. SufC: An unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 2003;22(3):427–437. doi: 10.1093/emboj/cdg061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277(32):28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 42.Lam JS, Taylor VL, Islam ST, Hao Y, Kocincova D. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front Microbiol. 2011;2:118. doi: 10.3389/fmicb.2011.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Penterman J, et al. Rapid evolution of culture-impaired bacteria during adaptation to biofilm growth. Cell Reports. 2014;6(2):293–300. doi: 10.1016/j.celrep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakayama K, et al. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol. 2000;38(2):213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 45.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33(3):966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Penterman J, Singh PK, Walker GC. Biological cost of pyocin production during the SOS response in Pseudomonas aeruginosa. J Bacteriol. 2014;196(18):3351–3359. doi: 10.1128/JB.01889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sano Y, Matsui H, Kobayashi M, Kageyama M. Molecular structures and functions of pyocins S1 and S2 in Pseudomonas aeruginosa. J Bacteriol. 1993;175(10):2907–2916. doi: 10.1128/jb.175.10.2907-2916.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akochy PM, Bernard D, Roy PH, Lapointe J. Direct glutaminyl-tRNA biosynthesis and indirect asparaginyl-tRNA biosynthesis in Pseudomonas aeruginosa PAO1. J Bacteriol. 2004;186(3):767–776. doi: 10.1128/JB.186.3.767-776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheppard K, et al. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36(6):1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kosono S, et al. Characterization of a multigene-encoded sodium/hydrogen antiporter (sha) from Pseudomonas aeruginosa: Its involvement in pathogenesis. J Bacteriol. 2005;187(15):5242–5248. doi: 10.1128/JB.187.15.5242-5248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hinz A, Lee S, Jacoby K, Manoil C. Membrane proteases and aminoglycoside antibiotic resistance. J Bacteriol. 2011;193(18):4790–4797. doi: 10.1128/JB.05133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cozzarelli NR, Koch JP, Hayashi S, Lin EC. Growth stasis by accumulated L-alpha-glycerophosphate in Escherichia coli. J Bacteriol. 1965;90(5):1325–1329. doi: 10.1128/jb.90.5.1325-1329.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merrikh H, Zhang Y, Grossman AD, Wang JD. Replication-transcription conflicts in bacteria. Nat Rev Microbiol. 2012;10(7):449–458. doi: 10.1038/nrmicro2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kimura S, Suzuki T. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2010;38(4):1341–1352. doi: 10.1093/nar/gkp1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raskin DM, Judson N, Mekalanos JJ. Regulation of the stringent response is the essential function of the conserved bacterial G protein CgtA in Vibrio cholerae. Proc Natl Acad Sci USA. 2007;104(11):4636–4641. doi: 10.1073/pnas.0611650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Potvin E, Sanschagrin F, Levesque RC. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev. 2008;32(1):38–55. doi: 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- 57.Wessel AK, Liew J, Kwon T, Marcotte EM, Whiteley M. Role of Pseudomonas aeruginosa peptidoglycan-associated outer membrane proteins in vesicle formation. J Bacteriol. 2013;195(2):213–219. doi: 10.1128/JB.01253-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goemans C, Denoncin K, Collet JF. Folding mechanisms of periplasmic proteins. Biochim Biophys Acta. 2014;1843(8):1517–1528. doi: 10.1016/j.bbamcr.2013.10.014. [DOI] [PubMed] [Google Scholar]
- 59.Brencic A, Lory S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol Microbiol. 2009;72(3):612–632. doi: 10.1111/j.1365-2958.2009.06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palmer KL, Aye LM, Whiteley M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol. 2007;189(22):8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schweizer HP, Choi KH. Characterization of molecular mechanisms controlling fabAB transcription in Pseudomonas aeruginosa. PLoS ONE. 2012;7(10):e45646. doi: 10.1371/journal.pone.0045646. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.