Significance
The limb skeletal elements that have unique morphology and distinct locations are developed from limb progenitors, derived from the lateral plate mesoderm. These skeletal elements arise during limb development. In this study, we show genetic evidence that function of Sall4 is essential prior to limb outgrowth for development of the anterior-proximal skeletal elements. Furthermore, genetic interaction between Sall4 and Gli3 is upstream of establishing Shh (Sonic hedgehog) expression, and therefore, Shh-dependent posterior skeletal elements. Our study identified early requirements of the Sall4-Gli3 system for two putative progenitor pools that develop into distinct sets of limb skeletal elements.
Keywords: Sall4, Gli3, limb progenitors, appendicular skeletal elements, Plzf-Hox
Abstract
Limb skeletal elements originate from the limb progenitor cells, which undergo expansion and patterning to develop each skeletal element. Posterior-distal skeletal elements, such as the ulna/fibula and posterior digits develop in a Sonic hedgehog (Shh)-dependent manner. However, it is poorly understood how anterior-proximal elements, such as the humerus/femur, the radius/tibia and the anterior digits, are developed. Here we show that the zinc finger factors Sall4 and Gli3 cooperate for proper development of the anterior-proximal skeletal elements and also function upstream of Shh-dependent posterior skeletal element development. Conditional inactivation of Sall4 in the mesoderm before limb outgrowth caused severe defects in the anterior-proximal skeletal elements in the hindlimb. We found that Gli3 expression is reduced in Sall4 mutant hindlimbs, but not in forelimbs. This reduction caused posteriorization of nascent hindlimb buds, which is correlated with a loss of anterior digits. In proximal development, Sall4 integrates Gli3 and the Plzf-Hox system, in addition to proliferative expansion of cells in the mesenchymal core of nascent hindlimb buds. Whereas forelimbs developed normally in Sall4 mutants, further genetic analysis identified that the Sall4-Gli3 system is a common regulator of the early limb progenitor cells in both forelimbs and hindlimbs. The Sall4-Gli3 system also functions upstream of the Shh-expressing ZPA and the Fgf8-expressing AER in fore- and hindlimbs. Therefore, our study identified a critical role of the Sall4-Gli3 system at the early steps of limb development for proper development of the appendicular skeletal elements.
How progenitor cells are spatially and temporarily organized to construct an organ is a central question in developmental biology. Limb skeletal elements develop from limb progenitors, which arise from the lateral plate mesoderm (LPM) that is originated from epithelial somatopleure (1). Limb progenitors initially form two paired protrusions, fore- and hindlimb buds, whose initiation occurs around embryonic day (E) 9.0 and E9.5, respectively, in mouse embryos. In the following steps, limb signaling centers, known as the zone of polarizing activity (ZPA) and apical ectodermal ridge (AER), are established. SHH (Sonic hedgehog) from the ZPA and FGF8 from the AER are major signal molecules that regulate proliferative expansion and patterning of early limb progenitor cells (reviewed in ref. 2). These processes lead to development of functional limbs with each skeletal element adopting a unique shape at a distinct location.
Several studies suggest that limb progenitors consist of two distinct pools, an anterior progenitor pool and a posterior progenitor pool. The posterior progenitor pool consists of cells that once expressed Shh and cells that received paracrine effects of SHH, which contribute to digit 2 (d2), d3, d4, and d5 and the posterior zeugopod (ulna, fibula) (3–6). Contrary to this, the anterior progenitors are not well characterized. However, a recent report suggested that the putative anterior progenitor pool contributes to d1 and anterior-proximal skeletal elements (tibia, femur) in hindlimbs. Inactivating Irx3 and Irx5 (Irx3/5), two homeodomain encoding genes expressed in the LPM and the anterior-proximal domain of developing fore- and hindlimb buds, resulted in failure to develop these anterior-proximal skeletons in hindlimbs, whereas forelimbs developed normally (7). The lack of the anterior-proximal hindlimb skeleton was observed only when Irx3/5 were inactivated before hindlimb initiation, suggesting that the putative anterior-progenitor pool is specified before hindlimb outgrowth.
In addition, limb anterior genes appear to have a critical role in establishing limb signaling centers. One of factors involved in this process is a zinc finger factor GLI3 (8). Gli3 functions for specifying anterior-posterior polarity in the nascent limb buds (9), and Gli3 null embryos develop polydactyly without polarity (10). Furthermore, simultaneous inactivation of Gli3 and Irx3/5 caused failure to express Shh and significantly reduced levels of Fgf8, which resulted in severe limb truncation (11). This result suggests that cooperation of anterior genes is upstream of establishing limb signaling centers, and therefore expansion of SHH-dependent posterior progenitors. This phenotype was observed only in the hindlimbs, although Irx3/5 and Gli3 are similarly expressed in both fore- and hindlimb buds. Despite these recent reports, factors that regulate the putative anterior progenitors are not well understood. Moreover, it is unknown whether similar or distinct mechanisms involving anterior genes regulate limb signaling center establishment in fore- and hindlimbs.
Sall genes encode zinc finger transcription factors that are vertebrate homologs of the Drosophila homeotic gene, spalt. They play diverse roles in embryonic development, including the development of limbs (12, 13). Among four Sall genes in mammals, Sall4 is a key regulator of stemness in stem cells and progenitor cells, such as embryonic stem cells, induced pluripotent stem cells, spermatogenial progenitor cells and cancer cells (14–19). Despite its robust expression during limb development, functions of Sall4 in limb development are not understood well. A previous report suggested a role of Sall4-Tbx5 interaction in most-anterior digit (d1) development in forelimbs (20). However, this study used embryos heterozygous for the Sall4 gene trap allele (Sall4GT), which exhibited a different phenotype from Sall4+/− mice (15), indicating that Sall4GT allele wound not be a simple loss-of-function model. Moreover, Sall4 null embryos die at periimplantation stages (15), excluding the possibility of identifying Sall4 functions in postimplantation stages. We show that inactivation of Sall4 in the mesoendoderm before limb outgrowth causes defects of proximal-anterior skeletal elements specifically in hindlimbs. We provide genetic evidence that Sall4 and Gli3 interact and regulate establishment of Shh-expression, and this regulation is common in fore- and hindlimb buds. Therefore, the Sall4-Gli3 system is an early and critical regulator acting on limb progenitor cells.
Results
Sall4 Is Required Before Outgrowth for Development of the Anterior-Proximal Skeletal Elements in Hindlimbs.
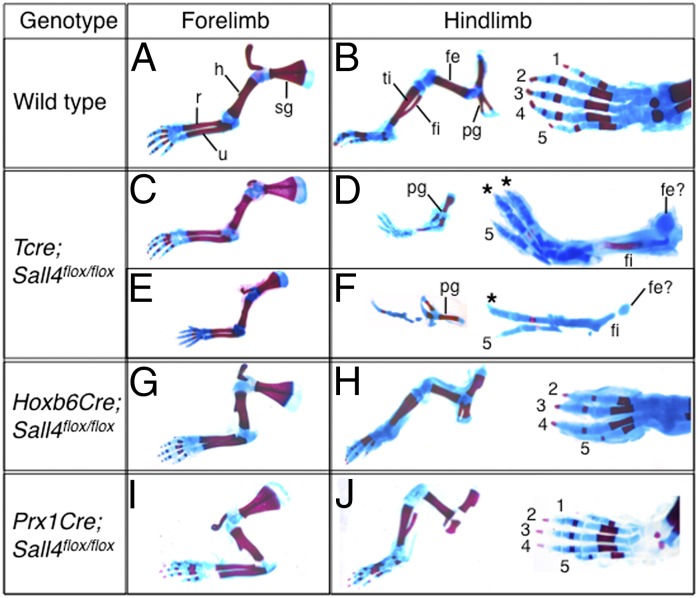
Sall4 is broadly expressed before limb outgrowth and is expressed in the developing limb bud (Fig. S1 A–C). To investigate the role of Sall4 in limb development, we conditionally inactivated Sall4 using three Cre deleters, with which recombination occurs at different timings (Fig. S2). Recombination by Tcre occurs in mesoendoderm lineages at E7.5, much earlier than limb outgrowth (21), and Sall4 transcripts were undetectable by E8.5 (Fig. S1D). Neonatal mutants (Tcre; Sall4flox/flox) exhibited severe skeletal defects specifically in hindlimbs (Fig. 1 A–F). In the stylopod, the femur was not formed, instead, a small cartilage aggregate was present (Fig. 1 D and F). In the zeugopod the tibia was missing, whereas the fibula developed with variable length. In the autopod, two to three digits were present in most of mutants (Table S1, 60.5% and 28.9%, respectively). Based on the morphology, the missing digits include the anterior d1, which is consistent with the expression of Hoxd12 and Hoxd13 in the entire autopod region of Tcre; Sall4 CKO hindlimbs (Fig. S3), whereas the remaining digits include d5 and the identity of other digits was difficult to determine. These skeletal defects were observed in both left and right hindlimbs.
Fig. 1.
Sall4 is required for development of the anterior-proximal skeletal elements in a temporally restricted manner. (A–J) Appendicular skeletal preparations of control (A and B) and Tcre; Sall4 CKO (C–F), Hoxb6Cre; Sall4 CKO (G and H) and Prx1Cre; Sall4 CKO (I and J) neonatal mice. (A, C, E, G, and I) Forelimbs develop without significant defects in mutants with size reduction in severely affected Tcre; Sall4 mutants. (D and F) In Tcre; Sall4 hindlimbs a small cartilage aggregate was present instead of the femur (fe). The fibula (fi) was present, but the tibia (ti) was missing. The pelvic girdle (pg) was small, but patterned normally. (H) In Hoxb6Cre; Sall4 CKO hindlimbs, d1 was missing in the autopod. (J) Hindlimbs of Prx1Cre; Sall4 CKO developed normally. fe, femur; fi, fibula; h, humerus; pg, pelvic girdle; r, radius; ti, tibia; u, ulna. Digits are numbered as 1–5. Asterisks mark digits whose identity is not completely determined.
A second Cre line, Hoxb6Cre, recombines in the LPM around E9.0 (22). Approximately half of the mutants (neonatal and >E16.5) exhibited milder hindlimb skeletal defects (Fig. 1 G and H and Table S1). The Hoxb6Cre; Sall4flox/flox mutants exhibited four (50%) or three (7.7%) digits. All mutants with reduced digit numbers developed the tibia, except for two hindlimbs (Fig. S4 A–C and Table S2). The expression pattern of Hoxd12 and Hoxd13 is consistent with the loss of d1 (Fig. S3). The loss of d1 has been reported in Hoxa13 mutants (23); however, Hoxa13 was detected in both Tcre; Sall4 and Hoxb6Cre; Sall4 CKO hindlimbs (Fig. S3), indicating that the loss of d1 is not caused by loss of Hoxa13 expression. A third deleter, Prx1Cre recombines at the time of limb initiation in the forelimbs and slightly after the onset of outgrowth in hindlimbs (24). All neonatal mutants exhibited normal skeletons in fore- and hindlimbs (Fig. 1 I and J).
These results indicate that Sall4 is required specifically for the development of the proximal and anterior hindlimb skeletons. Although forelimbs develop largely normal in the absence of Sall4, some severely affected mutants with tail truncation exhibited reduced forelimb size (Fig. 1E and Fig. S4 D and E). Tcre-dependent recombination occurs much earlier than that of Hoxb6Cre in the LPM (21, 22). Thus, the difference between the skeletal phenotype by inactivating Sall4 using Tcre and Hoxb6Cre suggests a temporal difference in Sall4 requirement before and after hindlimb initiation. Sall4 seems to act before hindlimb outgrowth for subsequent development of the femur and tibia, and at the early hindlimb outgrowth stage Sall4 seems to be required for anterior digit development. Moreover, development of normal hindlimbs in Prx1Cre; Sall4 mutant suggests that Sall4 is dispensable around E9.75 and later on. A recent study suggested that the anterior-proximal skeletons in hindlimbs are derived from the putative anterior progenitors that express Irx3/5 (7). Therefore, our data suggest that Sall4 functions in the putative anterior progenitors in hindlimbs.
Sall4 Regulates Anterior Genetic Programs for Proper Anterior–Posterior Patterning.
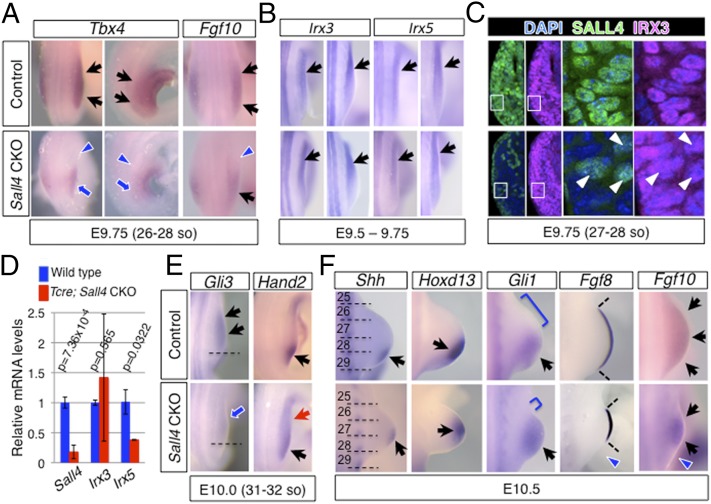
We focused our analyses on Tcre; Sall4flox/flox conditional knockout mutants (hereafter referred to as Sall4 CKO), given that all mutants exhibited strong phenotypes. The Sall4 CKO neonates were obtained at a significantly lower than expected ratio (Table S3), suggesting that a large portion of mutants died in utero. Due to the early requirement of Sall4, we first examined expression of genes that mark hindlimb progenitors at the onset of hindlimb outgrowth (E9.5–9.75). We detected weak expression of Tbx4 and Fgf10 (25, 26), and, in particular, their expression in the anterior portion exhibited significant down-regulation (Fig. 2A). Expression of Irx3/5, whose functions are required for development of the anterior-proximal skeletal elements (7), seemed not to be significantly affected in Sall4 CKO embryos (Fig. 2B). Although quantitative analysis indicated moderate reduction of Irx5 levels, we did not detect alteration of Irx3 levels (Fig. 2D). IRX3 immunoreactivity was also detected similarly in control and Sall4 CKO hindlimb progenitor cells. Costaining with an anti-SALL4 antibody showed that immunoreactive IRX3 signals were similar in wild-type cells, relative to cells that lost SALL4 or cells that escaped from Sall4 inactivation in Sall4 CKO embryos. (Fig. 2C). These results suggest that Sall4 and Irx3/5 might function independently to regulate development of anterior-proximal skeletal elements.
Fig. 2.
Loss of Sall4 caused defects of gene expression in hindlimb progenitors independent of Irx3/5. In situ hybridization (A, B, E, and F), immunofluorescence (C), and quantitative RT-PCR (D) of marker genes/proteins in control and Sall4 CKO hindlimb buds. (A) Dorsal views of Tbx4 and Fgf10, and lateral views of Tbx4 at E9.75. (B) Dorsal views of Irx3 and Irx5 at E9.5-E9.75. (C) Immunofluorescence of SALL4 (green) and IRX3 (magenta) on transverse sections of E9.75 anterior hindlimb buds. White arrowheads point to cells that escaped from Sall4 deletion in Tcre; Sall4 CKO hindlimb buds. (Right) Close up of the boxed areas in Left. (D) Quantitative analysis of Irx3 and Irx5 transcript levels. Relative mRNA levels of indicated genes are shown as average ± SD. Asterisks indicate statistical significance. P values for expression of each gene are included. (E) Dorsal views of Gli3 and Hand2 at E10.0. (F) Dorsal views of Shh, Hoxd13, Gli1, Fgf8 and Fgf10 at E10.5. (A, B, E, and F) Black arrows and blue arrows point to normal and reduced expression, respectively. Red arrows point to ectopic expression. Blue arrowheads denote loss of expression. In D, broken lines mark normal expression border of Gli3. In E, dotted lines mark somite boundaries in Shh expression panels. Brackets in Gli1 expression panels denote anterior domains that are free of Gli1 signals.
We detected down-regulation of Gli3, a critical anterior factor, in nascent hindlimb buds at E10. Consistent with mutual antagonism between Gli3 and Hand2, ectopic expression of Hand2, a posterior factor, was detected in the anterior portion of nascent hindlimb buds (Fig. 2E). HAND2 acts in concert with GLI3 to polarize the nascent limb mesenchyme and control transcriptional networks (27). Thus, the altered Hand2-Gli3 balance in nascent Sall4 CKO hindlimb buds might modify gene network and proliferation rates, which would contribute to Shh expression being shifted to the middle-distal region in hindlimb buds at E10.5 (Fig. 2F). A similar shift was also detected with the expression of Hoxd13, a target of SHH signaling (28) (Fig. 2F). This shift is correlated with anterior expansion of the expression domain of Gli1, a transcriptional target of SHH signaling (8), and with a reduction in size of the Gli1-free anterior domain. Fgf8 expressing AER was narrower and its posterior domain was missing in Sall4 CKO hindlimb buds, which is consistent with the positive feedback loop between the ZPA and AER (29, 30). Accordingly, expression of Fgf10, whose expression in developing limb buds is maintained by FGF8 signaling (26), exhibited a smaller expression domain (Fig. 2F). A similar but smaller degree of reduction of the SHH signaling-free domain was also observed in Sall4 CKO forelimb buds (Fig. S5C). Changes in the balance between Gli3 and Hand2 were subtle in forelimb buds compared with hindlimb buds (Fig. S5 A and B). We also examined whether Sall4 expression was altered in Gli3 mutants, but detected only mild up-regulation in forelimb buds (Fig. S6). The alterations of gene expression in the nascent and developing hindlimb buds in Sall4 CKO embryos support the idea that cells in the anterior portion, which are free of SHH signaling, are reduced in the absence of Sall4. A compatible interpretation is that limb progenitor cells might respond to the new geometry of ZPA and AER, which would cause fewer cells to be specified as anterior digit precursors. Such a reduction of the anterior cells would result in the lack of anterior digits in hindlimbs (Fig. 1) (7).
Sall4 Regulates Proximal Development Through the Plzf-Hox System.
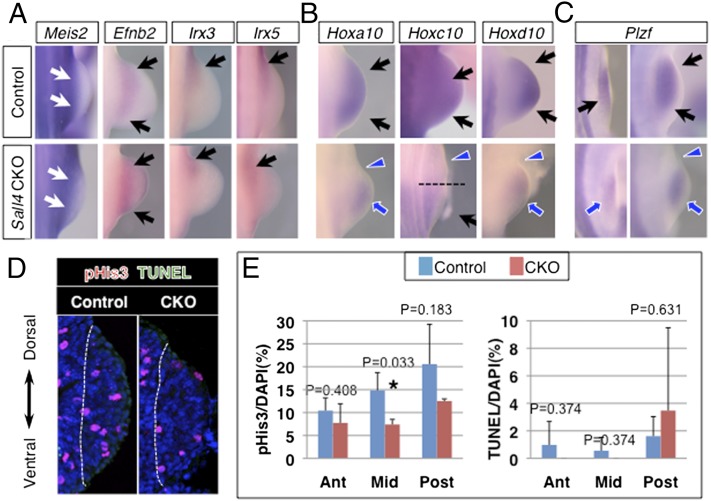
One of the notable phenotypes of Sall4 CKO hindlimbs is the failure to develop the femur. Despite the early defects in the proximal region, proximal marker genes, such as Meis2 and Efnb2, as well as Irx3 and Irx5 (7), were expressed similarly in both control and Sall4 CKO hindlimb buds (Fig. 3A and Fig. S7B). Instead, we observed an alteration in expression of Hox genes, which are regulators of skeletal patterning. More specifically, a genetic study has demonstrated that mice lacking all Hox10 genes (Hoxa10, c10, d10) exhibit a small cartilage condensation in the stylopod, specifically in the hindlimbs, whereas other skeletal elements in fore- and hindlimbs develop (31). In Sall4 CKO hindlimb buds, expression of Hoxa10 and Hoxd10 was undetectable in the proximal-anterior region, and was reduced in other regions (Fig. 3B). Expression of Hoxc10 was down-regulated in the anterior half of the hindlimb bud. These results indicate that Sall4 is an upstream regulator of Hox10 genes, and suggest that the proximal defects in Sall4 CKO hindlimbs involve reduced function of Hox10 genes in the anterior hindlimb bud.
Fig. 3.
Sall4 integrates Plzf-Hox10 system and proliferative expansion of limb progenitors. (A) Expression pattern of Meis2, Efnb2, Irx3, and Irx5 in control and Sall4 CKO hindlimbs at E10.5. (B) Expression pattern of Hoxa10, Hoxc10, and Hoxd10 in control and Sall4 CKO hindlimbs at E10.5. (C) Expression pattern of Plzf in control and Sall4 CKO hindlimbs at E9.75 and E10.5. Black arrows and blue arrows point to normal and reduced expression, respectively. In A, Meis2 expression is indicated by white arrows. Blue arrowheads denote loss of expression. In B, a broken line indicates the anterior boundary of Hoxc10 expression in Sall4 CKO hindlimb buds. (D) Representative images of pHis3 and TUNEL double staining of transverse sections of nascent hindlimb bud at E9.75 in control and Sall4 CKO embryos. Dotted lines indicate boundary between the nascent hindlimb bud and trunk. (E) Quantitative evaluation of cell proliferation and cell death in transverse sections of nascent hindlimb buds at E9.75 in control and Sall4 CKO embryos. P values are included.
Hox expression is known to be regulated by the Plzf zinc finger factor (32). We found that Plzf expression was also down-regulated in the anterior-proximal part of the hindlimb buds at E10.5 (Fig. 3C). This down-regulation was more significant in the nascent hindlimb bud at E10.0. A previous study showed that mice lacking both Gli3 and Plzf developed a small cartilage condensation in the stylopod only in the hindlimbs (33), similar to Sall4 CKO mice. Our results indicate that Sall4 acts upstream of Gli3 and the Plzf-Hox10 system and suggest that the proximal defects in Sall4 CKO hindlimbs are derived from, at least in part, reduced function of Gli3 and the Plzf-Hox10 system in the putative anterior progenitors.
Sall4 is known to regulate proliferation of stem cells (15), thus, we examined whether the proliferation of hindlimb progenitor cells is affected in Sall4 CKO embryos. Through serial section analysis (34), we found that the mesenchymal core of nascent hindlimb buds exhibited a reduced proliferation index (Fig. 3 D and E). In contrast, we did not detect alteration of cell death at E9.75 and E10.5 (Fig. 3 D and E and Fig. S7A). The reduced proliferation was detected only in the mesenchymal core, and cells in the anterior and posterior portions did not exhibit significant difference in cell proliferation. Reduced proliferation is associated with down-regulation of cell cycle progression related genes, such as Ccnd1, Ccne1, and Mycn (Fig. S8 A–L). These results suggest that loss of Sall4 caused failure to expand cells in a discrete region in the nascent hindlimb buds.
Interaction Between Sall4 and Gli3 Regulates Development of the Limb Signaling Centers in both Fore- and Hindlimbs.
A major difference between fore- and hindlimbs of Sall4 CKO mutants is Sall4 regulation of Gli3, which was evident in hindlimb buds but was subtle in forelimb buds (Fig. 2E and Fig. S5). To test whether hindlimb-specific skeletal defects in Sall4 CKO limbs involve reduced Gli3 rather than loss of Sall4 alone, we genetically removed Gli3 from the Sall4 CKO background. Because Sall4 CKO; Gli3+/− embryos did not survive beyond E10.5, we focused our analysis on Shh, whose expression is significantly affected in Sall4 CKO hindlimb buds (Fig. 2F). We also monitored Fgf8 expression, given a recent genetic study, involving Irx3/5 and Gli3, indicated that cooperation of anterior genes is necessary for establishing limb signaling centers, such as Shh-expressing ZPA and Fgf8-expressing AER (11).
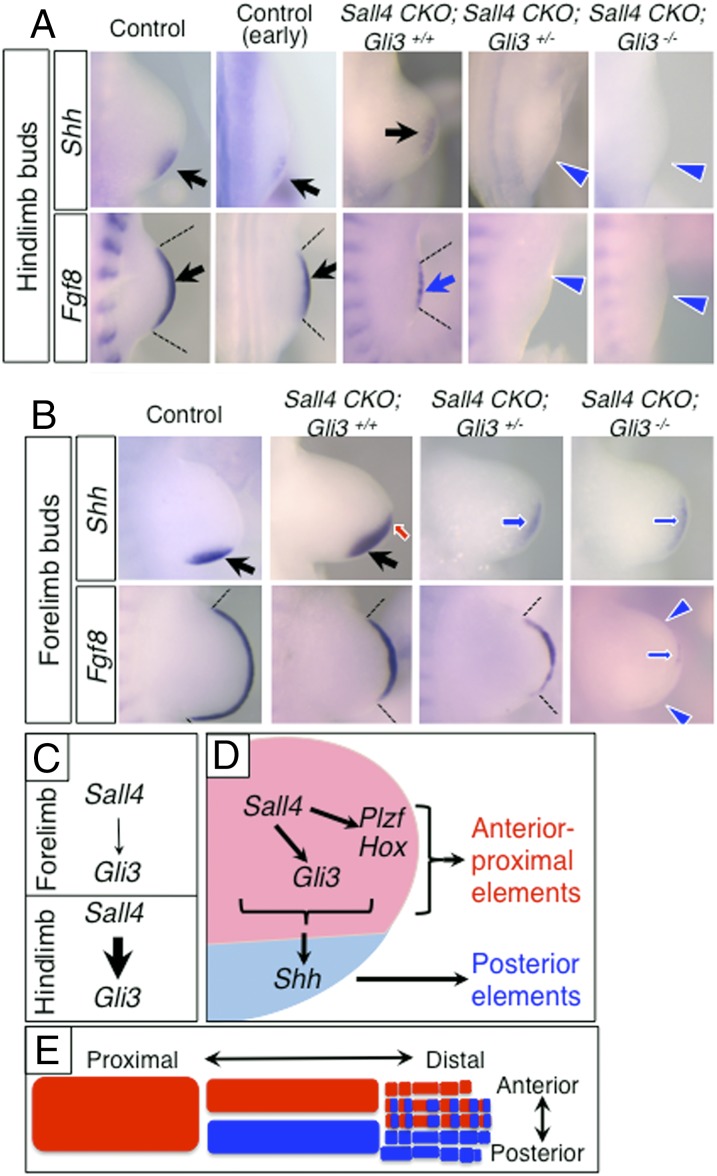
In hindlimbs, removing one allele of Gli3 from the Sall4 CKO background caused a failure to express both Shh and Fgf8 (Fig. 4A), which was also observed in Sall4 CKO; Gli3−/− double KO hindlimbs. Although Sall4 CKO; Gli3 double mutants exhibited smaller hindlimb buds, the lack of Shh and Fgf8 is unlikely simply due to small size of hindlimb buds, because wild-type hindlimbs with similar size (earlier stages) showed evident expression of both genes. Given that Gli3 is down-regulated in Sall4 CKO hindlimb buds, this result suggests that the Sall4 CKO; Gli3+/− genotype caused a condition in the hindlimb similar to the Sall4 CKO; Gli3−/− genotype.
Fig. 4.
Genetic interaction between Sall4 and Gli3 and a model of the role of Sall4-Gli3 system in limb progenitor cells. (A) Expression pattern of Shh and Fgf8 in hindlimbs of indicated genotypes. Due to reduced size of hindlimb buds in Sall4 CKO; Gli3 mutants, wild-type hindlimb buds at an earlier stage with similar size were included. (B) Expression pattern of Fgf8 and Shh in forelimbs of indicated genotypes. Black arrows and blue arrows point to normal and reduced expression, respectively. Red arrows point to ectopic expression. Blue arrowheads denote loss of expression. (C–E) A model of the role of Sall4 in the two-population model for the development of limb skeletal elements. (C) Sall4 regulation of Gli3 differs in fore- and hindlimb progenitor cells. (D) Model of Sall4 regulation of gene network in the putative anterior progenitors (red) and posterior progenitors (blue). (E) Schematic drawing of skeletal elements derived from the putative anterior progenitors (red) and posterior progenitors (blue).
In forelimb buds of Sall4 CKO; Gli3+/− mutants, Shh expression became weak, and its expression domain shifted to the middle-distal region (Fig. 4B). In addition, the Fgf8 expression domain was short. These expression patterns in forelimb buds resembled those of Sall4 CKO hindlimb buds. In Sall4 CKO; Gli3−/− double KO forelimbs, expression of Shh and Fgf8 was further down-regulated and only trace levels of signals were detected. The residual Shh and Fgf8 in Sall4 CKO; Gli3−/− forelimb buds implies that additional anterior factor(s) might participate in signaling center establishment in forelimb buds. Comparison of the expression pattern of Shh and Fgf8 between forelimb and hindlimb buds supports the idea that the hindlimb-specific phenotype in Sall4 CKO mutants involves reduced Gli3 levels. These results demonstrate that genetic interaction between two anterior factors, Sall4 and Gli3, is necessary to establish limb signaling centers in both fore- and hindlimbs. The results also suggest that the difference of Sall4 CKO skeletal phenotypes in fore- and hindlimbs would be derived from differences in Gli3 regulation by Sall4 (Fig. 2E and Fig. S5).
Because SALL4 and GLI3 are both nuclear proteins, they might physically interact. Indeed, we observed both SALL4a (long form) and SALL4b (short form) could interact with GLI3, when transfected in cultured cells (Fig. S8 M and N). This result suggests that SALL4 and GLI3 might directly interact, in addition to genetically, to regulate downstream developmental programs.
Discussion
Proper development of skeletal elements depends on specification, expansion and patterning of limb progenitor cells (35). Genetic experiments have shown that posterior skeletal elements are developed in a Shh-dependent manner (3, 4). However, how anterior skeletal elements are developed has been poorly understood. A recent report demonstrated that anterior progenitor cells, marked by Irx3/5, give rise to the femur, tibia, and d1 in the hindlimb (7). These skeletal elements are missing (d1, tibia) or are present as a small cartilage condensation (femur) in Sall4 CKO hindlimbs, suggesting that Sall4 function is required for the putative anterior progenitors. This phenotype was observed only when Tcre was used to inactivate Sall4, and Hoxb6Cre-dependent Sall4 inactivation only resulted in the lack of d1 in approximately half of the mutants. These different skeletal phenotypes from using Tcre and Hoxb6Cre suggest that the progenitors for the femur and tibia are specified before digits, and this specification occurs before hindlimb outgrowth.
The skeletal phenotype of Tcre; Sall4 CKO mutants are different from a previous study using Sall4+/GT mice, which exhibited elongated d1 in the forelimb autopod (20). The Sall4GT allele would generate N-terminal truncated SALL4, fused with βGeo protein, which could interact with other SALL proteins and interfere with their functions (36). Therefore, the Sall4GT allele would generate complex functional interference of all SALL proteins. In contrast, our approach could address functions specific to Sall4 during limb development.
Our data and a recent report (7) indicate that Sall4 CKO and Irx3/5 mutants share similar skeletal defects and altered gene expression in hindlimbs. We observed comparable expression patterns of Irx3/5 and immunoreactivities of IRX3 in Sall4 CKO embryos at E9.5–9.75, although Irx5 levels showed moderate reduction by qRT-PCR analysis. Given that one allele of Irx3/5 is sufficient for development of the anterior-proximal skeletons in hindlimbs (7), our data suggest that Sall4 and Irx3/5 function independently to regulate development of anterior-proximal skeletal elements. If so, it is conceivable that Sall4 and Irx3/5 share common downstream targets in the putative anterior progenitors. Alternatively, Irx3/5 might function upstream of Sall4 in the anterior progenitors.
Proximal skeletal defects are one of the notable phenotypes of Sall4 CKO hindlimbs. Our analysis suggests that Sall4 integrates Gli3 and the Plzf-Hox system for the development of the proximal skeletal elements. Because these genes are not completely down-regulated, additional mechanism(s) might be involved in the proximal defects. It has been suggested that development of a skeletal element requires expansion of precursors to sufficient levels, otherwise the element fails to form (37, 38). The reduced proliferation of the mesenchymal core of nascent hindlimb buds of Sall4 CKO embryos suggests that, in addition to regulation of Gli3 and the Plzf-Hox system, Sall4 regulation of proliferation of discrete populations of limb progenitors also contributes to the skeletal phenotype.
In the autopod, it has been suggested that the time and concentration that cells are exposed to SHH pattern the digits (3, 4). In this model, posterior d4 and d5 are derived from cells that expressed Shh, and d3 is partially derived from cells that expressed Shh and cells that received paracrine SHH, whereas d2 is derived from cells that received only paracrine SHH. In Sall4 CKO hindlimbs, ∼90% of mutants exhibited an absence of d2 or d2+d3 phenotype, in addition to the absence of d1. This digit phenotype was correlated with reduced Gli1-free domain, which suggested that the putative anterior progenitor-derived cells are reduced. Thus, the failure to expand the putative anterior progenitors in the absence of Sall4 would contribute to the loss of d2 and d3 through failure to provide enough digit precursor cells, even in the presence of precursors derived from the posterior progenitor cells. This idea suggests that development of d2 and d3 requires both the putative anterior progenitors and Shh-dependent posterior progenitors (Fig. 4E).
A recent report showed that cooperation of anterior genes at the beginning of limb development is necessary for establishing limb signaling centers. In particular, simultaneous inactivation of Irx3/5 and Gli3 caused failure to establish a Shh-expressing ZPA, and caused trace Fgf8 expression in the AER. However, this phenotype was hindlimb specific and forelimbs developed polydactyly, similar to Gli3 KO limbs (11). In significant contrast to this report, we found that Sall4-Gli3 cooperation regulates limb signaling center development in both fore- and hindlimbs. As discussed above, Sall4 CKO skeletal defects are specific to hindlimbs and forelimbs developed largely normal. However, this difference between two types of limbs is likely derived from difference in Sall4 regulation of Gli3 (Fig. 4 C and D). Due to reduction of Gli3 expression in Sall4 CKO hindlimb buds, the mutant hindlimb bud would be similar to Sall4 CKO; Gli3+/− condition. This idea is supported by genetic removal of Gli3 from Sall4 CKO background. Due to severe defects to establish expression of Fgf8 and Shh, we speculate that the Sall4-Gli3 system would regulate early genetic systems, including Fgf10 induction and/or its upstream mechanisms, such as expression of Hoxa and Hoxd cluster genes (39) in limb progenitor cells. Such mechanisms are to be investigated in the future to deepen our understanding of how limb progenitor cells are controlled to develop into functional limbs. Nonetheless, our genetic experiments indicate that the Sall4-Gli3 system is a common mechanism to regulate early limb progenitors in fore- and hindlimb buds.
The data presented here and a recent report (11) support the two-population model, in which the anterior and posterior progenitor pools contribute to specific limb skeletal elements, located in the anterior-proximal region and posterior region, respectively (Fig. 4 D and E). In this model the Sall4-Gli3 system is upstream of establishing Shh-expressing ZPA, which is a critical regulator of proliferative expansion of the posterior progenitor pool. Moreover, the Sall4, together with Gli3 and the Plzf-Hox system, regulates the putative anterior progenitor pool. Therefore, our data suggests that the Sall4-Gli3 system is a critical upstream regulator of both progenitor pools, and thus, development of the limb skeletal elements.
Materials and Methods
Mouse Mutants.
The mouse lines for Sall4 (15, 40), Gli3 (41), Tcre (21), Hoxb6Cre (22), and Prx1Cre (24) were maintained on a mixed genetic background. Skeletal preparation was done as published (42). Animal breeding and procedures were performed according to the approval by the Institutional Animal Care and Use Committee of the University of Minnesota.
Immunofluorescence and TUNEL Assay.
Immunofluorescence of IRX3 (43) and SALL4 (Santa Cruz, sc-101147, 1/300 dilution) was performed on cryo-sections according to standard procedures (42). Simultaneous cell proliferation and cell death analyses were performed by using anti-phospho histone H3 (Upstate, catalog no. 06–570) and the In Situ Cell Death Detection kit (Roche) as published (34). Cell death analysis at E10.5 was performed on coronal sections by using the In Situ Cell Death Detection kit. DAPI was used for nuclear staining. Fluorescent confocal images were obtained by using Zeiss LSM 710 laser scanning microscope system (Carl Zeiss Microscopy), and analyzed using ZEN2009 software (Carl Zeiss Microscopy).
Skeletal Preparation and in Situ Hybridization.
Preparation of skeletal staining and in situ mRNA detection were performed according to a standard procedure (42).
Quantitative RT-PCR for Irx3/5.
Relative expression of Irx3/5 in wild-type and Sall4 CKO was determined by isolating RNA from the hindlimb forming region/nascent hindlimb buds at E9.5–9.75 and performing quantitative PCR after reverse transcription. Details are provided in SI Materials and Methods.
Coimmunoprecipitation Assay.
HEK293T cells were transfected with expression constructs by using Polyethylenimine MAX (Polysciences). Cell lysates were prepared after two days and coimmunoprecipitation assay were performed by using protein G-Sepharose (GE Healthcare) and anti-Flag antibody (M2, Sigma). Proteins were resolved by SDS/PAGE, transferred to PVDF membranes (Millipore) and detected by anti-HA antibodies (4B2, Wako), HRP goat anti-mouse IgG, and a chemiluminescence system.
Supplementary Material
Acknowledgments
We thank Drs. Maria Barna, Juan Carlos Izpisua Belmonte, C-c Hui, Michael Kuehn, Mark Lewandoski, Jonathan Licht, Sridhar Rao, Sestuko Sahara, Toshihiko Shiroishi, and Stephanie Ware for sharing plasmids and/or mouse lines. We also thank Susan Morton for IRX3 antibody, Dr. Michael O’Connor for the use of LSM 710, Dr. David Zarkower for critical reading of the manuscript, and Drs. C-c Hui and Sevan Hopyan for sharing unpublished data. We thank Jessica Burlingame, Asha Elogna, Jenna Matson, Thu Quach, and Elizabeth West for their excellent technical support and Steve Pehoski for editorial assistance. This work is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of NIH (R01AR064195).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421949112/-/DCSupplemental.
References
- 1.Gros J, Tabin CJ. Vertebrate limb bud formation is initiated by localized epithelial-to-mesenchymal transition. Science. 2014;343(6176):1253–1256. doi: 10.1126/science.1248228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeller R, López-Ríos J, Zuniga A. Vertebrate limb bud development: Moving towards integrative analysis of organogenesis. Nat Rev Genet. 2009;10(12):845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- 3.Harfe BD, et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118(4):517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118(4):505–516. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 5.Kraus P, Fraidenraich D, Loomis CA. Some distal limb structures develop in mice lacking Sonic hedgehog signaling. Mech Dev. 2001;100(1):45–58. doi: 10.1016/s0925-4773(00)00492-5. [DOI] [PubMed] [Google Scholar]
- 6.Chiang C, et al. Manifestation of the limb prepattern: Limb development in the absence of sonic hedgehog function. Dev Biol. 2001;236(2):421–435. doi: 10.1006/dbio.2001.0346. [DOI] [PubMed] [Google Scholar]
- 7.Li D, et al. Formation of proximal and anterior limb skeleton requires early function of Irx3 and Irx5 and is negatively regulated by Shh signaling. Dev Cell. 2014;29(2):233–240. doi: 10.1016/j.devcel.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 8.Hui CC, Angers S. Gli proteins in development and disease. Annu Rev Cell Dev Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- 9.te Welscher P, Fernandez-Teran M, Ros MA, Zeller R. Mutual genetic antagonism involving GLI3 and dHAND prepatterns the vertebrate limb bud mesenchyme prior to SHH signaling. Genes Dev. 2002;16(4):421–426. doi: 10.1101/gad.219202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hui CC, Joyner AL. A mouse model of greig cephalopolysyndactyly syndrome: The extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat Genet. 1993;3(3):241–246. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- 11.Zhulyn O, et al. A switch from low to high Shh activity regulates establishment of limb progenitors and signaling centers. Dev Cell. 2014;29(2):241–249. doi: 10.1016/j.devcel.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 12.de Celis JF, Barrio R. Regulation and function of Spalt proteins during animal development. Int J Dev Biol. 2009;53(8-10):1385–1398. doi: 10.1387/ijdb.072408jd. [DOI] [PubMed] [Google Scholar]
- 13.Kawakami Y, et al. Sall genes regulate region-specific morphogenesis in the mouse limb by modulating Hox activities. Development. 2009;136(4):585–594. doi: 10.1242/dev.027748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao S, et al. Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol Cell Biol. 2010;30(22):5364–5380. doi: 10.1128/MCB.00419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakaki-Yumoto M, et al. The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development. 2006;133(15):3005–3013. doi: 10.1242/dev.02457. [DOI] [PubMed] [Google Scholar]
- 16.Hobbs RM, et al. Functional antagonism between Sall4 and Plzf defines germline progenitors. Cell Stem Cell. 2012;10(3):284–298. doi: 10.1016/j.stem.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yong KJ, et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med. 2013;368(24):2266–2276. doi: 10.1056/NEJMoa1300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buganim Y, et al. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 2014;15(3):295–309. doi: 10.1016/j.stem.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim CY, et al. Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem Cell. 2008;3(5):543–554. doi: 10.1016/j.stem.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Koshiba-Takeuchi K, et al. Cooperative and antagonistic interactions between Sall4 and Tbx5 pattern the mouse limb and heart. Nat Genet. 2006;38(2):175–183. doi: 10.1038/ng1707. [DOI] [PubMed] [Google Scholar]
- 21.Perantoni AO, et al. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132(17):3859–3871. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- 22.Lowe LA, Yamada S, Kuehn MR. HoxB6-Cre transgenic mice express Cre recombinase in extra-embryonic mesoderm, in lateral plate and limb mesoderm and at the midbrain/hindbrain junction. Genesis. 2000;26(2):118–120. doi: 10.1002/(sici)1526-968x(200002)26:2<118::aid-gene5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 23.Fromental-Ramain C, et al. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development. 1996;122(10):2997–3011. doi: 10.1242/dev.122.10.2997. [DOI] [PubMed] [Google Scholar]
- 24.Logan M, et al. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33(2):77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 25.Chapman DL, et al. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206(4):379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 26.Ohuchi H, et al. The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development. 1997;124(11):2235–2244. doi: 10.1242/dev.124.11.2235. [DOI] [PubMed] [Google Scholar]
- 27.Osterwalder M, et al. HAND2 targets define a network of transcriptional regulators that compartmentalize the early limb bud mesenchyme. Dev Cell. 2014;31(3):345–357. doi: 10.1016/j.devcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75(7):1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 29.Niswander L. Interplay between the molecular signals that control vertebrate limb development. Int J Dev Biol. 2002;46(7):877–881. [PubMed] [Google Scholar]
- 30.Zúñiga A, Haramis AP, McMahon AP, Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401(6753):598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]
- 31.Wellik DM, Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301(5631):363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- 32.Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25(2):166–172. doi: 10.1038/76014. [DOI] [PubMed] [Google Scholar]
- 33.Barna M, Pandolfi PP, Niswander L. Gli3 and Plzf cooperate in proximal limb patterning at early stages of limb development. Nature. 2005;436(7048):277–281. doi: 10.1038/nature03801. [DOI] [PubMed] [Google Scholar]
- 34.Akiyama R, et al. Distinct populations within Isl1 lineages contribute to appendicular and facial skeletogenesis through the β-catenin pathway. Dev Biol. 2014;387(1):37–48. doi: 10.1016/j.ydbio.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mariani FV, Martin GR. Deciphering skeletal patterning: Clues from the limb. Nature. 2003;423(6937):319–325. doi: 10.1038/nature01655. [DOI] [PubMed] [Google Scholar]
- 36.Kiefer SM, et al. Expression of a truncated Sall1 transcriptional repressor is responsible for Townes-Brocks syndrome birth defects. Hum Mol Genet. 2003;12(17):2221–2227. doi: 10.1093/hmg/ddg233. [DOI] [PubMed] [Google Scholar]
- 37.Wolpert L, Tickle C, Sampford M. The effect of cell killing by x-irradiation on pattern formation in the chick limb. J Embryol Exp Morphol. 1979;50:175–193. [PubMed] [Google Scholar]
- 38.Galloway JL, Delgado I, Ros MA, Tabin CJ. A reevaluation of X-irradiation-induced phocomelia and proximodistal limb patterning. Nature. 2009;460(7253):400–404. doi: 10.1038/nature08117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheth R, et al. Decoupling the function of Hox and Shh in developing limb reveals multiple inputs of Hox genes on limb growth. Development. 2013;140(10):2130–2138. doi: 10.1242/dev.089409. [DOI] [PubMed] [Google Scholar]
- 40.Yuri S, et al. Sall4 is essential for stabilization, but not for pluripotency, of embryonic stem cells by repressing aberrant trophectoderm gene expression. Stem Cells. 2009;27(4):796–805. doi: 10.1002/stem.14. [DOI] [PubMed] [Google Scholar]
- 41.Büscher D, Grotewold L, Rüther U. The XtJ allele generates a Gli3 fusion transcript. Mamm Genome. 1998;9(8):676–678. doi: 10.1007/s003359900845. [DOI] [PubMed] [Google Scholar]
- 42.Itou J, et al. Islet1 regulates establishment of the posterior hindlimb field upstream of the Hand2-Shh morphoregulatory gene network in mouse embryos. Development. 2012;139(9):1620–1629. doi: 10.1242/dev.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen JA, et al. Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron. 2011;69(4):721–735. doi: 10.1016/j.neuron.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.