Abstract
The tendency of many species to abandon migration remains a poorly understood aspect of evolutionary biology that may play an important role in promoting species radiation by both allopatric and sympatric mechanisms. Anadromy inherently offers an opportunity for the colonization of freshwater environments, and the shift from an anadromous to a wholly freshwater life history has occurred in many families of fishes. Freshwater-resident forms have arisen repeatedly among lampreys (within the Petromyzontidae and Mordaciidae), and there has been much debate as to whether anadromous lampreys, and their derived freshwater-resident analogues, constitute distinct species or are divergent ecotypes of polymorphic species. Samples of 543 European river lamprey Lampetra fluviatilis (mostly from anadromous populations) and freshwater European brook lamprey Lampetra planeri from across 18 sites, primarily in the British Isles, were investigated for 13 polymorphic microsatellite DNA loci, and 108 samples from six of these sites were sequenced for 829 bp of mitochondrial DNA (mtDNA). We found contrasting patterns of population structure for mtDNA and microsatellite DNA markers, such that low diversity and little structure were seen for all populations for mtDNA (consistent with a recent founder expansion event), while fine-scale structuring was evident for nuclear markers. Strong differentiation for microsatellite DNA loci was seen among freshwater-resident L. planeri populations and between L. fluviatilis and L. planeri in most cases, but little structure was evident among anadromous L. fluviatilis populations. We conclude that postglacial colonization founded multiple freshwater-resident populations with strong habitat fidelity and limited dispersal tendencies that became highly differentiated, a pattern that was likely intensified by anthropogenic barriers.
Keywords: anadromy, barriers to migration, Lampetra, life history, microsatellite, speciation
Introduction
Although the abandonment of migration remains a poorly understood aspect of evolutionary biology, there is evidence to suggest that this phenomenon might act as an initiator for adaptive radiation (Bell & Andrews 1997; Winker 2000; Räsänen & Hendry 2008; Langerhans & Riesch 2013). Differences in life history traits between resident and migrant individuals can be thought of as adaptive behaviours that act to increase growth, survival rate, fecundity and egg quality. This is reflected in the fitness outcomes of both life history strategies, with residency favoured when the cost of migration exceeds the benefits of doing so, particularly in terms of growth potential and mortality risk before reproduction (Fryxell & Sinclair 1988; Bell & Andrews 1997; Dingle 2006; Brönmark et al. 2008; Shaw & Couzin 2013).
Anadromy, which involves reproduction in freshwater and the majority of growth in the marine environment, is a distinctive migratory trait that is recognized in 18 fish families and 120 species (McDowall 1997; Chapman et al. 2012). Anadromy inherently offers an opportunity to colonize previously unexploited freshwater environments, and the shift from an anadromous to a wholly freshwater life history has occurred repeatedly in many taxa of fishes (e.g. Petromyzontiformes, Salmonidae, Gasterosteidae; Potter 1980; Taylor et al. 1996; Lucas & Baras 2001). Glacial cycles may have supported the evolution of wholly freshwater forms by either blocking migration routes and preventing anadromy or, upon deglaciation, making available new habitat and food resources that are inaccessible through freshwater but easily reached by anadromous fish (Bell & Andrews 1997; Lee & Bell 1999).
The extent to which anadromy is obligatory varies among species. Many populations of anadromous fishes contain a component that does not migrate to sea and instead remains in freshwater where they mature and spawn. In some cases, they may subsequently move little, but in other cases migrate between distinct freshwater habitats (potamodromy), often reproducing with their anadromous conspecifics (Lucas & Baras 2001; McDowall 2001). ‘Partial migration’ is the term coined for this resident-migratory dimorphism within populations (Chapman et al. 2011), and it is widespread in mammals, invertebrates, birds (Lundberg 1988; Jahn et al. 2010) and fishes (Olsson & Greenberg 2004; Brodersen et al. 2008; Kerr et al. 2009; Chapman et al. 2012).
Incipient speciation in these systems may be promoted through both allopatric and sympatric mechanisms (Chapman et al. 2011). Reduced gene flow between migrants and freshwater-residents breeding in allopatry could promote differentiation by genetic drift or local adaptation. Conversely, population differentiation is limited by the large-scale dispersal capacity of migrants, resulting in a greater chance of panmixia (Hoarau et al. 2002; Coltman et al. 2007). Migratory populations that exhibit philopatry, or habitat fidelity, however, can maintain discrete genetic differences between populations within species. For example, anadromous Atlantic salmon (Salmo salar) undergo extended oceanic migrations, yet exhibit significant local adaptation and substantial reproductive isolation between populations owing to precise philopatry and a high homing fidelity to their natal river or tributary (Taylor 1991).
In contrast to anadromous salmonids, anadromous lampreys (Petromyzontiformes) generally show very low interpopulation differentiation across geographically distant river systems (Almada et al. 2008; Goodman et al. 2008) and have been shown to use pheromones released by stream dwelling larvae as partial cues to find suitable spawning habitats (Fine et al. 2004). An evolutionary trend among lampreys is the occurrence in most genera of ‘paired species’ (Zanandrea 1959), whereby larvae are morphologically indistinguishable, while the adults of two putative species adopt either a nonparasitic freshwater-resident or a parasitic life history which can be either potamodromous or anadromous.
Nonparasitism has arisen repeatedly among lampreys (Docker 2009) and even within species (Espanhol et al. 2007), suggesting that feeding type is plastic and nonparasitic lineages may be polyphyletic (Docker 2009; Renaud et al. 2009). Nonetheless, there has been much controversy about the taxonomic status of many paired lamprey species (Zanandrea 1959; Hardisty 1986a; Schreiber & Engelhorn 1998; Youson & Sower 2001; Gill et al. 2003; Renaud et al. 2009; Docker et al. 2012). Although various studies have found little genetic differentiation between lamprey paired species (Docker et al. 1999; Yamazaki et al. 2006; Espanhol et al. 2007; Blank et al. 2008; Lang et al. 2009), Mateus et al. (2013b) found significant differentiation between sympatric European river and brook lamprey populations in Portugal based on nuclear genomic data, and Taylor et al. (2012) report differentiation between anadromous and freshwater-resident parasitic lampreys in British Columbia based on eight microsatellite DNA loci.
Here, we explore the population genetics of the anadromous European river lamprey (Lampetra fluviatilis L. 1758) and its nonparasitic freshwater-resident derivative the European brook lamprey (Lampetra planeri Bloch 1784), together with several L. fluviatilis populations that comprise potamodromous individuals that migrate within freshwater only (i.e. freshwater-residents; Maitland et al. 1994; Inger et al. 2010). We use a combination of mtDNA and microsatellite nuclear DNA markers to test the hypothesis that the postglacial expansion of anadromous L. fluviatilis during the Holocene prompted the establishment of multiple freshwater-resident L.planeri populations that subsequently became genetically differentiated. We also investigated the possibility that anthropogenic barriers are isolating lamprey populations and provide a robust quantitative assessment of this. In some freshwater fishes, the fragmentation of habitats by dams can promote genetic differentiation between the upstream and downstream populations resulting from the reduction of gene flow, often compounded by founder effects and subsequent genetic drift (Yamamoto et al. 2004; Palkovacs et al. 2008). Population divergence and dispersal at local to catchment scales were examined enabling inference about population connectivity and evolutionary viability, which may indicate important applications in conservation management (Latta 2008) and enhance our understanding of the systematics of these ancient fish.
Methods
Sampling and DNA isolation
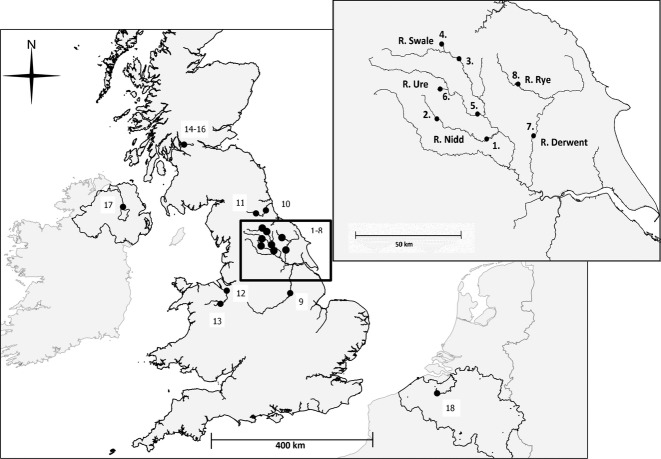
Tissue samples were collected across a total of 18 sites (Fig.1, Table S1, Supporting information). Unlike, for example, in some Baltic regions (Sjöberg 2011), there is no evidence or likelihood of historical stocking or translocation of lampreys at any of these sites. MtDNA loci were examined in n = 108 individuals from six sites including two paired sites (i.e. where Lampetra planeri and Lampetra fluviatilis were obtained from the same river; Table S1, Supporting information, Table 1, Fig.1). For microsatellite loci, 543 samples were collected from 18 sites, including seven paired sites (PS; Table S1, Supporting information, Fig.1). One of these paired sites also included a freshwater-resident L. fluviatilis population (PS7; Loch Lomond, Scotland, Table S1, Supporting information). Three additional sites for L. fluviatilis were also included in the analysis (sites 9, 17, 18, Table S1, Supporting information, Fig.1); one of which is a freshwater-resident population of L. fluviatilis in the River Bann (site 17). In Loch Lomond (PS7 in Table S1, Supporting information; Scotland), all three ‘ecotypes’ (i.e. L. planeri, L. fluviatilis and freshwater-resident L. fluviatilis) are truly sympatric; however, in all other paired sites, L. planeri samples were obtained upstream (within the same river) of anadromous L. fluviatilis populations, which were usually separated by migration barriers (Table S2, Supporting information). It should also be noted that the location from which the River Swale L. planeri samples were obtained is a spawning site for both L. planeri and sometimes L. fluviatilis.
Fig 1.
Map showing location of sampling sites 1–18 (see Table S1, Supporting information for detail). Inset is a detailed map of part of the Ouse subcatchment of the Humber catchment, showing sampling locations. Only sampled rivers are shown.
Table 1.
Diversity indices for MtDNA ATPase
| Population | Site no. | Country | N | H | π | h | D | DP | Fs | FsP |
|---|---|---|---|---|---|---|---|---|---|---|
| Bann (Lf Res) | 17 | N. Ireland | 20 | 3 | 0.0002 ± 0.0003 | 0.1947 | −1.5128 | 0.059 | −1.1801 | 0.015 |
| Scheldt (Lf) | 18 | Belgium | 20 | 10 | 0.0012 ± 0.0009 | 0.7105 | −2.0976 | 0.006 | −8.7029 | 0 |
| Nidd (Lf) | 1 | England | 17 | 2 | 0.0001 ± 0.0002 | 0.1176 | −1.1639 | 0.125 | −0.7484 | 0.092 |
| Nidd (Lp) | 2 | England | 18 | 1 | 0 | 0 | 0 | N.A. | 0 | N.A. |
| Dee (Lf) | 12 | Wales | 16 | 4 | 0.0006 ± 0.0006 | 0.3500 | −1.8309 | 0.015 | −1.7904 | 0.014 |
| Dee (Lp) | 13 | Wales | 17 | 2 | 0.0003 ± 0.0004 | 0.2206 | −0.4913 | 0.264 | 0.0353 | 0.255 |
| All | 108 | 16 | 0.0007 ± 0.0006 | 0.4907 | −2.1898 | 0 | −18.4452 | 0 |
MtDNA analysis was performed on only a subset of the 543 lampreys and 18 sites used for the microsatellite analysis. The ‘Site No.’ column corresponds to the site numbers in Fig.1 and Table S1 (Supporting information).
N = Sample size, H = number of haplotypes, π = nucleotide diversity, h = haplotype diversity, D = Tajima's D, DP = Tajima's D P-value, Fs = Fu's F, FsP = Fu's F P-value, and Lf = L. fluviatilis, Lp = L. planeri, and Lf Res = freshwater-resident population of L. fluviatilis.
Samples were obtained by hand-netting, electro-fishing and the utilization of static double-funnel traps to capture spawning, and upstream-migrating lampreys (Table S1, Supporting information). Both L. fluviatilis and L. planeri were sampled where they were found to be locally abundant prior to the spawning period and so were, in most cases, captured in the vicinity of their spawning grounds. L. planeri were normally captured in the upstream reaches of rivers where they were abundant, and in all cases, except at the Endrick Water, Loch Lomond were sampled upstream of the L. fluviatilis spawning areas. Only adult and juvenile lampreys, unambiguously identifiable to species, were included in this study. Adult anadromous, and freshwater-resident L. fluviatilis (e.g. Loch Lomond, Morris 1989; and the R. Bann, Goodwin et al. 2006), as well as nonparasitic L. planeri can be separated using standard lamprey taxonomic characteristics (Renaud 2011). Individuals were identified and measured under anaesthesia (MS-222, 0.1 g/L) using a field key (Gardiner 2003), and fin clips taken from the second dorsal fin were stored in 20% DMSO saturated NaCl solution (Amos & Hoelzel 1991). Total genomic DNA was extracted from samples using a proteinase K digestion procedure followed by the standard phenol–chloroform method and stored at −20 °C.
Amplification and sequencing of mitochondrial DNA
The PCR primers ATPfor and ATPrev (Espanhol et al. 2007) were used to amplify 838 bp of the mitochondrial gene ATPase subunits 6 and 8. This locus was chosen to facilitate comparison with previous data from Espanhol et al. (2007) and Mateus et al. (2011). Each 20 μL reaction contained 1.2 μL (final conc. 1.5 mm) MgCl2, 2 μL dNTPs (2.0 mm), 0.2 μL of each primer (10 mm), 4 μL of Colorless GoTaq® Reaction Buffer (Promega), 0.1 μL GoTaq DNA polymerase (Promega) and 1 μL of template DNA. Cycle conditions were as follows: initial denaturation at 94 °C for 3 min, followed by 30 cycles of; denaturation at 94 °C for 1 min, annealing temperature 57.1 °C for 1 min and extension at 72 °C for 2 min; and followed by a final extension at 72 °C for 2 min. The resulting PCR products were purified using the Qiagen PCR Purification kit and sequenced using an ABI PRISM 3730 DNA Analyser (DBS genomics Durham University).
Amplification and genotyping of microsatellites
Thirteen recently developed polymorphic microsatellite loci were used to examine genetic differentiation among and between all L. fluviatilis and L. planeri populations. Eight microsatellite primers developed for European Lampetra (Lp-003, Lp-006, Lp-009, Lp-018, Lp-027, Lp-028, Lp-046 and Lp-045; Gaigher et al. 2013), one primer set developed for Lampetra richardsoni (Lri-5; Luzier et al. 2010), and four microsatellite primers developed in this study (using the protocol described in White et al. 2010) and optimized for European Lampetra species (Lamper_1, Lamper_2, Lamper_3, Lamper_4) were included (Table S3, Supporting information).
Microsatellite loci were multiplex amplified using a Qiagen Multiplex kit. Thermal cycler conditions were as follows: initial denaturation at 95 °C for 15 min; followed by 35 cycles of denaturation at 94 °C for 30 s, annealing temperature 60 °C for 90 s and extension at 72 °C for 60 s; and followed by a final extension at 60 °C for 30 min. PCR products were genotyped on a 3730 ABI DNA Analyser (DBS Genomics, Durham, UK) and visualized with Geneious VR6 (Biomatters). Microsatellite loci were tested for null alleles, large allele dropout and scoring errors due to stutter peaks using microchecker 2.2.3 (van Oosterhout et al. 2004). The program arlequin 3.5 (Excoffier & Lischer 2010) was then used to test deviation from Hardy–Weinberg equilibrium. Tests for linkage disequilibrium were carried out for each pair of loci using an exact test based on a Markov chain method as implemented in genepop 4.2 (Raymond & Rousset 1995; Rousset 2008). The program Lositan (Antao et al. 2008) was used to test for outliers indicating positive or balancing selection (using a forced neutral mean FST, a confidence interval of 0.99 and false discovery of 0.1), and no loci with evidence for selection were found.
Genetic diversity and structure
MtDNA sequences were aligned manually using geneious vR6 (Biomatters). The program dnasp 10.4.9 (Rozas et al. 2003) was then used to calculate mitochondrial DNA polymorphism estimated as haplotypic diversity (Nei & Tajima 1981) and nucleotide diversity (Nei 1987). To determine the level of genetic differentiation between pairs of populations, F-statistics (Weir & Cockerham 1984) were calculated for mtDNA and microsatellite DNA loci using arlequin version 3.5. Significance was tested using 1000 permutations. arlequin was also used to calculate Fu's F, Tajima's D and mismatch distributions. We estimated the putative time of population expansion from the mismatch distribution using the statistic tau (τ; Rogers & Harpending 1992). Substitution rate was estimated after Ho et al. (2007) who suggest an average of ∼50% per site per million years for the control region, based on recent evolutionary time frames, although of course this varies among species. The substitution rate for the control region can be ten times faster than the rest of the mitochondrial genome (McMillan & Palumbi 1997). Therefore, 5% per site per million years was used as a rough estimate for ATPase. Mutation rates of 1% and 10% per million years were also used to illustrate the effect that the rate of divergence will have in the expansion times. The relationship between haplotypes was investigated using a median-joining network (MJN) constructed using the program network 3.1.1.1 (Bandelt et al. 1999) and epsilon values of 0, 10, 20 and 30 were tested.
For microsatellite DNA data, allelic richness for each locus and population and FIS (inbreeding coefficient) were calculated using the program fstat 2.9.3 (Goudet 1995). structure 2.0 was used to assign individuals by genotype to a putative number of populations (K; Pritchard et al. 2000). ΔK, a measure of the second order rate of change in the likelihood of K (Evanno et al. 2005), was calculated using structure Harvester (Earl & vonHoldt 2012) to assess the highest hierarchical level of structure. Four independent runs for each K value were performed at 2 000 000 Markov chain Monte Carlo (MCMC) repetitions and 500 000 burn-in using no population prior information and assuming correlated allele frequencies and admixture. structure was also used with a location prior (LOCPRIOR) to clarify population structure within the Loch Lomond system (Hubisz et al. 2009). Burn-in and run lengths were the same as for runs without prior population information. Due to the large number of putative population subdivisions, subsamples were compared by region to increase resolution, in addition to an analysis involving all regions. Full-sibling pairs within a sampling site (for the five localities where there are populations of both putative species: Wear, Dee, Derwent, Nidd & Ure) were identified using the maximum-likelihood method in colony version 2.0.1.1 with male and female polygamy permitted and a medium run length (Jones & Wang 2010).
Patterns of microsatellite differentiation were subsequently examined using a factorial correspondence analysis (FCA) implemented in genetix 4.05.2 (Belkhir et al. –20041996), which gives a visual representation of individual genotype clustering. A test for a positive association between genetic [FST/(1 − FST)] and geographic distances [Isolation by distance (IBD)] based on microsatellite DNA loci was carried out using a Mantel test (10 000 permutations) in genepop v4.2. Geographic distances were calculated between sample sites using linear referencing tools in Quantum GIS (Lisboa). A Mantel test was also carried out to test for association between genetic distances and number of physical barriers (defined as any anthropogenic feature larger than 0.5 m height at base river level which reaches the full width of the river) between sample sites. The 0.5 m value was subjective, based on the fact that many structures of this height or greater generate discrete water level differences (upstream–downstream) at base flows, on published and unpublished data on the impact of different height potential barriers on lamprey movement (L. fluviatilis, Lucas et al. 2009; L. planeri, M. C. Lucas personal observation), and on our ability to identify potential barriers in field surveys and databases. Only river systems for which information on barriers was available were utilized in the Mantel tests (including the Dee, Wear and all rivers within the Ouse subcatchment, excluding the Swale due to the low sample size attained for L. planeri).
migrate-n (v 3.2.6) was used to estimate levels of historical gene flow between populations (Beerli & Felsenstein 2001; Beerli 2006; Beerli & Palczewski 2010). Pairwise comparisons were carried out between putative species (i.e. L. fluviatilis and L. planeri) at six locations (Wear, Dee, Lomond, Nidd, Ure, Derwent), of which the latter three are all tributaries in the same river catchment, where samples from both species were available. To implement Bayesian inference in MIGRATE-N, the Brownian motion approximation was selected with an MCMC search of 100 000 burn-in steps followed by 5 000 000 steps with parameters recorded every 100 steps; exponential prior on theta (min: 0, mean: 30, max: 60); and an exponential prior on migration (min: 0, mean: 650 max: 1300). migrate-n was run with parameter values starting from FST-based estimates, and the distribution of parameter values was compared across runs to ensure overlap of 95% CI. bayesass 1.3 (Wilson & Rannala 2003) was used to estimate the magnitude and directionality of contemporary gene flow between L. fluviatilis and L. planeri. Pairwise comparisons were carried out for the same six locations that were used in the migrate-n analysis. In contrast to migrate-n, bayesass estimates all pairwise migration rates rather than a user-defined migration matrix and provides unidirectional estimates of migration for each population pair. bayesass does not assume a migration–drift equilibrium, an assumption that is frequently violated in natural populations (Whitlock & McCauley 1999). A total of 10 000 000 MCMC iterations were run of which 1 000 000 were for the burn-in. All other options were left at their default settings. Five to 10 runs with a different starting point were performed for each population pair and results are given as means. The program tracer version 1.5 (Rambaut & Drummond 2007) was used as a method to qualitatively assess MCMC convergence.
Results
MtDNA
ATPase subunits 6 and 8 were sequenced and haplotypes determined for 108 lampreys (Lampetra fluviatilis and Lampetra planeri) from six sampling sites (Table 1). Over all populations, haplotype and nucleotide diversity were low, with freshwater-resident populations of both L. planeri and L. fluviatilis generally exhibiting lower haplotype and nucleotide diversity than the anadromous L. fluviatilis populations. Both Tajima's D and Fu's F were negative and highly significant (Table 1), consistent with a population expansion (e.g. after a bottleneck) or a selective sweep. Using the value of tau, which was 0.673 (Fig. S1, Supporting information), an expansion time of 16 263 (10 182–26 952; 95% CI) years ago was calculated using the mutation rate of 5% per million years. Using mutation rates of 1% and 10%, expansion times would be 81 182 and 8118 years ago, respectively. Sixteen haplotypes were observed, with private haplotypes found only in the L. planeri population from the River Nidd and no species-specific lineages (see median-joining network in Fig.2a). FST values between sites ranged from 0.01955 to 0.94093 with only FST values associated with the Nidd (L. planeri) being statistically significant (P < 0.0001; Table 2).
Fig 2.
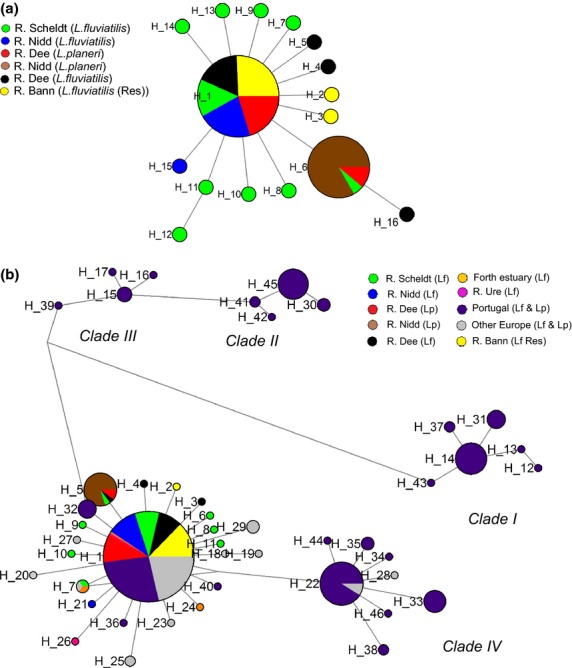
(a) Median-joining network showing 16 haplotypes found from 108 samples of Lampetra at six sampling locations. Note that Bann (Lf) is a freshwater-resident L. fluviatilis population. Lf = anadromous L. fluviatilis, Lp = L. planeri and Lf Res = freshwater-resident population of L. fluviatilis. Details of the sample locations are given in Table S1 (Supporting information). (b) Forty-six haplotypes from combined studies comprising of both L. fluviatilis and L. planeri. Circled groups show correspondence with clades identified in Mateus et al. (2011). Clades I–III consist of freshwater-resident L. planeri (but see Mateus et al. 2013b) with restricted distribution, and clade IV contains both freshwater-resident Lp and anadromous Lf with a wider distribution along with haplotypes identified in Espanhol et al. (2007) from France, Sweden and Germany (Lp and Lf H22) and France (Lp H28). Please see the open-access online paper for a colour version of this figure.
Table 2.
Matrix of pairwise FST values for mtDNA analysis of six populations of Lampetra
| Bann (Lf Res) | Dee (Lf) | Nidd (Lp) | Scheldt (Lf) | Nidd (Lf) | |
|---|---|---|---|---|---|
| Dee (Lf) | 0.0033 | ||||
| Nidd (Lp) | 0.8534 | 0.7717 | |||
| Scheldt (Lf) | 0.0074 | −0.0001 | 0.5702 | ||
| Nidd (Lf) | −0.0038 | 0.0024 | 0.9409 | 0.0082 | |
| Dee (Lp) | −0.0258 | −0.0196 | 0.8676 | 0.0024 | 0.0417 |
Significant FST values [i.e. all FST values associated with Nidd (Lp)] are highlighted in grey (P < 0.0001).
Lf = Lampetra fluviatilis, Lp = Lampetra planeri and Lf Res = freshwater-resident population of L. fluviatilis.
A network showing the European haplotype distribution, incorporating data from Espanhol et al. (2007) and Mateus et al. (2011), revealed 46 haplotypes with Portuguese populations being visibly further removed from the majority of other samples (Fig.2b). Identified lineages were concordant with those reported by Mateus et al. (2011), and as observed by Espanhol et al. (2007), not species specific. Clades I, II and III were considered to be composed of adult L. planeri (Mateus et al. 2011; now regarded as three cryptic species, L. alavariensis, L. auremensis and L. lusiticanica, Mateus et al. 2013a) and larvae of unknown specific status, while clade IV comprises L. planeri, anadromous and freshwater-resident L. fluviatilis adults and larvae.
Microsatellite analysis
A total of 543 lampreys were genotyped at thirteen loci. All loci were in Hardy–Weinberg equilibrium and not impacted by null alleles for most populations, and there were no consistent issues for any given population (Table S4, Supporting information). A total of 112 of the 136 FST values (82.4%) were statistically significant (P < 0.05; Fig.3, Table S5, Supporting information). All FST values between L. planeri populations were significant with a range from 0.06045 to 0.191 (Wear vs. Nidd); however, only 45.4% of FST values for L. fluviatilis populations were significant, with a range from −0.00524 to 0.11945. When the freshwater-resident L. fluviatilis populations were not included, the FST values ranged from −0.00524 to 0.02537. FST values between L. fluviatilis and L. planeri populations ranged from 0.011 to 0.18554. Average allelic richness per locus ranged from 2.43 (Lp_003) to 14.9 (Lamper_4). Average FIS per site ranged from −0.095 [Wear (L. planeri)] to 0.028 [Lomond (anadromous L. fluviatilis)].
Fig 3.
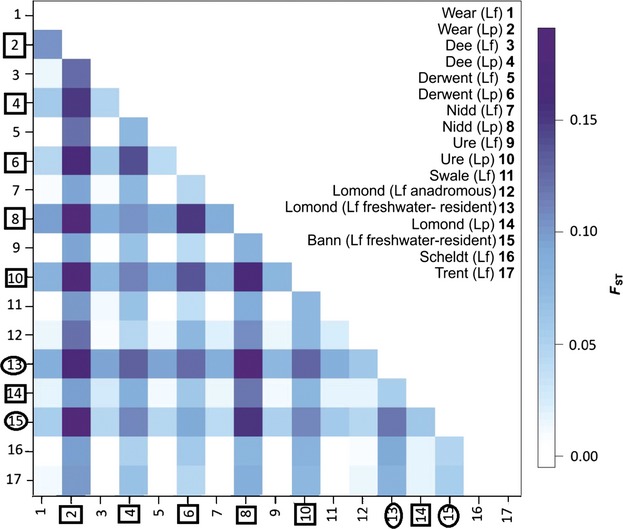
Matrix of pairwise FST values using 13 microsatellite loci, for all Lampetra populations sampled. Lf = anadromous L. fluviatilis, Lp = L. planeri and Lf Res = freshwater-resident population of L. fluviatilis. Table showing the actual values is included in Supporting information (Table S5). Numbers on axes are marked with a square to represent L. planeri and a circle to represent freshwater-resident L. fluviatilis.
In COLONY, tests for the proportion of putative full-siblings (as an indicator of close kin) in populations of either species showed this to be rare, 0% in some cases for both species, and no higher than 0.74%. One randomly chosen individual of each full-sibling pair was excluded, and analysis was repeated. There were no differences that affected inference in the results with full-siblings included or excluded, so all individuals were included in the analysis.
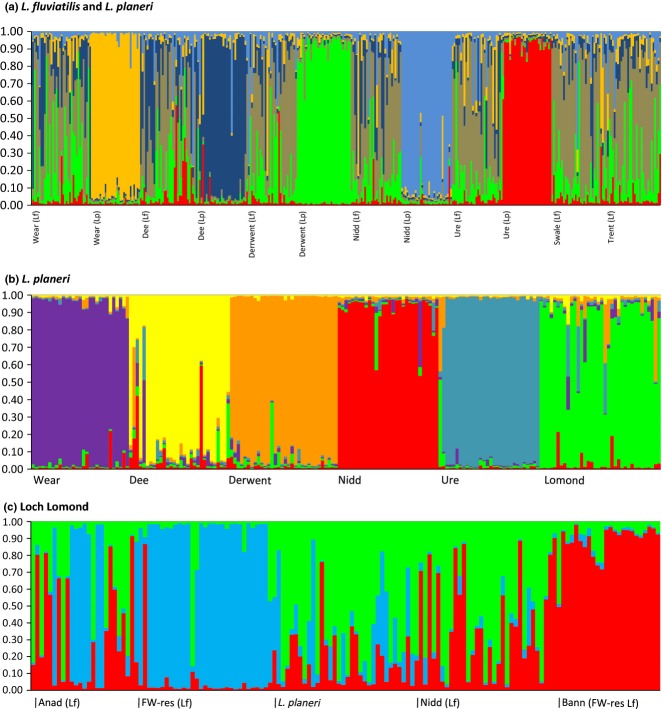
structure analyses consistently identified L. planeri populations as being separate from L. fluviatilis populations (anadromous and freshwater-resident) and from each other (Fig.4). The only exception was the small sample of L. planeri on the Swale compared to the L. fluviatilis population downstream on the same river (Fig. S2a, Supporting information). Figure4a shows the most likely population structure among 12 sampling locations in England and Wales (excluding the Scottish Loch Lomond system) incorporating both species, where K = 6 showed the highest LnP(D) (Fig. S3, Supporting information). Lampetra fluviatilis samples appear as a single mixed population. Representing a higher hierarchical level, ΔK = 2 primarily supports separation of L. fluviatilis and L. planeri (Fig. S3 and S4, Supporting information). Figure S4c (Supporting information) shows a comparison across all populations where K = 9 [the highest LnP(D) outcome]. There were several peaks for ΔK at K = 2, 5 and 8 (Fig. S4d, Supporting information), but the maximum LnP(D) result (K = 9) was most informative, distinguishing all L. planeri and L. fluviatilis freshwater-resident populations (with the exception of the L. planeri population in the Swale).
Fig 4.
structure bar plot generated from microsatellite data for three population clusters of lampreys. (a) Comparison between Lampetra fluviatilis and Lampetra planeri populations (K = 6); (b) L. planeri populations (K = 6); (c) Loch Lomond populations compared to a population of L. fluviatilis from the Humber catchment and freshwater-resident L. fluviatilis populations from the R. Bann in N. Ireland (K = 3). Please see the open-access online paper for a colour version of this figure.
When only L. planeri populations were compared, the highest likelihood result identified all populations as distinct (Fig.4b). In this case, ΔK was 4 (Fig. S3 and S4, Supporting information); however, this linked samples from the Nidd with the Dee, and Loch Lomond with the Derwent, in each case populations on opposite sides of British Isles (see Fig.1; Fig. S4b, Supporting information). When only anadromous L. fluviatilis populations were compared, the outcome was K = 1 (not shown). The Loch Lomond system (which contains anadromous L. fluviatilis, freshwater-resident L. fluviatilis and L. planeri populations) was compared to an anadromous L. fluviatilis population (Nidd) and another freshwater-resident L. fluviatilis population (Bann). structure identified three populations with highest likelihood, while ΔK was 2 (Fig.4c; Fig. S3, Supporting information). Using prior location information for Loch Lomond, five populations were identified. However, ΔK = 2, showing differentiation at a higher hierarchical level between the freshwater-resident L. fluviatilis population in Loch Lomond and the other populations (Fig. S2b,c, Supporting information). Location priors did not provide any useful additional inference for other analyses.
The FCA plots support essentially the same clusters as identified in structure showing L. fluviatilis as being dominated by one large grouping, with the freshwater-resident populations differentiated (Fig. S5a, Supporting information) and L. planeri populations as all being separated from each other (Fig. S5b, Supporting information). Mantel tests for correlation between genetic and geographic distance showed a significant negative trend for L. planeri populations (R² = 0.2963; P < 0.05; Fig.5a) and a weak but significant positive linear relationship for all L. fluviatilis populations (R2 = 0.0841; P < 0.05). However, when freshwater-resident L. fluviatilis populations were excluded (Bann and Loch Lomond), the positive relationship was much stronger (R2 = 0.40, P < 0.0001; Fig.5b). Mantel tests examining correlations between genetic distance and the number of barriers along migration/dispersal routes for L. fluviatilis and L. planeri (populations included as described in methods) showed a highly significant positive correlation (R2 = 0.8256, P < 0.0001; Fig.5c).
Fig 5.
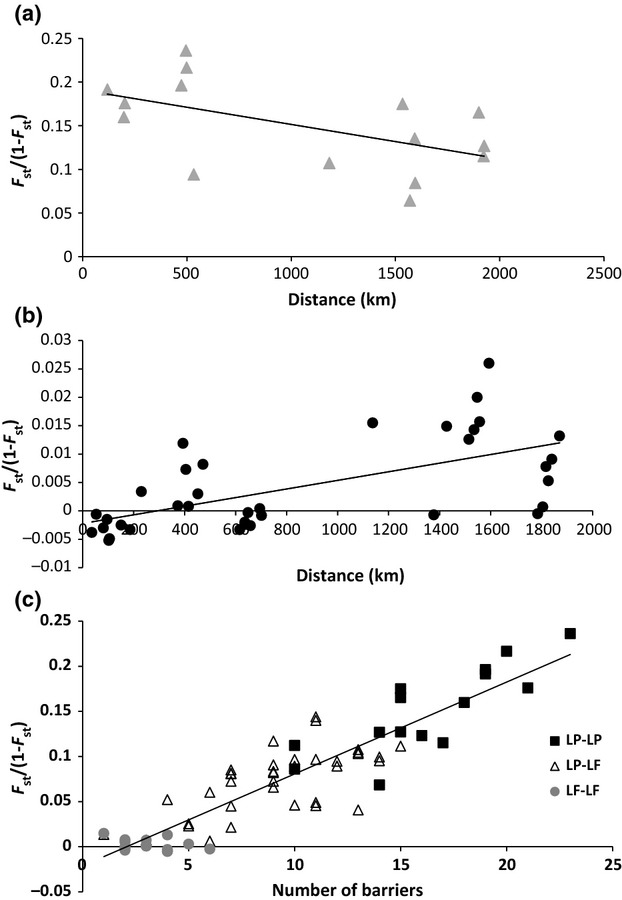
Isolation by distance tests for correlation between genetic differentiation (based on microsatellites) showing (a) geographic distance between freshwater-resident Lampetra planeri populations (R2 = 0.30, P < 0.05) and (b) geographic distance between anadromous Lampetra fluviatilis populations (R2 = 0.40, P < 0.0001; i.e. excluding freshwater-resident Bann and Lomond Lf). Inclusion of freshwater-resident Lf populations in the analysis reduced the strength of the correlation (R2 = 0.0841, P < 0.05)—not shown. (c) number of barriers between samples sites (R2 = 0.8256, P < 0.0001) where LP–LP signifies comparison of numbers of barriers between L. planeri sampling sites, LP–LF is number of barriers between L. planeri and L. fluviatilis sampling sites, and LF–LF is the number of barriers between L. fluviatilis sampling sites. Only sites for which barrier information was available were included in the analysis (i.e. Lf and Lp for Wear, Dee, Derwent, Nidd, Ure, and Swale Lf only).
Migration rate estimates between species (using migrate-n) ranged from 3.73 to 10.43 migrants/generation from L. fluviatilis to L. planeri, and from 4.18 to 16.28 from L. planeri to L. fluviatilis (Table S6, Supporting information). The six pairwise comparisons all suggested asymmetric gene flow greater in the direction from L. planeri to L. fluviatilis (which apart from Loch Lomond was always in the downstream direction), but 95% confidence intervals were large and overlapping. The bayesass analysis indicated low-level contemporary gene flow between the putative species, and some comparisons also suggest the downstream direction from L. planeri to L. fluviatilis (especially in the Derwent; Table S7a,b, Supporting information). It also indicated ongoing gene flow between the three forms in the Loch Lomond system (Table S7b, Supporting information).
Discussion
Population history
This study was based in a geographic region that has undergone profound cyclical changes over the course of the Pleistocene (2.58 Ma–11 700 years ago), with suitable riverine habitat available only during interglacial periods (Hays et al. 1976). For our study sites in the UK, mtDNA failed to show any differentiation between the two putative Lampetra species or among populations, which is consistent with data for some other northern European populations (Espanhol et al. 2007). While this may suggest ongoing gene flow or the incomplete sorting of ancestral polymorphisms, it is also consistent with recent founder events establishing these populations. The network analyses and neutrality tests support this, indicating small founder populations and subsequent expansion. Conversely, for populations in southern Europe where the climate has been more stable over time, there are far higher nucleotide diversities and significant mtDNA phylogeographic structuring (Pereira et al. 2010; Mateus et al. 2011).
Espanhol et al. (2007) suggested that Lampetra planeri in Europe may be polyphyletic and have originated within at least two evolutionary lineages, possibly the result of independent divergence events from Lampetra fluviatilis with the repeated loss of anadromy. Pereira et al. (2010) have since found several Portuguese populations of L. planeri which are isolated among themselves and also from the anadromous lamprey population. These populations had only private haplotypes, suggesting that a significant amount of time had passed to establish independent evolutionary histories. The fact that genetically distinct non-migratory Lampetra populations are found in many Portuguese rivers (Pereira et al. 2010; Mateus et al. 2011, 2013a) suggests lamprey were once more abundant and widespread in Iberia. The higher levels of divergence shown in our mtDNA median-joining network that included Portuguese lampreys (Fig.2), compared to other populations examined across Europe, also suggests that sufficient time may have passed to establish a complex of incipient freshwater-resident species, although further nuclear DNA data would help resolve this question. Similar processes generating multiple origins have been suggested, for example, in the marine to freshwater transitions of three-spine sticklebacks (Gasterosteus aculeatus; Hohenlohe et al. 2010).
Our study estimates the expansion time of L. planeri and L. fluviatilis populations in the British Isles and northern Europe as 16 236 (10 182–26 952) years ago using tau and a mutation rate of 5% per million years, which roughly coincides with the last glacial maximum (19–26 000 years ago; Clark et al. 2009). The Pleistocene climatic fluctuations impacted much of Europe (Hays et al. 1976; Webb & Bartlein 1992) and significantly influenced the distribution and genetic diversity of plants and animals (Hofreiter & Stewart 2009). In addition to cycles of habitat loss and release as glaciers extended and receded, the ‘refugium theory’ proposes that temperate species survived the glacial maxima in southern refugia and colonized northern latitudes during interglacial periods (Taberlet et al. 1998; Hewitt 2000). The results shown here, coupled with data from the Iberian Peninsula, suggest that southern latitudes served as an important refugium for Lampetra during the Pleistocene glaciations, intermittently acting as a point of dispersal for postglacial expansion (Espanhol et al. 2007; Mateus et al. 2012, 2013b).
Therefore, there may have been a tendency during interglacial periods, while anadromous Lampetra were expanding northwards, for populations at lower latitudes to abandon anadromy and eventually become restricted to freshwater. This is consistent with the findings of a recent study utilizing restriction site associated DNA sequencing (RAD seq.) that identified strong genetic differentiation between sympatric L. fluviatilis and L. planeri in the Iberian Peninsula with numerous fixed and diagnostic single nucleotide polymorphisms (SNPs) between the two putative species, some associated with genes related to osmoregulation (Mateus et al. 2013b). A study using RAD sequencing to compare Pacific lamprey (Entosphenus tridentatus) geographic populations also found evidence consistent with local adaptation (Hess et al. 2013). Our median-joining network in Fig.2 shows that for the available samples, only clade IV shares a haplotype with the lineage representing the northern expansion, suggesting a possible link between these lineages (with clade IV providing the ancestor of the anadromous group that founded the postglacial population in northern Europe). With expansion into previously unoccupied territory, it is expected that genetic diversity should decrease from the south to the north (Hewitt 2000), consistent with our findings.
Population structure
In contrast to mtDNA, we found considerable structure at microsatellite DNA loci between L. fluviatilis and L. planeri populations, especially among populations of L. planeri, but much less among anadromous L. fluviatilis populations. Anadromous lampreys (Lethenteron spp.) in Japan (Yamazaki et al. 2011) and Petromyzon mari-nus in North America (Bryan et al. 2005) and Europe (Almada et al. 2008) exhibit similar levels of panmixia, with little or no genetic structure, despite their widespread distribution. Spice et al. (2012) found that Pacific lamprey along the west coast of North America showed low but significant differentiation among locations. However, instead of being philopatric like many other anadromous fish species (McDowall 2001), differentiation was suggested to be due to greater restrictions to dispersal at sea compared to other anadromous lamprey species. The lack of population structure found in our study was, therefore, consistent with the general lack of natal homing seen for other anadromous lamprey species.
The absence of a clear genetic signal for species-level differences between anadromous and freshwater-resident populations is consistent with findings for other paired lamprey species (Espanhol et al. 2007; Hubert et al. 2008; Docker 2009; April et al. 2011; Mateus et al. 2011; Boguski et al. 2012; Docker et al. 2012). Greater differentiation among populations within L. planeri, than between L. planeri and L. fluviatilis, suggests the unexpected pattern of greater gene flow between the putative species than within L. planeri (while the greatest gene flow occurs among populations of L. fluviatilis). Gene flow between the putative species may be possible owing to a combination of interspecific nest association (Huggins & Thompson 1970; Lasne et al. 2010) and sneaker male behaviour (Malmqvist 1983; Hume et al. 2013). As larvae of both species tend to move downstream through voluntary and involuntary drift behaviour (Hardisty & Potter 1971a; Moser et al. 2015), the distribution and overlap of spawning adults of the two species ultimately depends on a combination of the degree of downstream drift of L. planeri from upstream tributaries where they predominate, towards L. fluviatilis-dominated zones, and the subsequent upstream movements of freshwater-resident L. planeri and anadromous or freshwater-resident L. fluviatilis (Hardisty & Potter 1971b; Malmqvist 1980).
Both assignment (bayesass) and coalescent (migrate-n) methods suggested directionality in genetic migration, favouring the direction of L. planeri to L. fluviatilis, although the confidence limits were broad. Asymmetric gene flow occurring in these types of freshwater systems can significantly influence the distribution of genetic variation, with downstream populations typically exhibiting higher genetic diversity than headwater populations (Caldera & Bolnick 2008; Morrissey & de Kerckhove 2009; Julian et al. 2012). Yamazaki et al. (2011) found gene flow to exist at multitemporal scales between ‘potentially sympatric’ lamprey populations and suggested ongoing gene flow was the result of imperfect size-assortative mating and the plastic determination of life histories. The observed increase in genetic diversity as one moves downstream towards the lower reaches of the river could result from historical patterns of colonization, with contemporary dispersal reflecting movement bias, fragmented habitat or the presence of dispersal barriers (Morrissey & de Kerckhove 2009; Dehais et al. 2010). Asymmetric gene flow would be expected if L. planeri populations remain primarily resident further up the catchments with occasional migrants moving further downstream to where they may encounter spawning L. fluviatilis.
Connectivity and anthropogenic factors
Mantel tests for isolation by distance revealed a positive correlation between geographic and genetic distance for anadromous L. fluviatilis, and a counterintuitive negative correlation among L. planeri populations (Fig.5). However, while the correlation for L. fluviatilis was significant (especially when freshwater-resident L. fluviatilis were omitted), and consistent with expectations (implying that long-range dispersal is less common), the correlation with L. planeri was weak and showed a broad range of values for a given distance (see Fig.5a). The L. planeri correlation may, therefore, simply reflect a stochastic pattern or ancestral relationships.
The number of anthropogenic barriers between populations was found to be significantly positively correlated with genetic distance, and such barriers have been shown to limit the upstream migration of L. fluviatilis (Lucas et al. 2009). Anthropogenic barriers could therefore be amplifying (beyond natural processes) the isolation of L. planeri populations by inhibiting the upstream movement of anadromous L. fluviatilis and preventing geneflow mediation in this manner between populations. Meldgaard et al. (2003) also detected a statistically significant increase of FST with the number of weirs between grayling (Thymallus thymallus) populations in a Danish river system. Similar decreases of genetic diversity from downstream towards upstream populations have been observed in other fish species in relation to anthropogenic barriers (Yamamoto et al. 2004; Caldera & Bolnick 2008; Raeymaekers et al. 2009). Yamazaki et al. (2011) found freshwater-resident nonparasitic lamprey populations in the upper regions of dammed rivers to be genetically divergent from seasonally sympatric, anadromous, parasitic populations. This pattern is consistent with a scenario where barriers amplify the asymmetry of gene flow from upstream towards downstream sites by allowing some passive downstream drift, while obstructing active upstream migration. Spice et al. (2012) also found that larvae from an anadromous population of E. tridentatus at a spawning site upstream of nine dams (which only a small number of adults successfully pass each year) exhibited higher genetic differentiation (i.e. higher FST values) than most other population comparisons.
When a freshwater-resident lamprey population is physically isolated from anadromous parasitic populations (which may mediate gene flow between freshwater-resident populations), acceleration in genetic divergence may result in the subsequent establishment of allopatric speciation (Yamazaki & Goto 2000). It is probable, however, that freshwater-resident L. planeri populations would have become, and tended to remain, isolated without the added anthropogenic hurdles, as there is a degree of population separation that is due to the natural extent of upstream migration in anadromous L. fluviatilis. As previous studies have shown, this is usually limited to higher order channels, and individuals do not generally penetrate the smaller streams even where access is unhindered by barriers (Hardisty & Potter 1971c; Hardisty 1986b).
The system in Loch Lomond offers evidence of the potential for gene flow between morphologically differentiated ecotypes, indicating that where they are found sympatrically, gene flow between L. fluviatilis and L. planeri can occur. This scenario is also supported by the lack of evidence for differentiation between the geographically proximate L. fluviatilis and L. planeri populations on the River Swale, although the sample size for the latter population was small (Fig. S2, Supporting information). Similarly, Docker et al. (2012) found no genetic differentiation between silver (Ichthyomyzon unicuspis) and northern brook (I. fossor) lampreys occurring sympatrically (also using microsatellite loci), but did find differentiation among parapatric populations. Yamazaki et al. (2011) also found a lack of differentiation between sympatric populations of Arctic lamprey (Lethenteron camtschaticum) and its nonparasitic derivatives in the Ohno River, Japan.
The bayesass analysis suggests that contemporary gene flow is occurring between all three populations in Loch Lomond, consistent with a tendency for interbreeding when there are no environmental barriers to limit connectivity. The divergence of the freshwater-resident L. fluviatilis population would then suggest a period of differentiation in isolation. Therefore, in Loch Lomond, the anadromous strategy is also paralleled by a population component with potamodromous behaviour, with some fish apparently showing migration mostly between the loch and spawning streams. While all three Loch Lomond populations were significantly differentiated from each other (Fig.3, Table S5, Supporting information), there were also data indicating contemporary gene flow among them, and the anadromous L. fluviatilis and sympatric L. planeri populations both showed evidence of connectivity with the wider L. fluviatilis populations.
Conclusions
Alternative life history strategies are common among fishes inhabiting postglacial lakes, often resulting from adaptation to different foraging strategies or environments (Robinson & Parsons 2002). This is one of the best supported mechanisms for speciation in sympatry, for example among cichlid species in Holocene lakes (Barluenga et al. 2006). The divergence of multiple independent populations is a common trend in the evolution of diversity for diadromous fish (Schluter & Nagel 1995; Waters & Wallis 2001), and a number of studies have shown the influence of glacial movement within the Holocene on the phylogeographical structure of freshwater fishes (Harris & Taylor 2010; Boguski et al. 2012). However, in our study, the geographic scale is small for the extent of differentiation observed. It is apparent that at an initial stage, there was a postglacial expansion of anadromous Lampetra fluviatilis from southern refugia and the subsequent establishment of multiple freshwater-resident Lampetra planeri populations. These may have been relatively small founder groups that retained some degree of reproductive isolation that was likely intensified, although perhaps not entirely determined, by the anthropogenic introduction of barriers. Moreover, it was ascertained that there is gene flow between L. fluviatilis and L. planeri in both long-term and contemporary timescales and the pattern of gene flow is apparently asymmetric. This has significant implications for the management of L. planeri populations and the extent to which this is underpinned by natural processes will have important evolutionary implications with respect to the mechanisms that generate diversity. Our data emphasizes the importance of founder events in the evolution of diversity among populations and as a frequent component of the speciation process (Templeton 2008). These data also strongly support a scenario of multitemporal and multispatial radiation. In contrast to higher levels of Lampetra divergence present in the Iberian Peninsula, the northern European populations appear to have been established relatively recently, and the process of differentiation is still ongoing. There may be a natural tendency towards speciation in freshwater-resident populations that remain environmentally stable over time, but a dynamic process instead at higher latitudes experiencing a cycle of habitat loss and release.
Acknowledgments
We are grateful to colleagues listed in Table 1 for provision of tissue samples. FB was funded by awards from the Freshwater Biological Association award and EPSRC (via Durham Energy Institute CDT). JH was funded by the University of Glasgow and a Scottish Natural Heritage award.
Data accessibility
DNA sequences: GenBank accessions: KP722176–KP722191; microsatellite DNA genotypes and mtDNA sequence alignment file: Dryad doi:10.5061/dryad.v105s.
Supporting information
Additional supporting information may be found in the online version of this article.
Table S1 Numbers collected and location of origin for all genetic samples of Lampetra planeri and Lampetra fluviatilis and by whom they were collected.
Table S2 Number of barriers (distance, km) between sampling locations.
Table S3 Details of microsatellite loci and primers utilised in the study.
Table S4 Table outlining diversity indices for microsatellite loci by population, and by locus.
Table S5 Pairwise FST values for 13 microsatellite loci.
Table S6 migrate-n analysis (Bayesian) showing posterior distributions for six paired sites (95% confidence intervals in brackets).
Table S7 Pairwise estimation of current gene flow, M, between (a) populations of anadromous Lampetra fluviatilis (Lf) and Lampetra planeri (Lp) from within the same river, and (b) populations within Loch Lomond by bayesass.
Fig. S1 Mismatch distribution (demographic expansion) with Tau 0.673, showing an expansion pattern for the six populations of Lampetra fluviatilis and Lampetra planeri presented in Table 1.
Fig. S2 structure bar plot generated from microsatellite data showing (a) ΔK = 2 (LnP(D) = −2294.8) where Swale Lp is compared to another Lampetra planeri population and Lampetra fluviatilis form the same river (b) ΔK = 2 when prior location information is used to analyse the Loch Lomond system which shows the freshwater-resident Lomond population to be differentiated and (c) ΔK = 5 (LnP(D) = −4681) when a location prior is used.
Fig. S3 Posterior probability of the data (Ln[P(D|K)]) and values of ΔK (Evanno et al. 2005) as a function of K (number of clusters), associated with the results shown in Fig. 4 in the main text.
Fig. S4 (a) Structure for 12 populations where ΔK = 2. (b) Structure for Lampetra planeri only where ΔK = 4. (c) Structure for all samples included when K = 10 (Lf = Lampetra fluviatilis, Lp = L. planeri, LL = Loch Lomond; anad = anadromous; res = resident). (d) ΔK plot for the structure plot in part c.
Fig. S5 FCA analysis for (a) Lampetra fluviatilis and (b) Lampetra planeri population.
References
- Almada VC, Pereira JI, Fonseca JP, Levy A, Maia C, Valenta A. Mitochondrial DNA fails to reveal genetic structure in sea lampreys along European shores. Molecular Phylogenetics and Evolution. 2008;46:391–396. doi: 10.1016/j.ympev.2007.05.024. [DOI] [PubMed] [Google Scholar]
- Amos B, Hoelzel AR. Long-term preservation of whale skin for DNA analysis. Report of the International Whaling Commission Special Issue. 1991;13:99–103. [Google Scholar]
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. BMC Bioinformatics. 2008;9:323. doi: 10.1186/1471-2105-9-323. LOSITAN: A workbench to detect molecular adaptation based on a Fst-outlier method. [DOI] [PMC free article] [PubMed] [Google Scholar]
- April J, Mayden RL, Hanner RH, Bernatchez L. Genetic calibration of species diversity among North America's freshwater fishes. Proceedings of the National Academy of Sciences. 2011;108:10602–10607. doi: 10.1073/pnas.1016437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barluenga M, Stölting KN, Salzburger W, Muschick M, Meyer A. Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature. 2006;439:719–723. doi: 10.1038/nature04325. [DOI] [PubMed] [Google Scholar]
- Beerli P. Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics. 2006;22:341–345. doi: 10.1093/bioinformatics/bti803. [DOI] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using coalescent approach. Proceedings of the National Academy of Sciences. 2001;98:4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli P, Palczewski M. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics. 2010;185:313–326. doi: 10.1534/genetics.109.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, Logiciel Sous Windows TM Pour la Génétique des Populations. Montpellier, France: Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II; 1996. –2004. [Google Scholar]
- Bell MA, Andrews CA. Evolutionary consequences of postglacial colonization of fresh water by primitively anadromous fishes. In: Streit B, Staedler T, Lively CM, editors. Evolutionary Ecology of Freshwater Animals. Switzerland: Birkhaeuser Verlag; 1997. pp. 322–363. [Google Scholar]
- Blank M, Jurss K, Bastrop R. A mitochondrial multigene approach contributing to the systematics of the brook and river lampreys and the phylogenetic position of Eudontomyzon mariae. Canadian Journal of Fisheries and Aquatic Sciences. 2008;65:2780–2790. [Google Scholar]
- Boguski DA, Reid SB, Goodman DH, Docker MF. Genetic diversity, endemism and phylogeny of lampreys within the genus Lampetra sensu stricto (Petromyzontiformes: Petromyzontidae) in western North America. Journal of Fish Biology. 2012;81:1891–1914. doi: 10.1111/j.1095-8649.2012.03417.x. [DOI] [PubMed] [Google Scholar]
- Bryan MB, Zalinski D, Filcek KB, Libants S, Li W, Scribner KT. Patterns of invasion and colonization of the sea lamprey (Petromyzon marinus) in North America as revealed by microsatellite genotypes. Molecular Ecology. 2005;14:3757–3773. doi: 10.1111/j.1365-294X.2005.02716.x. [DOI] [PubMed] [Google Scholar]
- Brodersen J, Ådahl E, Brönmark C, Hansson L-A. Ecosystem effects of partial fish migration in lakes. Oikos. 2008;117:40–46. [Google Scholar]
- Brönmark C, Skov C, Brodersen J, Nilsson PA, Hansson LA. Seasonal migration determined by a trade-off between predator avoidance and growth. PLoS One. 2008;3:e1957. doi: 10.1371/journal.pone.0001957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldera EJ, Bolnick DI. Effects of colonization history and landscape structure on genetic variation within and among threespine stickleback (Gasterosteus aculeatus) populations in a single watershed. Evolutionary Ecology Research. 2008;10:575–598. [Google Scholar]
- Chapman BB, Brönmark C, Nilsson J, Hansson LA. The ecology and evolution of partial migration. Oikos. 2011;120:1764–1775. [Google Scholar]
- Chapman BB, Skov C, Hulthén K, et al. Partial migration in fishes: definitions, methodologies and taxonomic distribution. Journal of Fish Biology. 2012;81:479–499. doi: 10.1111/j.1095-8649.2012.03349.x. [DOI] [PubMed] [Google Scholar]
- Clark PU, Dyke AS, Shakun JD, et al. The last glacial maximum. Science. 2009;325:710–714. doi: 10.1126/science.1172873. [DOI] [PubMed] [Google Scholar]
- Coltman DW, Stenson G, Hammill MO, Haug T, Davis CS, Fulton TL. Panmictic population structure in the hooded seal (Cystophora cristata. Molecular Ecology. 2007;16:1639–1648. doi: 10.1111/j.1365-294X.2007.03229.x. [DOI] [PubMed] [Google Scholar]
- Dehais C, Eudeline R, Berrebi P, Argillier C. Microgeographic genetic isolation in chub (Cyprinidae: Squalius cephalus) population of the Durance River: estimating fragmentation by dams. Ecology of Freshwater Fish. 2010;19:267–278. [Google Scholar]
- Dingle H. Animal migration: is there a common migratory syndrome? Journal of Ornithology. 2006;147:212–220. [Google Scholar]
- Docker MF. A review of the evolution of nonparasitism in lampreys and an update of the paired species concept. In: Brown LR, Chase SD, Mesa MG, Beamish RJ, Moyle PB, editors. Biology, Management, and Conservation of Lampreys in North America. Bethesda, Maryland: 2009. pp. 293–309. American Fisheries Society Symposium 72. [Google Scholar]
- Docker MF, Youson JH, Beamish RJ, Devlin RH. Phylogeny of the lamprey genus Lampetra inferred from mitochondrial cytochrome b and ND3 gene sequences. Canadian Journal of Fisheries and Aquatic Sciences. 1999;56:2340–2349. [Google Scholar]
- Docker M, Mandrak N, Heath D. Contemporary gene flow between “paired” silver (Ichthyomyzon unicuspis) and northern brook (I. fossor) lampreys: implications for conservation. Conservation Genetics. 2012;13:823–835. [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Espanhol R, Almeida PR, Alves MJ. Evolutionary history of lamprey paired species Lampetra fluviatilis (L.) and Lampetra planeri (Bloch) as inferred from mitochondrial DNA variation. Molecular Ecology. 2007;16:1909–1924. doi: 10.1111/j.1365-294X.2007.03279.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fine JM, Vrieze LA, Sorensen PW. Evidence that petromyzontid lampreys employ a common migratory pheromone that is partially comprised of bile acids. Journal of Chemical Ecology. 2004;30:2091–2110. doi: 10.1023/b:joec.0000048776.16091.b1. [DOI] [PubMed] [Google Scholar]
- Fryxell JM, Sinclair ARE. Causes and consequences of migration by large herbivores. Trends in Ecology & Evolution. 1988;3:237–241. doi: 10.1016/0169-5347(88)90166-8. [DOI] [PubMed] [Google Scholar]
- Gaigher A, Launey S, Lasne E, Besnard A-L, Evanno G. Characterization of thirteen microsatellite markers in river and brook lampreys (Lampetra fluviatilis and L. planeri. Conservation Genetics Resources. 2013;5:141–143. [Google Scholar]
- Gardiner R. 2003. Identifying lamprey. A field key for sea, river and brook lamprey. Conserving Natura 2000 rivers. Conservation techniques No. 4. English Nature, Peterborough.
- Gill HS, Renaud CB, Chapleau F, Mayden RL, Potter IC. Phylogeny of living parastic lampreys (Petromyzontiformes) based upon morphological data. Copeia. 2003;4:687–703. [Google Scholar]
- Goodman DH, Reid SB, Docker MF, Haas GR, Kinziger AP. Mitochondrial DNA evidence for high levels of gene flow among populations of a widely distributed anadromous lamprey Entosphenus tridentatus (Petromyzontidae) Journal of Fish Biology. 2008;72:400–417. [Google Scholar]
- Goodwin CE, Griffiths D, Dick JTA, Elwood RW. A freshwater-feeding Lampetra fluviatilis L. population in Lough Neagh, Northern Ireland. Journal of Fish Biology. 2006;68:628–633. [Google Scholar]
- Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Hardisty MW. A general introduction to lampreys. In: Holcík J, editor. The Freshwater Fishes of Europe. Part 1. Petromyzontiformes. Wiesbaden: Aula-Verlag; 1986a. pp. 19–84. [Google Scholar]
- Hardisty MW. Lampetra fluviatilis (Linnaeus, 1758) and Lampetra planeri (Bloch. 1784) In: Holcík J, editor. The Freshwater Fishes of Europe. Part 1. Petromyzontiformes. Wiesbaden: Aula-Verlag; 1986b. pp. 249–304. [Google Scholar]
- Hardisty MW, Potter IC. The behaviour, ecology and growth of larval lampreys. In: Hardisty MW, Potter IC, editors. The Biology of Lampreys. London: Academic Press; 1971a. pp. 83–126. [Google Scholar]
- Hardisty MW, Potter IC. The general biology of adult lampreys. In: Hardisty MW, Potter IC, editors. The Biology of Lampreys. London: Academic Press; 1971b. pp. 127–206. [Google Scholar]
- Hardisty MW, Potter IC. Paired species. In: Hardisty MW, Potter IC, editors. The Biology of Lampreys. London: Academic Press; 1971c. pp. 249–278. [Google Scholar]
- Harris LN, Taylor EB. Pleistocene glaciations and contemporary genetic diversity in a Beringian fish, the broad whitefish, Coregonus nasus (Pallas): inferences from microsatellite DNA variation. Journal of Evolutionary Biology. 2010;23:72–86. doi: 10.1111/j.1420-9101.2009.01858.x. [DOI] [PubMed] [Google Scholar]
- Hays JD, Imbrie J, Shackleton NJ. Variations in the earth's orbit: pacemaker of the ice ages. Science. 1976;194:1121–1132. doi: 10.1126/science.194.4270.1121. [DOI] [PubMed] [Google Scholar]
- Hess JE, Campbell NR, Close DA, Docker MF, Narum SR. Population genomics of Pacific lamprey: adaptive variation in a highly dispersive species. Molecular Ecology. 2013;22:2898–2916. doi: 10.1111/mec.12150. [DOI] [PubMed] [Google Scholar]
- Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- Ho SYW, Kolokotronis S-O, Allaby RG. Elevated substitution rates estimated from ancient DNA sequences. Biology Letters. 2007;3:702–705. doi: 10.1098/rsbl.2007.0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoarau G, Rijnsdorp AD, van der Veer HW, Stam WT, Olsen JL. Population structure of plaice (Pleuronectes platessa L.) in northern Europe: microsatellites revealed large-scale spatial and temporal homogeneity. Molecular Ecology. 2002;11:1165–1176. doi: 10.1046/j.1365-294x.2002.01515.x. [DOI] [PubMed] [Google Scholar]
- Hofreiter M, Stewart J. Ecological change, range fluctuations and population dynamics during the Pleistocene. Current biology. 2009;19:R584–R594. doi: 10.1016/j.cub.2009.06.030. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genetics. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert N, Hanner R, Holm E, et al. Identifying Canadian freshwater fishes through DNA barcodes. PLoS One. 2008;3:e2490. doi: 10.1371/journal.pone.0002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins RJ, Thompson A. Communal spawning of brook and river lampreys, Lampetra planeri Bloch and Lampetra fluviatilis L. Journal of Fish Biology. 1970;2:53–54. [Google Scholar]
- Hume JB, Adams CE, Mable B, Bean CW. Sneak male mating tactics between lampreys (Petromyzontiformes) exhibiting alternative life-history strategies. Journal of Fish Biology. 2013;82:1093–1100. doi: 10.1111/jfb.12047. [DOI] [PubMed] [Google Scholar]
- Inger R, McDonald RA, Rogowski D, et al. Do non-native invasive fish support elevated lamprey populations? Journal of Applied Ecology. 2010;47:121–129. [Google Scholar]
- Jahn AE, Levey DJ, Hostetler JA, Mamani AM. Determinants of partial bird migration in the Amazon Basin. Journal of Animal Ecology. 2010;79:983–992. doi: 10.1111/j.1365-2656.2010.01713.x. [DOI] [PubMed] [Google Scholar]
- Jones OR, Wang J. COLONY: a program for parentage and sibship inference from multilocus genotype data. Molecular Ecology Resources. 2010;10:551–555. doi: 10.1111/j.1755-0998.2009.02787.x. [DOI] [PubMed] [Google Scholar]
- Julian J, Armin P, Catherine EW, et al. River fragmentation increases localized population genetic structure and enhances asymmetry of dispersal in bullhead (Cottus gobio. Conservation Genetics. 2012;13:545–556. [Google Scholar]
- Kerr LA, Secor DH, Piccoli PM. Partial migration of fishes as exemplified by the estuarine-dependent white perch. Fisheries. 2009;34:114–123. [Google Scholar]
- Lang NJ, Roe KJ, Renaud CB. Novel relationships among lampreys (Petromyzontiformes) revealed by a taxonomically comprehensive molecular data set. In: Brown LR, Chase SD, Mesa MG, Beamish RJ, Moyle PB, et al., editors. Biology, Management, and Conservation of Lampreys in North America. Bethesda, Maryland: American Fishereies Society; 2009. pp. 41–56. [Google Scholar]
- Langerhans RB, Riesch R. Speciation by selection: a framework for understanding ecology's role in speciation. Current Zoology. 2013;59:31–52. [Google Scholar]
- Lasne E, Sabatié MR, Evanno G. Communal spawning of brook and river lampreys (Lampetra planeri and L. fluviatilis) is common in the Oir River (France) Ecology of Freshwater Fish. 2010;19:323–325. [Google Scholar]
- Latta RG. Conservation genetics as applied evolution: from genetic pattern to evolutionary process. Evolutionary Applications. 2008;1:84–94. doi: 10.1111/j.1752-4571.2007.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CE, Bell MA. Causes and consequences of recent freshwater invasions by saltwater animals. Trends in Ecology & Evolution. 1999;14:284–288. doi: 10.1016/S0169-5347(99)01596-7. [DOI] [PubMed] [Google Scholar]
- Lucas MC, Baras E. Migration of Freshwater Fishes. Oxford: Blackwell; 2001. [Google Scholar]
- Lucas MC, Bubb DH, Jang MH, Ha K, Masters JEG. Availability of and access to critical habitats in regulated rivers: effects of low-head barriers on threatened lampreys. Freshwater Biology. 2009;54:621–634. [Google Scholar]
- Lundberg P. The evolution of partial migration in birds. Trends in Ecology and Evolution. 1988;3:172–175. doi: 10.1016/0169-5347(88)90035-3. [DOI] [PubMed] [Google Scholar]
- Luzier CW, Docker MF, Whitesel TA. Characterization of ten microsatellite loci for western brook lamprey Lampetra richardsoni. Conservation Genetics Resources. 2010;2:71–74. [Google Scholar]
- Maitland PS, Morris KH, East K. The ecology of lampreys (Petromyzonidae) in the Loch-Lomond area. Hydrobiologia. 1994;290:105–120. [Google Scholar]
- Malmqvist B. The spawning migration of the brook lamprey, Lampetra planeri Bloch, in a south Swedish stream. Journal of Fish Biology. 1980;16:105–114. [Google Scholar]
- Malmqvist B. The feeding, breeding and population ecology of the brook lamprey (Lampetra planeri. Dissertation Abstracts International C European Abstracts. 1983;44:537. [Google Scholar]
- Mateus CS, Almeida PR, Quintella BR, Alves MJ. MtDNA markers reveal the existence of allopatric evolutionary lineages in the threatened lampreys Lampetra fluviatilis (L.) and Lampetra planeri (Bloch) in the Iberian glacial refugium. Conservation Genetics. 2011;12:1061–1074. [Google Scholar]
- Mateus CS, Rodríguez-Muñoz R, Quintella BR, Alves MJ, Almeida PR. Lampreys of the Iberian Peninsula: distribution, population status and conservation. Endangered Species Research. 2012;16:183–198. [Google Scholar]
- Mateus CS, Alves MJ, Quintella BR, Almeida PR. Three new cryptic species of the lamprey genus Lampetra Bonnaterre, 1788 (Petromyzontiformes: Petromyzontidae) from the Iberian Peninsula. Contributions to Zoology. 2013a;82:37–53. [Google Scholar]
- Mateus CS, Stange M, Berner D, et al. Strong genome-wide divergence between sympatric European river and brook lampreys. Current Biology. 2013b;23:R649–R650. doi: 10.1016/j.cub.2013.06.026. [DOI] [PubMed] [Google Scholar]
- McDowall RM. The evolution of diadromy in fishes (revisited) and its place in phylogenetic analysis. Reviews in Fish Biology and Fisheries. 1997;7:443–462. [Google Scholar]
- McDowall RM. Anadromy and homing: two life-history traits with adaptive synergies in salmonid fishes? Fish and Fisheries. 2001;2:78–85. [Google Scholar]
- McMillan WO, Palumbi SR. Rapid rate of control-region evolution in Pacific butterflyfishes (Chaetodontidae) Journal of Molecular Evolution. 1997;45:473–484. doi: 10.1007/pl00006252. [DOI] [PubMed] [Google Scholar]
- Meldgaard T, Nielsen E, Loeschcke V. Fragmentation by weirs in a riverine system: a study of genetic variation in time and space among populations of European grayling (Thymallus thymallus) in a Danish river system. Conservation Genetics. 2003;4:735–747. [Google Scholar]
- Morris KH. A multivariate morphometric and meristic description of a population of freshwater-feeding river lampreys, Lampetra fluviatilis (L), from Loch-Lomond, Scotland. Zoological Journal of the Linnean Society. 1989;96:357–371. [Google Scholar]
- Morrissey MB, de Kerckhove DT. The maintenance of genetic variation due to asymmetric gene flow in dendritic metapopulations. The American Naturalist. 2009;174:875–889. doi: 10.1086/648311. [DOI] [PubMed] [Google Scholar]
- Moser ML, Jackson AD, Lucas MC, Mueller RP. Behaviour and potential threats to survival of migrating lamprey ammocoetes and juveniles. Reviews in Fish Biology and Fisheries. 2015;25:103–116. [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nei M, Tajima F. DNA polymorphism detectable by restriction endonucleases. Genetics and Molecular Research. 1981;97:145–163. doi: 10.1093/genetics/97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson IC, Greenberg LA. Partial migration in a landlocked brown trout population. Journal of Fish Biology. 2004;65:106–121. [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Palkovacs EP, Dion KB, Post DM, Caccone A. Independent evolutionary origins of landlocked alewife populations and rapid parallel evolution of phenotypic traits. Molecular Ecology. 2008;17:582–597. doi: 10.1111/j.1365-294X.2007.03593.x. [DOI] [PubMed] [Google Scholar]
- Pereira AM, Robalo JI, Freyhof J, et al. Phylogeographical analysis reveals multiple conservation units in brook lampreys Lampetra planeri of Portuguese streams. Journal of Freshwater Ecology. 2010;77:311–434. doi: 10.1111/j.1095-8649.2010.02675.x. [DOI] [PubMed] [Google Scholar]
- Potter IC. Ecology of larval and metamorphosing lampreys. Canadian Journal of Fisheries and Aquatic Sciences. 1980;37:1641–1657. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeymaekers JAM, Raeymaekers D, Koizumi I, Geldof S, Volckaert FAM. Guidelines for restoring connectivity around water mills: a population genetic approach to the management of riverine fish. Journal of Applied Ecology. 2009;46:562–571. [Google Scholar]
- Rambaut A, Drummond A. 2007. Tracer, version 1.5. Available from http://beast.bio.ed.ac.uk/Tracer.
- Räsänen K, Hendry AP. Disentangling interactions between adaptive divergence and gene flow when ecology drives diversification. Ecology letters. 2008;11:624–636. doi: 10.1111/j.1461-0248.2008.01176.x. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Renaud CB. Lampreys of the World. An Annotated and Illustrated Catalogue of Lamprey Species Known to Date. Rome: FAO; 2011. FAO Species Catalogue for Fishery Purposes. No. 5. [Google Scholar]
- Renaud CB, Docker MF, Mandrak NE. Taxonomy, distribution, and conservation of lampreys in Canada. In: Brown LR, Chase SD, Mesa MG, Beamish RJ, Moyle PB, editors. Biology, Management, and Conservation of Lampreys in North America. Bethesda, Maryland: American Fishereies Society; 2009. pp. 71–114. [Google Scholar]
- Robinson BW, Parsons KJ. Changing times, spaces and faces: tests and implications of adaptive morphological plasticity in the fishes of northern postglacial lakes. Canadian Journal of Fisheries and Aquatic Sciences. 2002;59:1819–1833. [Google Scholar]
- Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Rousset F. Genepop'007: a complete reimplementation of the Genepop software for Windows and Linux. Molecular Ecology Resources. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Schluter D, Nagel LM. Parallel speciation by natural selection. The American Naturalist. 1995;146:292. [Google Scholar]
- Schreiber A, Engelhorn R. Population genetics of a cyclostome species pair, river lamprey (Lampetra fluviatilis L.) and brook lamprey (Lampetra planeri Bloch) Journal of Zoological Systematics and Evolutionary Research. 1998;36:85–99. [Google Scholar]
- Shaw AK, Couzin ID. Migration or residency? The evolution of movement behavior and information in seasonal environments. The American Naturalist. 2013;181:114–124. doi: 10.1086/668600. [DOI] [PubMed] [Google Scholar]
- Sjöberg K. River lamprey Lampetra fluviatilis (L.) fishing in the area around the Baltic Sea. Journal of Northern Studies. 2011;5:51–86. [Google Scholar]
- Spice EK, Goodman DH, Reid SB, Docker MF. Neither philopatric nor panmictic: microsatellite and mtDNA evidence suggests lack of natal homing but limits to dispersal in Pacific lamprey. Molecular Ecology. 2012;21:2916–2930. doi: 10.1111/j.1365-294X.2012.05585.x. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Fumagalli L, Wust S, Cosson JF. Comparative phylogeography and postglacial colonization routes in Europe. Molecular Ecology. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- Taylor EB. A review of local adaptation in Salmonidae, with particular reference to Pacific and Atlantic salmon. Aquaculture. 1991;98:185–207. [Google Scholar]
- Taylor EB, Foote CJ, Wood CC. Molecular genetic evidence for parallel life-history evolution within a Pacific salmon (Sockeye salmon and Kokanee, Oncorhynchus nerka. Evolution. 1996;50:401–416. doi: 10.1111/j.1558-5646.1996.tb04502.x. [DOI] [PubMed] [Google Scholar]
- Taylor EB, Harris LN, Spice EK, Docker MF. Microsatellite DNA analysis of parapatric lamprey (Entosphenus spp.) populations: implications for evolution, taxonomy and conservation of a Canadian endemic. Canadian Journal of Zoology. 2012;90:291–303. [Google Scholar]
- Templeton AR. The reality and importance of founder speciation in evolution. BioEssays. 2008;30:470–479. doi: 10.1002/bies.20745. [DOI] [PubMed] [Google Scholar]
- Waters JM, Wallis GP. Cladogenesis and the loss of the marine life-history phase in freshwater Galaxiid fishes (Osmeriformes: Galaxiidae) Evolution. 2001;55:587–597. doi: 10.1554/0014-3820(2001)055[0587:calotm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Webb T, Bartlein PJ. Global changes during the last 3 million years—climatic controls and biotic responses. Annual Review of Ecology and Systematics. 1992;23:141–173. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- White TA, Fotherby HA, Hoelzel AR. Nineteen new microsatellite loci for the blue hake (Antimoura rostrata. Conservation Genetics Resources. 2010;2:249–251. [Google Scholar]
- Whitlock MC, McCauley DE. Indirect measures of gene flow and migration: F ≠ 1/(4Nm + 1) Heredity. 1999;82:117–125. doi: 10.1038/sj.hdy.6884960. [DOI] [PubMed] [Google Scholar]
- Wilson GA, Rannala B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics. 2003;163:1177–1191. doi: 10.1093/genetics/163.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winker K. Evolution: migration and speciation. Nature. 2000;404:36. doi: 10.1038/35003651. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Morita K, Koizumi I, Maekawa K. Genetic differentiation of white-spotted charr (Salvelinus leucomaenis) populations after habitat fragmentation: spatial–temporal changes in gene frequencies. Conservation Genetics. 2004;5:529–538. [Google Scholar]
- Yamazaki Y, Goto A. Present status and perspectives on the phylogenetic systematics and speciation of lampreys. Japanese Journal of Ichthyology. 2000;47:1–28. [Google Scholar]
- Yamazaki Y, Yokoyama R, Nishida M, Goto A. Taxonomy and molecular phylogeny of Lethenteron lampreys in eastern Eurasia. Journal of Fish Biology. 2006;68:251–269. [Google Scholar]
- Yamazaki Y, Yokoyama R, Nagai T, Goto A. Formation of a fluvial non-parasitic population of Lethenteron camtschaticum as the first step in petromyzontid speciation. Journal of Fish Biology. 2011;79:2043–2059. doi: 10.1111/j.1095-8649.2011.03150.x. [DOI] [PubMed] [Google Scholar]
- Youson JH, Sower SA. Theory on the evolutionary history of lamprey metamorphosis: role of reproductive and thyroid axes. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2001;129:337–345. doi: 10.1016/s1096-4959(01)00341-4. [DOI] [PubMed] [Google Scholar]
- Zanandrea G. Speciation among lampreys. Nature. 1959;184:380. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Numbers collected and location of origin for all genetic samples of Lampetra planeri and Lampetra fluviatilis and by whom they were collected.
Table S2 Number of barriers (distance, km) between sampling locations.
Table S3 Details of microsatellite loci and primers utilised in the study.
Table S4 Table outlining diversity indices for microsatellite loci by population, and by locus.
Table S5 Pairwise FST values for 13 microsatellite loci.
Table S6 migrate-n analysis (Bayesian) showing posterior distributions for six paired sites (95% confidence intervals in brackets).
Table S7 Pairwise estimation of current gene flow, M, between (a) populations of anadromous Lampetra fluviatilis (Lf) and Lampetra planeri (Lp) from within the same river, and (b) populations within Loch Lomond by bayesass.
Fig. S1 Mismatch distribution (demographic expansion) with Tau 0.673, showing an expansion pattern for the six populations of Lampetra fluviatilis and Lampetra planeri presented in Table 1.
Fig. S2 structure bar plot generated from microsatellite data showing (a) ΔK = 2 (LnP(D) = −2294.8) where Swale Lp is compared to another Lampetra planeri population and Lampetra fluviatilis form the same river (b) ΔK = 2 when prior location information is used to analyse the Loch Lomond system which shows the freshwater-resident Lomond population to be differentiated and (c) ΔK = 5 (LnP(D) = −4681) when a location prior is used.
Fig. S3 Posterior probability of the data (Ln[P(D|K)]) and values of ΔK (Evanno et al. 2005) as a function of K (number of clusters), associated with the results shown in Fig. 4 in the main text.
Fig. S4 (a) Structure for 12 populations where ΔK = 2. (b) Structure for Lampetra planeri only where ΔK = 4. (c) Structure for all samples included when K = 10 (Lf = Lampetra fluviatilis, Lp = L. planeri, LL = Loch Lomond; anad = anadromous; res = resident). (d) ΔK plot for the structure plot in part c.
Fig. S5 FCA analysis for (a) Lampetra fluviatilis and (b) Lampetra planeri population.
Data Availability Statement
DNA sequences: GenBank accessions: KP722176–KP722191; microsatellite DNA genotypes and mtDNA sequence alignment file: Dryad doi:10.5061/dryad.v105s.