Abstract
Herpesviruses exemplified by herpes simplex virus-1 (HSV-1) attach to cell surface heparan sulfate (HS) for entry into host cells. However, during a productive infection the HS moieties on parent cells can trap newly exiting viral progenies and inhibit their release. Here, we demonstrate that a HS-degrading enzyme of the host, heparanase (HPSE), is upregulated through NF-kB and translocated to the cell surface upon HSV-1 infection for the removal of HS to facilitate viral release. We also find a significant increase in HPSE release in vivo during infection of murine corneas and that knockdown of HPSE in vivo inhibits virus shedding. Overall, we propose that HPSE acts as a molecular switch for turning a virus-permissive “attachment mode” of host cells to a virus-deterring “detachment mode”. Since many human viruses use HS as an attachment receptor, the HPSE-HS interplay may delineate a common mechanism for virus release.
Introduction
Herpesviruses are among the most prevalent human infections worldwide, with almost all eight viruses infecting a majority of the world population [1]. Among a number of other pathologies, HSV-1 is known to cause distressing infection of the human cornea, with HSV keratitis as the leading cause of infectious blindness in developed nations. Although acyclovir and its analogs are currently used for treatment of active herpes infections, these drugs fail to prevent permanent establishment of viral latency, allowing the virus to reactivate and cause clinical disease at a later time [2]. In order to generate more effective therapies, there is a critical need to understand the mechanism of viral spread to other cells and parts of the body after initial inoculation.
The mechanism of HSV egress from cells largely remains a mystery, but it is known that HSV-1 uses heparan sulfate (HS) as an attachment as well as an entry receptor for infection of host cells [1,3]. HS is an evolutionarily conserved glycosaminoglycan present ubiquitously at the cell surface and extracellular matrix (ECM) of a wide range of cell types [4]. Viral glycoproteins gB, gC, and gD are primarily responsible for viral binding to HS chains that are present on host cell surfaces as part of many proteoglycans including syndecans [1,5–6]. During viral egress, however, this important interaction has not been studied. Viral progenies have functional gB, gC, and gD, and interactions of these glycoproteins with HS on parent cells during egress could significantly hamper their release. In this paper, we provide novel evidence that surface HS levels of infected cells are dramatically decreased by the infection-induced host enzyme heparanase-1 (HPSE). In normal cell physiology, HPSE plays a homeostatic role in regulating the turnover of cell-associated HS [7].
Here, we show that the upregulation of HPSE upon infection serves as a means for newly produced virions to avoid re-attachment to HS and re-entry into parent cells, thus increasing viral spread. Our evidence suggests that unlike influenza viruses, which encode neuraminidase for the removal of their attachment receptor during exit, herpesviruses are supported by a host enzyme for this key step in their life cycle.
Results
Loss of HS from cell surface after infection
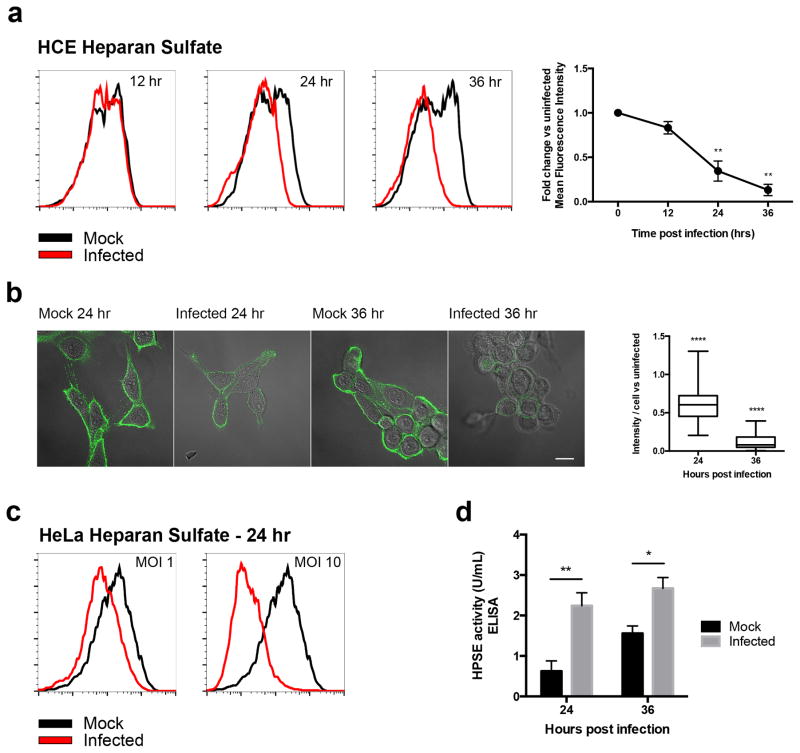
During our quest to develop a new intervention against HSV infection, we attempted to use our newly discovered anti-HS peptides for preferentially targeting infected cells, and examined HS levels on the host cell surface [5]. We have previously shown that HS expression is increased during the initial stages of HSV-1 infection, and that this initial increase in HS expression enhances virus attachment to cells [6]. Quite unexpectedly, when we extended our study to longer time points, a dramatic decrease in HS was seen during later stages of infection (24 and 36 hrs). Both flow cytometry and immunofluorescence microscopy results demonstrated a continuous loss of HS from the surface of human corneal epithelial (HCE) cells, using HS antibody 10E4, US Biological (Fig. 1a, 1b). This phenomenon was also observed in HeLa cells (Fig. 1c). To examine the cause for decreased HS expression, an ELISA kit was used to measure HS degradation after infection. At 24 and 36 hours post infection (hpi), HS in infected cells was degraded at higher rates than in uninfected cells (Fig. 1d). This result implicated the only known mammalian HS-degrading enzyme, heparanase (HPSE), in the loss of HS seen late during HSV-1 infection.
Figure 1. Loss of HS from cell surface after infection.
a. Left- HS expression is decreased on HCE cell surface at different time points post-infection, measured by flow cytometry. Cells were infected with KOS-WT at MOI 0.1 and were stained for HS at 12, 24, and 36 hrs post-infection (hpi) with HS Antibody 10E4. Right- Fluorescence intensity measurements based on flow cytometry results. Integrated mean fluorescence intensity of the whole population was measured and fold change was normalized to uninfected mock samples for each time point.
b. Left- Representative immunofluorescence microscopy images show decrease in HS expression on HCE cell surface of 24 and 36 hpi samples when compared to mock images. Cells were infected with KOS-WT at MOI 0.1 and were stained with HS Antibody 10E4. Green represents HS expression. Images are merged with brightfield to show cell borders. Scale bar 10 μm. Right- Images were quantified for HS intensity. Eighty cells were picked at random from each sample and intensity of HS was measured for each cell using IMAGE J.
c. HS expression is decreased on HeLa cell surface after infection, measured by flow cytometry. Cells were infected with KOS-WT at MOIs of 1 and 10 for 24 hrs, and then stained for HS using HS antibody 10E4.
d. ELISA measurement of HPSE activity indicates increased degradation of HS upon infection. Infected cells showed more HS degradation activity when compared to uninfected cells at each time point. Increased HPSE activity was observed in uninfected cells at 36 hrs due to expansion of the number of cells in culture; the assay provides a measurement of the total amount of HPSE produced rather than on a per cell basis. All data are presented as mean ± s.e.m. of three independent experiments (n=3). Asterisks denote a significant difference as determined by Student’s t-test; *P<0.05, **P<0.01, ****P<0.0001.
HPSE is upregulated after HSV-1 infection
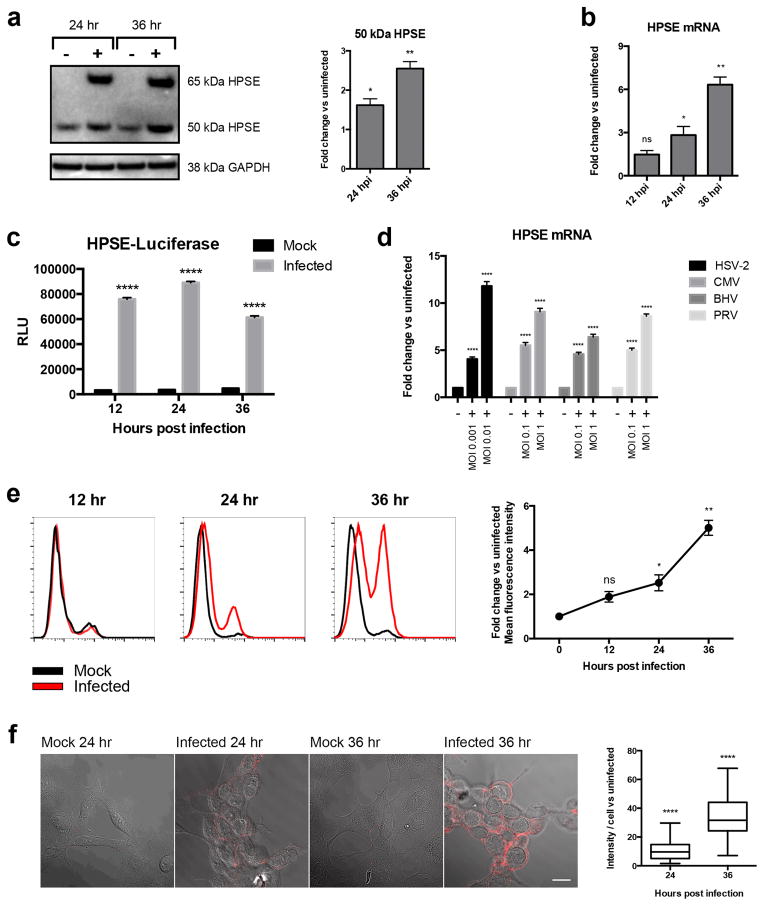
Since HS loss was increased after infection, we investigated HPSE expression in infected cells. Western blot analysis revealed that expression of active HPSE (50 kDa) was significantly greater at 24 and 36 hpi than in uninfected cells (Fig. 2a). Quantitative PCR (qPCR) revealed that HPSE mRNA was significantly elevated at 24 and 36 hpi (Fig. 2b). Furthermore, when HCE cells were transfected with HPSE promoter fused to luciferase reporter, an increase in luciferase expression was observed at 12, 24, and 36 hpi, when compared to uninfected mock-treated cells (Fig. 2c). We thus concluded that HPSE expression is regulated at the transcriptional level, with an increase in active HPSE protein produced upon HSV-1 infection. Interestingly, we observed similar increases in HPSE transcript levels (Fig. 2d) and protein expression levels (Supplementary Fig. 1a) after infection of HCE cells with other herpesviruses, namely herpes simplex virus type-2 (HSV-2), cytomegalovirus (CMV), bovine herpes virus (BHV) and pseudorabies virus (PRV). In addition, an increase in HPSE was observed in various cell types when infected with HSV-1 KOS-WT (Supplementary Fig. 1b). HPSE upregulation seems to be a phenomenon common to multiple herpes viral infections and various cell types. As HPSE expression is increased inside the cells, we asked if HPSE is translocated to the surface, the primary location of its substrate HS. Complementary to our HS expression results, a significant increase in HPSE was observed by flow cytometry and immunofluorescence microscopy at 24 and 36 hpi (Fig. 2e, 2f). It is known that both active HPSE (50 kDa) and inactive HPSE (65 kDa) can be found on the cell surface [7]. By isolating cell surface proteins and probing for HPSE by western blot, we observed that only active 50 kDa HPSE is present at the cell surface at 24 and 36 hpi (Supplementary Fig. 2a). Together, these results suggest that infected cells upregulate active HPSE, which is then translocated to the cell surface, producing the decreased HS expression observed on the cell surface after infection.
Figure 2. HPSE is upregulated after HSV-1 infection.
a. Left- Western blot shows increased HPSE expression of latent (65kDa) and active (50kDa) after 24 and 36 hr infection of HCE cells at MOI 0.1. Right- Densitometry quantification of 50 kDa active HPSE normalized to GAPDH. Fold change of infected over uninfected cells is shown for each time point.
b. Increase in HPSE transcripts in HCE cells after infection with KOS-WT at MOI 0.1. Results are shown as fold change of infected over uninfected cells at each time point, normalized to GAPDH.
c. Increase in promoter activity of HPSE gene upon infection in HCE cells. KOS-WT was used to infect cells at MOI 0.1 for 12, 24, and 36 hrs. Shown is the average fold increase over uninfected control. Experimental values are normalized to those obtained with pGL3 as a control for transfection efficiency.
d. Increase in HPSE transcripts when cells were infected with herpes simplex virus type-2 (HSV-2), cytomegalovirus (CMV), bovine herpes virus (BHV), and pseudorabies virus (PRV). HCE cells were used for HSV-2, BHV, and PRV infections whereas primary human foreskin fibroblasts (HFF) were used for CMV infections. HCE cells were infected for 24 hrs. HFF were infected with CMV for 7 days at indicated MOIs. Results are shown as fold change of infected over uninfected cells at each time point, normalized to GAPDH.
e. Left- HPSE expression is increased on cell surface after infection, measured by flow cytometry. HCE cells were infected with KOS-WT at MOI 0.1 and were stained with HPSE antibody HP130. Right- Integrated mean fluorescence intensity of the whole population was measured and fold change was normalized to mock at each time point.
f. Left- Immunofluorescence microscopy images show increased HPSE expression on surface of infected HCE cells. Red represents HPSE expression. Images are merged with brightfield to show cell borders. Scale bar 10 μm. Right- Images were quantified for HPSE intensity. Eighty cells were picked at random for each sample and intensity of HPSE was measured for each cell using IMAGE J. All data are presented as mean ± s.e.m. of three independent experiments (n=3). Asterisks denote a significant difference as determined by Student’s t-test; *P<0.05, **P<0.01, ****P<0.0001, ns, not significant. See Supplementary Figure 8 for full-length images of blots.
Mechanism of HPSE upregulation upon infection
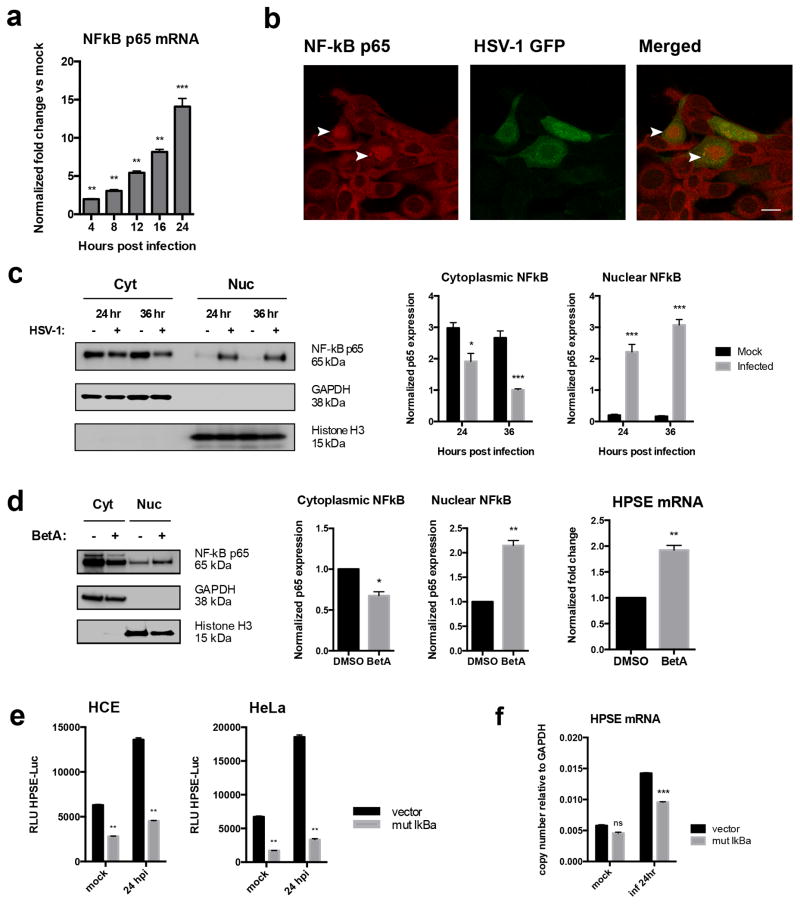
In our search for the mechanism of HPSE upregulation upon infection, we learned from prior studies that a host factor, nuclear factor (NF)-kB, can transcriptionally increase HPSE expression [8–11]. A number of reports have also described an increased expression of the transcription factor NF-kB after HSV-1 infection [12–15]. Therefore, while studying a possible connection between NF-kB and HPSE up-regulation during HSV-1 infection, we also observed a steady increase in NF-kB p65 levels (Fig. 3a), and increased nuclear translocation of the p65 subunit at 24 and 36 hpi (Fig. 3b, 3c). Thus, based on previous reports [8–11] and our observations, it is likely that NF-kB activation during infection plays a role in HPSE induction. Adding support to this possibility, a similar phenomenon was reproduced when uninfected HCE cells were treated with an NF-kB inducer, Betulinic acid (BetA) [16]. Quite evidently, NF-kB nuclear translocation and more importantly, HPSE mRNA upregulation were observed (Fig. 3d). The role of NF-kB was further evaluated by overexpressing a plasmid encoding mutant IkBa (S32A/S36A) in HCE cells. This dominant-negative mutant is incapable of being phosphorylated and degraded [17]. Clearly, expression of the IkBa mutant inhibited NF-kB activation and its translocation to the nucleus, and we noted a decrease in HPSE expression by luciferase assay and qPCR (Fig. 3e, 3f). Collectively, our results indicate that NF-kB or one of its downstream effectors is directly involved in regulation of HPSE expression. In parallel studies, to probe any direct involvement of viral proteins in HPSE upregulation, we examined several known transcriptional regulators encoded in the HSV genome. HCE cells were separately transfected with expression plasmids for ICP0, ICP4, ICP22, and VP16, incubated for 24 hrs, and harvested for RNA isolation. No significant change in HPSE expression was observed by qPCR (Supplementary Fig. 3a). Together, these findings suggest that the upregulation of HPSE is likely a host response to infection that the virus takes advantage of to enhance its release from the parent cell.
Figure 3. Mechanism of HPSE upregulation upon infection.
a. Increase in NF-kB p65 transcript levels after infection with KOS-WT at MOI 0.1 at various time points. Results are normalized to GAPDH and reported as fold change of infected over uninfected cells.
b. Nuclear translocation of NF-kB p65 upon HSV-1 infection, observed by immunofluorescence microscopy. HCE cells were infected with GFP-KOS at MOI 0.1 for 12 hrs. Translocation of NF-kB to the nucleus can be seen in GFP-positive cells (arrowheads). Scale bar 10 μm.
c. Nuclear translocation of NF-kB p65 upon HSV-1 infection measured by western blot shows an increase in NF-kB translocation from cytoplasm to nucleus. Left- HCE cells were infected with KOS-WT at MOI 0.1 for 24 and 36 hrs, and nuclear/cytoplasmic extractions were performed. Right- Densitometry quantification of NF-kB p65 in cytoplasm and nucleus, normalized to GAPDH and Histone H3, respectively.
d. Induction of NF-kB activation by Betulinic acid (BetA) increases HPSE mRNA. Left- HCE cells were treated with BetA at 10 μg/mL or DMSO vehicle for 24 hrs. NF-kB translocation into nucleus with BetA treatment is shown in HCE cells. Also shown is densitometry quantification of NF-kB p65 in cytoplasm and nucleus, normalized to GAPDH and Histone H3, respectively. Right- HPSE mRNA is increased with BetA treatment of HCE cells.
e. Inhibition of NF-kB activation results in decreased HPSE promoter activity. HCE cells (left) and HeLa cells (right) were transfected with mutant IkBa incapable of degradation (S32A/S36A), thereby specifically inhibiting NF-kB activation and nuclear translocation. Cells were infected with KOS-WT at MOI 0.1 for 24 hrs. Cell lysates were isolated, and luciferase assay was performed. Results shown are normalized to empty pGL3 vector as a control for transfection efficiency.
f. Inhibition of NF-kB activation results in decreased HPSE transcript levels upon infection. HCE cells were transfected with mutant IkBa as described above, and infected with KOS-WT at MOI 0.1 for 24 hrs. All data are presented as mean ± s.e.m. of three independent experiments (n=3). Asterisks denote a significant difference as determined by Student’s t-test; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, ns, not significant. See Supplementary Figure 8 for full-length images of blots.
Functional phenotype of HPSE on viral egress and attachment
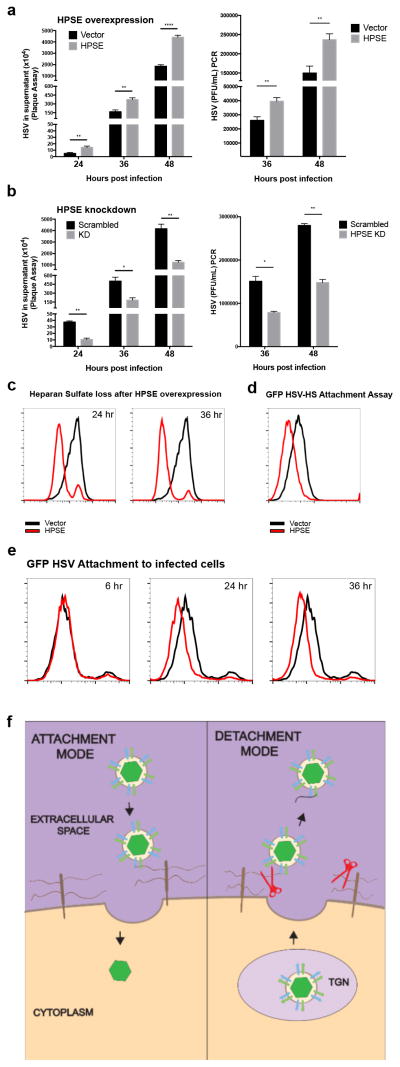
To elucidate the role of higher HPSE production and associated enzymatic activity in viral egress, we transfected HCE cells with a HPSE expression plasmid and quantified the virus particles released in the cell culture supernatants at different time points by a plaque assay and viral DNA PCR. It was evident that viral release was increased in HPSE-transfected cells at 24, 36, and 48 hpi (Fig. 4a). To verify that the DNA PCR assay is suitable to detect viral titer, a standard curve was calculated (Supplementary Fig. 4a). Conversely, using the same assays we observed a loss of virus in the supernatant when HPSE was knocked down using lentiviral shRNA constructs (Fig. 4b). Loss of HPSE expression in knockdown cells is shown in Supplementary Fig. 5a. Furthermore, in support of a direct role in egress, we found that active, functional HPSE is required for efficient viral egress, as transfection of various nonfunctional HPSE mutants produced a defect in viral release (Supplementary Fig. 6a). According to our proposed mechanism, active HPSE is overexpressed inside cells and is translocated to the cell surface during viral egress to cleave HS, which facilitates increased viral shedding from cells. To prove that higher HPSE expression is the cause of HS loss at the surface, we transfected HCE cells with HPSE plasmid in the absence of any infection and found that HS levels start to decrease at 24 hrs post transfection (Fig. 4c). To delve deeper into the role of HPSE activity in viral release, we performed a viral attachment assay, which confirmed that a loss in HS decreases viral binding at the surface (Fig. 4d). Next, to further understand HSV egress under physiological conditions, we tested binding of exogenous GFP HSV-1 to cells that had already been infected with wildtype virus. We hypothesized that if HS levels decrease during the later stages of infection, any exogenous virions added to already infected cells should demonstrate a loss of binding to the surface. Indeed, compared to uninfected cells, the binding of exogenous GFP virus to the surface of wildtype virus infected cells (24 and 36 hpi) was decreased (Fig. 4e). Together these results suggest that HSV-1 infected cells become less prone to virus attachment as they start to release their progeny virions. Given our findings of an inverse relationship between HS and HPSE expression, we propose that HPSE is a switch needed to convert the cell from a virus permissive “attachment mode” to a virus deterring “detachment mode” during the productive period of infection (Fig. 4f).
Figure 4. Functional phenotype of HPSE on viral egress and attachment.
a. Viral egress is increased in HPSE overexpressing cells. Left- HCE cells were infected with KOS-WT at MOI 0.1 24 hrs after transfection with HPSE or control vector. Supernatant was collected at indicated time points post-infection and virus was quantified by titering on Vero cells in a plaque assay. Right- HCE cells were transfected and infected as mentioned above and culture supernatant containing viruses was collected and quantified by PCR using primers against glycoprotein-D (gD).
b. Viral egress is decreased in HPSE deficient cells. Stably transduced HCE cell lines were generated using lentiviral HPSE shRNA and scrambled shRNA constructs, and these were then infected with KOS-WT at MOI 0.1. As described above, supernatant was collected to perform plaque assay (left) and viral DNA was quantified by PCR for gD (right).
c. HS is lost from the cell surface upon HPSE overexpression. HCE cells were transfected with HPSE plasmid or control vector, incubated at 37°C for 24 hrs. Cells were then stained for HS using Antibody 10E4 at 24 and 36 hpi and analyzed by flow cytometry.
d. Decreased attachment of HSV-1 after HPSE overexpression. HCE cells were transfected with HPSE plasmid or control vector, and 24 hrs later incubated with GFP-KOS for 2 hrs at 4°C to allow viral binding. GFP fluorescence intensity from the virus was measured by flow cytometry.
e. Decreased attachment of GFP-KOS after previous infection with KOS-WT at specified time points. HCE cells were infected with KOS-WT for 6, 24, or 36 hrs, then incubated with GFP-KOS for 2 hrs at 4°C to allow viral binding. GFP fluorescence intensity from the virus was measured by flow cytometry. Black curve represents 0 hr time point.
f. Model of HSV egress, showing role of HPSE at the cell surface. This model shows the attachment and detachment modes of the HSV lifecycle. During attachment, virus is bound to HS. However, during detachment HPSE (red) is upregulated and translocated to the cell surface, cleaving HS during viral exit. All data are presented as mean ± s.e.m. of three independent experiments (n=3). Asterisks denote a significant difference as determined by Student’s t-test; *P<0.05, **P<0.01, ****P<0.0001.
Significance of HPSE and HSV egress in vivo
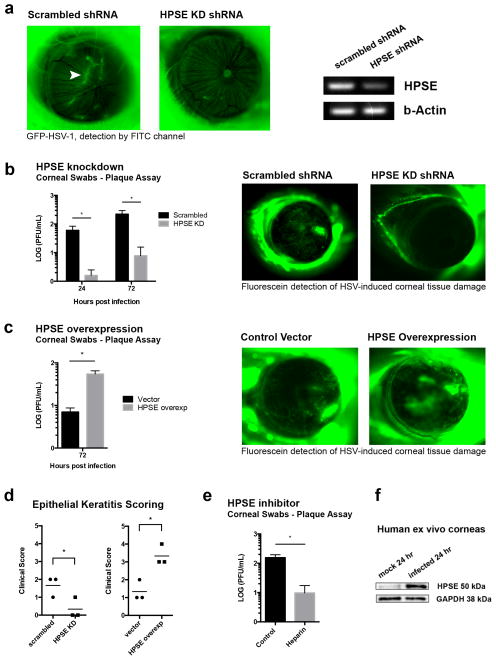
Mice corneas were injected with vectors containing shRNA against murine HPSE or scrambled sequence, and then infected with GFP HSV. Decreased viral spread through the cornea was observed with HPSE knockdown (Fig. 5a). Swabs collected from knockdown corneas at 24 and 72 hpi produced fewer plaques when compared to eyes with scrambled shRNA (Fig. 5b, left). This suggests that HPSE activity promotes viral egress in the epithelium of the cornea. Furthermore, using fluorescein dye we detected decreased tissue damage in HPSE knockdown corneas infected with wildtype virus, as compared to corneas of mice that received scrambled sequence (Fig. 5b, right). Conversely, when mice eyes were transfected with HPSE and then infected with HSV-1, more virus was present as measured by corneal swabs (Fig. 5c, left). Also, these HPSE-overexpressing corneas displayed increased corneal ulceration and tissue damage when compared to empty vector transfected corneas (Fig. 5c, right). To further establish HPSE’s significance in disease, the four aforementioned groups of mice were observed by slit lamp biomicroscope 7 days post ocular infection and assigned clinical scores according to pre-established criteria [18]. HPSE knockdown produced fewer signs of keratitis, while HPSE overexpression resulted in exaggerated disease (Fig. 5d). Additionally, treating mice with heparin, a HPSE inhibitor, resulted in less virus present in corneal swabs (Fig. 5e). Together these results indicate that HPSE activity promotes viral egress in the corneal epithelium, contributing to the tissue damage characteristic of herpetic keratitis. Lastly, we aimed to determine if HSV could upregulate HPSE expression in human tissue. Epithelial cells of human donor corneoscleral buttons infected with HSV showed increased levels of active HPSE (Fig. 5f). This suggests that the virus-HPSE association is relevant in human pathogenesis as well.
Figure 5. Significance of HPSE and HSV egress in vivo.
a. Knockdown of HPSE in mice eyes decreases viral spread through cornea. Mice corneas were injected with shRNA against HPSE or with scrambled shRNA and infected with GFP-KOS. Left- Fluorescence detection 72 hrs post-infection. Arrowhead indicates GFP-HSV. Right- Knockdown of HPSE in mouse cornea demonstrated by reverse transcriptase PCR.
b. Left- Corneal eye swabs collected at 24 and 72 hpi for plaque assays show decreased HSV titer in HPSE knockdown corneas (n=5 for each group). Right- Decreased corneal tissue damage in HPSE knockdown mice corneas after HSV-1 infection. HPSE was knocked down in mice corneas as described above, and 7 days post-infection with KOS-WT, fluorescein was applied to mice eyes to highlight corneal damage.
c. Left- Mice corneas were transfected with HPSE plasmid in corneal epithelium and infected with KOS-WT. Corneal swabs collected at 72 hpi for plaque assays show an increase in HSV titer in cornea transfected with HPSE plasmid (n=3 for each group). Right- Increased corneal tissue damage in HPSE overexpressing mouse corneas after HSV-1 infection. Fluorescein was applied to mice eyes to highlight tissue damage 7 days post-infection.
d. Seven days post ocular infection, mouse corneas were examined for tissue damage by slit lamp biomicroscope and scored. Left- Decreased tissue damage in HPSE shRNA knockdown corneas when compared to scrambled shRNA controls. Right- Increased tissue damage in HPSE overexpressed corneas when compared to vector transfected controls (n=3 for each group).
e. Decreased viral egress after heparin treatment, measured by plaque assay. Mice corneas were infected with KOS-WT at 105 PFU. After infection, 5 μL of 25 μg/μL heparin solution was applied directly to corneas at 24 and 48 hpi and corneal swabs for plaque assay were collected at 72 hpi.
f. Human corneo-scleral buttons from the same subject were cultured and infected with KOS-WT at 105 pfu. The epithelium was extracted at 24 hpi and an increase in HSPE (50kDa) was observed. The experiment was repeated three times and representative western blot is shown. All data are presented as mean ± s.e.m. of three independent experiments (n=3). Asterisks denote a significant difference as determined by Student’s t-test; *P<0.05. See Supplementary Figure 8 for full-length images of gels and blots.
Discussion
HS is a ubiquitously expressed glycosaminoglycan that is used as a docking site by HSV-1. Glycoproteins gB, gC, and gD mediate this initial contact of virus with host cell HS, which is followed by fusion, entry, and eventually production of viral progeny [1]. These newly formed viral progeny have functional glycoproteins that could interact with HS during egress. Influenza virus also faces a similar predicament in which viral haemagglutinin binds to host sialic acid chains during egress. To avoid being trapped by parent host cells, these chains are cleaved with the help of the enzyme neuraminidase, which is encoded in the genome of the influenza virus [19]. In contrast, HSV-1 is not known to encode any enzymes in its genome that cleave HS chains. Hence, HSV and other herpesviruses could have found support from a host-derived enzyme, namely HPSE, to perform the task of cleaving HS chains.
In addition to the cleavage of HS during infection, it is possible that entire proteoglycans are also being shed. It has been described previously that increased HPSE activity results in an increased rate of syndecan-1 shedding on the cell surface [20]. In this study, using slot blot of cell culture supernatants, we have observed a moderate increase in syndecan-1 shedding after HSV-1 infection (Supplementary Fig. 7a). Although the exact mechanism is not known, shedding of syndecan-1 due to HPSE could be attributable to HPSE-induced upregulation of host cell proteases such as MMP-9 [21]. In this way, HPSE could play a dual role in viral egress by cleaving HS and the proteoglycan to which it is attached. However, we believe that the HPSE-HS association plays the major role in viral egress because viral dispersion is limited when bound to an entire proteoglycan rather than small HS chains. Additionally, any shedding of syndecan proteoglycans probably plays a relatively minor role during viral egress since the bulky proteoglycan core would likely need to be removed prior to progeny entry into neighboring cells. Another proteoglycan that plays a minor role in viral entry is chondroitin sulfate (CSPG) [22]. However, since the vast majority of literature shows that HSPGs are the primary receptors when compared to CSPGs, our current study is focused on the former. Future studies will aim to elucidate the mechanism of egress in relation to CSPGs.
With the new evidence from this study, we propose two distinct modes in the life of infected cells, viral attachment and detachment modes, which are regulated by the host enzyme heparanase (Fig. 4f). During the attachment mode, HSV glycoproteins bind to HS at the cell surface, an interaction that facilitates viral entry. With the upregulation of HPSE upon infection, which coincides well with viral maturation and egress phases, the host cell transforms into detachment mode by translocating HPSE to the plasma membrane, removing HS from the surface, and aiding in viral exit. Quite notably, HPSE is found to have 4–5 times increased enzymatic activity in an acidic environment of pH 6.4 when compared to pH 7.2 [23]. Interestingly, the trans-Golgi network (TGN) also has a low pH value of 5.91 [24]. Hence, when the TGN or TGN-derived vesicles fuse with the plasma membrane, it creates an acidic microenvironment promoting activation of HPSE, which in the case of infected cells can specifically increase the effectiveness of viral egress. All such events discussed above allow HPSE to act efficiently as a switch in infected cells, making them less prone to secondary infection by aggressive HS cleavage.
Herpes viral glycoproteins use other cell surface receptors for entry, the most common of which being nectin-1. However, it has been shown that expression of nectin-1 is diminished before cell death in neuronal cells of infected mice and human corneal epithelial cells during egress [25–26]. It is likely, though not well studied, that the surface expression of other HSV entry receptors is also diminished with infection. A more significant problem, therefore, is likely posed by the presence of HS on cell surfaces. Given the high abundance on cell surfaces and presence of highly charged groups, HS proteoglycans are considered most important for virus attachment to cells [1], and are hence most likely to trap egressing virions. Adding further complexity, a single HS chain can bind multiple viral ligands. As an example, HS chains on human HEp-2 cells are on average 105 kDa in apparent molecular mass, which corresponds to 420 sugar residues per chain or a chain length of 190 nm. Thousands of such chains may be present on the surface of single cells. Hence, for the entire length of the proteoglycan, there exist multiple chances for it to be trapped by HS on the same cell [26–27]. Therefore, removal of HS moieties is likely to clear a big hurdle in viral release.
HS proteoglycans are ubiquitously expressed glycosaminoglycans and are used by many viruses such as adeno-associated viruses [28], dengue viruses [29], hepatitis-C virus [30], foot-and-mouth disease virus [31], vaccinia virus [32], yellow fever virus [33], sindbis virus [34], HIV [35], papilloma virus [36], cytomegalovirus [37], human respiratory syncytial virus [38], and virtually all herpesviruses [1] for initial attachment to cells [28–39]. It is thus reasonable to assume that HPSE activity could play a role in the pathogenesis of these viruses as well. In fact, it has been reported that HPSE expression was significantly higher in Hepatitis C virus (HCV) related hepatocellular carcinoma (HCC) patients when compared to HCV-negative HCC patient population [40]. It is likely that future studies will directly link heparanase activity to the release of virtually all viruses that rely on HS for attachment to their host cells. Interestingly, in an unrelated study, a potent heparanase inhibitor that is currently in human clinical trials for cancer treatment, PI-88, successfully decreased cell-to-cell spread of HSV-1 and HSV-2 [41].
In ocular infection, distressing lesions caused by HSV-1 infection can result in vision impairment and blindness resulting from a chronic inflammatory reaction in the cornea causing neovascularization by limbal blood vessels [42]. Recent studies have implicated HPSE overexpression in multiple pathologic processes, including inflammation, angiogenesis, tumor metastasis, and atherosclerosis [7,43–46]. By breaking down epithelial and endothelial basement membranes, HPSE produces increased vascular permeability and leukocyte extravasation [47], and liberates HS-bound cytokines and growth factors normally sequestered in the ECM, making them available for angiogenesis and inflammation in the surrounding area [7,45,48–50]. Vision loss from HSV keratitis is predominantly due to corneal clouding caused by inflammation. In light of these recent findings, heparanase may be a unique therapeutic target since heparanase inhibitors could reduce viral release and control the host inflammatory response.
Methods
Cells and viruses
Human corneal epithelial cell line (RCB1834 HCE-T) was obtained from Kozaburo Hayashi (National Eye Institute, Bethesda, MD) and was cultured in MEM (Life Technologies, Carlsbad, CA) with 10% Fetal Bovine Serum (FBS) and 1% penicillin/streptomycin. For convenience this cell line is referred to as HCE cells throughout the paper. All infections were done with HSV-1 KOS-WT at MOI of 0.1 on HCE cells unless mentioned otherwise. Primary human foreskin fibroblasts for CMV infections were provided by Dr. Nell Lurain (Rush University, Chicago, IL). The African green monkey kidney (Vero), HEK293T, HeLa, and mouse embryonic fibroblast (MEF) cell lines were obtained from Dr. Patricia G. Spear (Northwestern University, Chicago, IL) and cultured in DMEM (Life Technologies) with 10% FBS and 1% penicillin/streptomycin. Betulinic Acid was purchased from R&D Systems (Minneapolis, MN) and was used at a concentration of 10 μg/mL in DMSO to induce NF-kB activation in cell cultures.
The viruses used in this study, HSV-1 (KOS-WT) [51], GFP-KOS [52], PRV [53], and BHV [54] were provided by Dr. Patricia G. Spear (Northwestern University, Chicago, IL). CMV strain AD169 [55] was provided by Dr. Nell Lurain (Rush University Medical Center, Chicago, IL). All virus stocks except for CMV were propagated in Vero cells and stored at −80°C. CMV was propagated in human foreskin fibroblast cells.
Antibodies and plasmids
Heparanase antibody HP130 (Advanced Targeting Systems, San Diego, CA) was used for western blot (1:200), imaging (1:100) and flow cytometry studies (1:100). Histone H3 antibody (Cell Signaling Technology, Danvers, MA) was used at a dilution of 1:2000 for western blot. Anti-human heparan sulfate mAb 10E4 (US Biological, Salem, MA) was used for flow cytometry (1:50) and cell imaging (1:100). NF-kB p65 antibody C-20 (Santa Cruz Biotechnology) was used for imaging (1:500) and western blot (1:2000). GAPDH (Santa Cruz Biotechnology) was used for western blot at dilution 1:2000. Na+/K+-ATPase (Cell Signaling) was used for western blot at dilution 1:20,000.
Human heparanase expression plasmid pIRES2 EGFP-HPSE1 (HPSE) and the control empty vector pIRES2 EGFP plasmid [56] were provided by Dr. Ralph Sanderson (University of Alabama at Birmingham, Birmingham, AL). Mutant IkBa (S32/36A) plasmid [17] was provided by Dr. Michael Karin (University of California, San Diego, La Jolla, CA). Expression constructs for viral transcriptional regulators ICP0 [57], and VP16 were obtained from Dr. Bernard Roizman (University of Chicago, Chicago, IL). VP16 plasmid was kindly prepared by Grace Zhou. VP16 coding sequence driven by CMV promoter was inserted in MCS of pcDNA 3.1(+). The whole fragment was flanked by VP16 gene flanking sequences for recombination purposes. ICP4 plasmid [58] was obtained from Dr. Neal DeLuca (University of Pittsburg, PA). ICP22 plasmid was kindly provided by Dr. Kumar Penmetcha (National institute of Advanced Industrial Science and Technology, Ibaraki, Japan). ICP22 coding sequence driven by CMV promoter was inserted in MCS of pcDNA 3.1(+). The whole fragment was flanked by ICP22 gene flanking sequences for recombination purposes. HPSE expression constructs GS-3, TIMB, C-domain, and Delta 17 were a kind gift from Dr. Israel Vlodavsky (Rappaport Institute, Haifa, Israel) [59]. All overexpression transfections were performed using Lipofectamine-2000 transfection reagent (Life Technologies), according to the manufacturer’s specifications.
Western blot
Proteins from samples in this study were collected using Radio Immunoprecipitation Assay (RIPA) buffer (Sigma-Aldrich) according to manufacturer’s protocol. After gel electrophoresis, membranes were blocked in 5% milk/TBS-T for 1 hour followed by incubation with primary antibody overnight. After washes and incubation with respective horseradish peroxidase conjugated secondary antibodies (anti-mouse 1:10,000, anti-rabbit 1:20,000) for 1 hr, protein bands were visualized using SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific, Waltham, MA) with ImageQuant LAS 4000 biomolecular imager (GE Healthcare Life Sciences, Pittsburg, PA). The densities of the bands were quantified using ImageQuant TL image analysis software (GE Healthcare Life Sciences).
Supernatant slot blot
Cell culture supernatants of infected or mock-treated HCE cells were vacuum filtered through Immobilon-Ny+ nylon membrane (Millipore Corporation, Billerica, MA) using Bio-Dot SF apparatus (Bio-Rad, Hercules, CA). Membranes were blotted overnight with antibody specific for Syndecan-1 (Santa Cruz Biotechnology, sc-6532) followed by species-specific secondary antibody and chemiluminescence detection using ImageQuant LAS 4000 biomolecular imager (GE Healthcare Life Sciences), as described above.
Nuclear protein extraction
Cells were washed with cold PBS and collected using a cell scraper. Cells were then treated with hypotonic buffer (20 M Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2) and 10% NP-40 and centrifuged. The pellet was treated with cell extraction buffer (Life Technologies) according to company’s protocol to collect nuclear extract. Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific) was added to the samples and stored in −80°C until analysis.
Cell surface protein extraction
Cell Surface Protein Isolation Kit (Thermo Scientific) was used to isolate proteins on the surface of the cell membrane. Briefly, cell surface proteins were labeled with sulfo-NHS-SS-Biotin and lysed. Biotinylated proteins from the remaining lysates were isolated using NeutrAvidin agarose columns, eluted, and analyzed by western blot.
PCR
RNA was extracted from cells using Trizol (Life Technologies) according to manufacturer’s protocol. RNA was then transcribed to DNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Real time quantitative PCR was performed using Fast SYBR Green Master Mix (Applied Biosystems) using QuantStudio 7 Flex (Applied Biosystems). The primers used in this study are: HPSE forward primer CTCGAAGAAAGACGGCTA and reverse primer GTAGCAGTCCGTCCATTC; viral glycoprotein gD forward primer TACAACCTGACCATCGCTTC and reverse primer GCCCCCAGAGACTTGTTGTA; NF-kB p65 forward primer TGGGGACTACGACCTGAATG and reverse primer GGGGGCACGATTGTCAAAGA; GAPDH forward primer TCCACTGGCGTCTTCACC and reverse primer GGCAGAGATGATGACCCTTTT. Semi-quantitative RT-PCR was performed using PCR Master Mix (Thermo Scientific) according to the manufacturer’s protocol. Human HPSE forward primer TTCGATCCCAAGAAGGAATCAAC and reverse primer GTAGTGATGCCATGTAACTGAATC for 25 cycles. Human GAPDH forward primer TCCACTGGCGTCTTCACC and reverse primer GGCAGAGATGATGACCCTTTT for 25 cycles. Mouse HPSE forward primer CAAGAACAGCACCTACTCAAG and reverse primer AGCAGTAGTCAAGGAGAAGC for 28 cycles. Mouse β-actin forward primer CGGTTCCGATGCCCTGAGGCTCTT and reverse primer CGTCACACTTCATGATGGAATTGA for 28 cycles.
Luciferase assay
pHep1(0.7kb)-luc plasmid [60] expressing firefly luciferase driven by 0.7 kb promoter area upstream of the transcription start site of human HPSE was provided by Dr. Xiulong Xu (Rush University, Chicago, IL). Empty vector, pGL3-Basic (Promega, Madison, WI), was used as a control for transfection efficiency. Luciferase activity was measured using Berthold Luminometer.
Flow cytometry
Heparan sulfate and heparanase cell surface expression were detected after HSV-1 KOS-WT infection. Monolayers of HCE cells were infected at MOI 0.1 and harvested at different times post infection. The following procedure for detecting heparan sulfate expression has been described previously [6]. Briefly, cells were harvested and incubated with primary antibody diluted in PBS with 1% BSA for 20 min at 4°C followed by 30 min incubation with FITC-conjugated anti-mouse IgM diluted to 1:100. For detection of heparanase on the cell surface, cells were harvested and incubated with (3 μg per sample) primary antibody diluted in PBS with 1% BSA for 1 hr at 4°C and 30 min incubation with FITC-conjugated anti-mouse IgG diluted to 1:100. Cells stained with respective FITC-conjugated secondaries were used as background controls. Entire cell populations were used for mean fluorescence intensity calculations.
For GFP HSV attachment assay, cells were counted and incubated with GFP-KOS at MOI 1000. Cells were allowed to adsorb the virus for 2 hrs at 4°C in order to facilitate attachment to cell surface but prevent entry. Viral attachment with GFP-KOS virus was quantified using flow cytometry. Entire cell populations were used for mean fluorescence intensity calculations. Flow cytometry was performed on a CyAn ADP Analyzer (Beckman Coulter, Brea, CA) and data was analyzed using FlowJo analysis software.
Immunofluorescence microscopy
HCE cells were cultured in glass bottom dishes (MatTek Corporation, Ashland, MA). Cells were fixed in 10% paraformaldehyde for 10 min and permeabilized with 0.1% Triton-X for 10 min for intracellular labeling. This was followed by incubation with primary antibody for 2 hrs at 4°C. When a secondary antibody was needed, cells were incubated with FITC-conjugated secondary antibody at dilution 1:150 for 1 hr and were examined under Zeiss Confocal 710, Germany using a 63x oil objective lens. Pinhole was set to 1 Airy Unit. For cell surface staining, cells were incubated with respective antibodies prior to fixation with paraformaldehyde for imaging. Fluorescence intensity of images was calculated using Image J.
Plaque assay
Viral egress was measured using plaque assay. Monolayers of HCE cells were plated in 6 well plates and infected with KOS-WT virus at MOI 0.1. Media was collected at different time points post infection and titered on Vero cells. Briefly, primary incubation of collected media was performed with Opti-MEM (Life Technologies) for 2 hrs. Vero cells were then incubated with growth media containing 1% methylcellulose for 72 hrs followed by fixing with 100% methanol and staining with crystal violet solution.
Virus titer by PCR
Supernatant was collected from infected samples and viral DNA was purified using QIAamp MinElute Virus Spin Kit (Qiagen, Venlo, Netherlands). The extracted DNA was eluted in 20 μl and was subjected to qPCR (2 μl of DNA/sample) using HSV gD primers. To verify that this assay is suitable to detect changes in HSV, a standard curve was calculated (Supplementary Fig. 4a). A virus of known titer was serially diluted to five different dilutions, and the viral DNA was extracted to perform qPCR. The coefficient of determination (R2) of the standard curve was 0.999 and the equation calculated was used for determining the amount of virus present in supernatant.
Heparanase activity ELISA
HCE cells were infected with KOS-WT virus (MOI 0.1). At different times post infection, infected samples were used to measure HPSE activity using Heparan Degrading Enzyme Assay Kit (Takara Bio Inc., Shiga, Japan). Briefly, infected samples were mixed 1:1 with reaction buffer and incubated for 45 min at 37°C. This mix was transferred to wells coated with FGF, which binds undegraded HS, and incubated for 15 min at 37°C, after which Avidin phenacetin O-deethylase (POD) conjugate was added and incubated for 45 min. POD substrate was added and absorbance at 450 nm was measured with a microplate reader. HPSE degradation activity (U/mL) of samples was measured utilizing a standard curve. One unit is defined as the activity that can degrade 0.063 ng of biotinylated HS when reacted at pH 5.8 at 37°C for 1 min.
Mouse cornea nucleic acid delivery and ocular infection
Animals were cared for and procedures were performed in accordance with institutional and NIH guidelines and approved by the Animal Care Committee at University of Illinois. Male and female Balb/c mice from Jackson Laboratory (Bar Harbor, ME) were used for animal experiments. Plasmids containing murine HPSE shRNA (Sigma-Aldrich, TRCN0000098111), scrambled shRNA, active HPSE GS-3, or empty vector were introduced into the epithelium using a previously established protocol [61–62]. Briefly, a nick in the epithelium and anterior stroma of mice eyes were made with a 30-gauge needle. 2 μL of DNA was injected into the stroma using 33-gauge Hamilton syringe (Hamilton, Reno, NV). Fluorescence images of the eye were captured using a stereoscope (StereoLumar V.12, Carl Zeiss, GmbH, Hamburg, Germany) according to previously mentioned protocol [63]. Briefly, an anesthetized mouse was placed on the stereoscope stage with mouse adaptor (RWD Lifesciences, San Diego, CA, Cat. #900-0068-010). 5 μL of proparacaine (0.5%, Bausch & Lomb, Tampa, FL) was applied for 3 minutes, and the pupil was constricted with 5 μL 0.01% carbachol (Miostat Alcon, Fort Worth, TX) for 5 minutes prior to acquiring images. Mice eyes were infected at Day 3 post transfection with KOS-WT virus or GFP-KOS (1x105 pfu in 5 μL), according to the described protocol [64]. Swabs were collected on appropriate days according to previously mentioned protocol [65] and plaque assay was performed on Vero cells. Briefly, eye swabs were collected by anesthetizing mice and gently proptosing each eye and wiping a sterile cotton swab three times around the eye in a circular motion and twice over the center of the cornea in an ‘X’ pattern. The swabs were place in 1 mL of DMEM containing 10% FBS, 1% penicillin/streptomycin, and stored at −80°C until titered. Before titering, the swabs were thawed and vortexed for 30 s at max speed. Heparin (H3393) was purchased from Sigma-Aldrich (St. Louis, MO) and 5 μL of 25 μg/μL was applied directly to mice eyes as a HPSE inhibitor when applicable.
Clinical scoring of epithelial keratitis
7 days post ocular infection, the same observer applied fluorescein dye strips to the sclera of mouse eyes, examined corneas with a handheld slit-lamp biomicroscope, and scored the severity of epithelial lesions according to the following established criteria [18]: 0, no epithelial lesion or punctate epithelial erosion; 1, stellate keratitis or residue of dendritic keratitis; 2, dendritic keratitis occupying less than one quarter of the cornea; 3, dendritic keratitis occupying one quarter to one half of the cornea; 4, dendritic keratitis extending over more than one half of the cornea.
Human corneas
Human donor corneoscleral buttons were obtained from the Illinois Eye Bank, Chicago, IL. The corneas were cultured and infected with 104 pfu HSV-1 as previously described [66] with some modifications. Briefly, corneoscleral buttons were placed in a dish with medium, and virus diluted in 10 μL medium was added to the corneal apex. Virus diluted media did not mix with the media in the dish. Corneal epithelial cells were harvested and lysed by scraping into 200 μL RIPA buffer with a scalpel blade. To minimize variability, eyes of the same donor were used as controls.
Statistics
Error bars of all figures represent s.e.m. of three independent experiments (n=3), unless otherwise specified. Asterisks denote a significant difference as determined by Student’s t-test; *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, ns, not significant.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants AI081869 (DS), EY023058 (DS), AI103754 (DS), and CA138340 (RS), a core grant (EY001792), and unrestricted funds from Research to Prevent Blindness Inc. We thank Dr. Patricia G. Spear (Northwestern University, Chicago, IL) for providing viruses used in this study. We are grateful to Dr. Sandeep Jain and Dr. Joy Sarkar (University of Illinois at Chicago, Chicago, IL) for providing their expertise with StereoLumar microscope. We thank Dr. Israel Vlodavsky (Rappaport Institute, Haifa, Israel) for providing HPSE mutant expression plasmids. Heparanase luciferase constructs were a kind gift from Dr. Xiulong Xu (Rush University, Chicago, IL). We also thank Dr. Michael Karin (University of California, San Diego, La Jolla, CA) for providing mut IkBa expression constructs, and Dr. Bernard Roizman (University of Chicago, Chicago, IL) and Dr. Neal DeLuca (University of Pittsburgh, Pittsburgh, PA) for access to several viral transcription factor expression constructs.
Footnotes
Author Contributions:
D.S., S.H., A.A. designed experiments; S.H., A.A., G.A.K., T.A., A.Y., V.C.R. performed experiments and analyzed data; S.H., A.A., D.S., R.S., A.R.D. wrote the manuscript.
Conflicts of Interest Statement:
The authors declare no conflict of interest.
References
- 1.Shukla D, Spear PG. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farooq AV, Shukla D. Herpes simplex epithelial and stromal keratitis: an epidemiological update. Surv Ophthalmol. 2012;57:448–462. doi: 10.1016/j.survophthal.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shukla D, et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 4.Xu D, Esko JD. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem. 2014;83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiwari V, Liu J, Valyi-Nagy T, Shukla D. Anti-heparan sulfate peptides that block herpes simplex virus infection in vivo. J Biol Chem. 2011;286:25406–25415. doi: 10.1074/jbc.M110.201103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacsa S, et al. Syndecan-1 and syndecan-2 play key roles in herpes simplex virus type-1 infection. J Gen Virol. 2011;92:733–743. doi: 10.1099/vir.0.027052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest. 2001;108:341–347. doi: 10.1172/JCI13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andela VB, Schwarz EM, Puzas JE, O’Keefe RJ, Rosier RN. Tumor metastasis and the reciprocal regulation of prometastatic and antimetastatic factors by nuclear factor kappaB. Cancer Res. 2000;60:6557–6562. [PubMed] [Google Scholar]
- 9.Cao HJ, et al. Tumor metastasis and the reciprocal regulation of heparanase gene expression by nuclear factor kappa B in human gastric carcinoma tissue. World J Gastroenterol. 2005;11:903–907. doi: 10.3748/wjg.v11.i6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu W, Pan C, Yu H, Gong H, Wang Y. Heparanase expression in gallbladder carcinoma and its correlation to prognosis. J Gastroenterol Hepatol. 2008;23:491–497. doi: 10.1111/j.1440-1746.2007.04945.x. [DOI] [PubMed] [Google Scholar]
- 11.Wu W, et al. Hypoxia activates heparanase expression in an NF- kappaB dependent manner. Oncol Rep. 2010;23:255–261. [PubMed] [Google Scholar]
- 12.Santoro MG, Rossi A, Amici C. NF-kappa B and virus infection: who controls whom. EMBO J. 2003;22:2552–2560. doi: 10.1093/emboj/cdg267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gimble JM, et al. Activation of the human immunodeficiency virus long terminal repeat by herpes simplex virus type 1 is associated with induction of a nuclear factor that binds to the NF-kappa B/core enhancer sequence. J Virol. 1988;62:4104–4112. doi: 10.1128/jvi.62.11.4104-4112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel A, et al. Herpes simplex type 1 induction of persistent NF-kappa B nuclear translocation increases the efficiency of virus replication. Virology. 1998;247:212–222. doi: 10.1006/viro.1998.9243. [DOI] [PubMed] [Google Scholar]
- 15.Cai M, et al. The herpes simplex virus 1- encoded envelope glycoprotein B activates NF-kB through the Toll-like receptor 2 and MyD88/TRAF6- dependent signaling pathway. PLoS One. 2013;8:e54586. doi: 10.1371/journal.pone.0054586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasperczyk H, et al. Betulinic acid as new activator of NF-kappaB: molecular mechanisms and implications for cancer therapy. Oncogene. 2005;24:6945–6956. doi: 10.1038/sj.onc.1208842. [DOI] [PubMed] [Google Scholar]
- 17.DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IkappaB kinase that activates the transcription factor NF-kappaB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 18.Inoue T, et al. Preventive effect of local plasmid DNA vaccine encoding gD or gD-IL-2 on herpetic keratitis. Invest Ophthalmol Vis Sci. 2000;41:4209–4215. [PubMed] [Google Scholar]
- 19.Air GM, Laver WG. The neuraminidase of influenza virus. Proteins. 1989;6:341–356. doi: 10.1002/prot.340060402. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, et al. Heparanase enhances syndecan-1 shedding: a novel mechanism for stimulation of tumor growth and metastasis. J Biol Chem. 2007;282:13326–13333. doi: 10.1074/jbc.M611259200. [DOI] [PubMed] [Google Scholar]
- 21.Purushothaman A, Chen L, Yang Y, Sanderson RD. Heparanase stimulation of protease expression implicates it as a master regulator of the aggressive tumor phenotype in myeloma. J Biol Chem. 2008;283:32628–32636. doi: 10.1074/jbc.M806266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shieh MT, WuDunn D, Montgomery RI, Esko JD, Spear PG. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J Cell Biol. 1992;116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilat D, et al. Molecular behavior adapts to context: heparanase functions as an extracellular matrix-degrading enzyme or as a T cell adhesion molecule, depending on the local pH. J Exp Med. 1995;181:1929–1934. doi: 10.1084/jem.181.5.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc Natl Acad Sci. 1998;95:6803–6808. doi: 10.1073/pnas.95.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukla D, et al. Expression of nectin-1 in normal and herpes simplex virus type 1-infected murine brain. Appl Immunohistochem Mol Morphol. 2006;14:341–347. doi: 10.1097/00129039-200609000-00014. [DOI] [PubMed] [Google Scholar]
- 26.Shah A, et al. HSV-1 infection of human corneal epithelial cells: receptor-mediated entry and trends of re-infection. Mol Vis. 2010;16:2476–2486. [PMC free article] [PubMed] [Google Scholar]
- 27.Shukla D, Spear PG. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Summerford C, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, et al. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 30.Barth H, et al. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278:41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 31.Jackson T, et al. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J Virol. 1996;70:5282–5287. doi: 10.1128/jvi.70.8.5282-5287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung CS, Hsiao JC, Chang YS, Chang W. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J Virol. 1998;72:1577–1585. doi: 10.1128/jvi.72.2.1577-1585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Germi R, et al. Heparan sulfate-mediated binding of infectious dengue virus type 2 and yellow fever virus. Virology. 2002;292:162–168. doi: 10.1006/viro.2001.1232. [DOI] [PubMed] [Google Scholar]
- 34.Klimstra WB, Ryman KD, Johnston RE. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol. 1998;72:7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roderiquez G, et al. Mediation of human immunodeficiency virus type 1 binding by interaction of cell surface heparan sulfate proteoglycans with the V3 region of envelope gp120-gp41. J Virol. 1995;69:2233–2239. doi: 10.1128/jvi.69.4.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. Human papillomavirus infection requires cell surface heparan sulfate. J Virol. 2001;75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Compton T, Nowlin DM, Cooper NR. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology. 1993;193:834–841. doi: 10.1006/viro.1993.1192. [DOI] [PubMed] [Google Scholar]
- 38.Feldman SA, Audet S, Beeler JA. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J Virol. 2000;74:6442–6447. doi: 10.1128/jvi.74.14.6442-6447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Thorp SC. Cell surface heparan sulfate and its roles in assisting viral infections. Med Res Rev. 2002;22:1–25. doi: 10.1002/med.1026. [DOI] [PubMed] [Google Scholar]
- 40.El-Assal ON, Yamanoi A, Ono T, Kohno H, Nagasue N. The clinicopathological significance of heparanase and basic fibroblast growth factor expressions in hepatocellular carcinoma. Clin Cancer Res. 2001;7:1299–1305. [PubMed] [Google Scholar]
- 41.Nyberg K, et al. The low molecular weight heparan sulfate-mimetic, PI-88, inhibits cell-to-cell spread of herpes simplex virus. Antiviral Res. 2004;63:15–24. doi: 10.1016/j.antiviral.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Giménez F, Suryawanshi A, Rouse BT. Pathogenesis of herpes stromal keratitis—a focus on corneal neovascularization. Prog Retin Eye Res. 2013;33:1–9. doi: 10.1016/j.preteyeres.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meirovitz A, et al. Heparanase in inflammation and inflammation-associated cancer. FEBS J. 2013;280:2307–2319. doi: 10.1111/febs.12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldberg R, et al. Versatile role of heparanase in inflammation. Matrix Biol. 2013;32:234–240. doi: 10.1016/j.matbio.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elkin M, et al. Heparanase as a mediatory of angiogenesis: mode of action. FASEB J. 2001;280:1661–1663. doi: 10.1096/fj.00-0895fje. [DOI] [PubMed] [Google Scholar]
- 46.Vlodavsky I, Blich M, Li JP, Sanderson RD, Ilan N. Involvement of heparanase in atherosclerosis and other vessel wall pathologies. Matrix Biol. 2013;32:241–251. doi: 10.1016/j.matbio.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edovitsky E, et al. Role of endothelial heparanase in delayed-type hypersensitivity. Blood. 2006;107:3609–3616. doi: 10.1182/blood-2005-08-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schönherr E, Hausser HJ. Extracellular matrix and cytokines: a functional unit. Dev Immunol. 2000;7:89–101. doi: 10.1155/2000/31748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parish CR, Freeman C, Hulett MD. Heparanase: a key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:99–108. doi: 10.1016/s0304-419x(01)00017-8. [DOI] [PubMed] [Google Scholar]
- 50.Taipale J, Keski-Oja J. Growth factors in the extracellular matrix. FASEB J. 1997;11:51–59. doi: 10.1096/fasebj.11.1.9034166. [DOI] [PubMed] [Google Scholar]
- 51.Smith KO. Physicial and biological observations on herpesvirus. J Bacteriol. 1963;86:999–1009. doi: 10.1128/jb.86.5.999-1009.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shukla D, Rowe CL, Dong Y, Racaniello VR, Spear PG. The murine homolog (Mph) of human herpesvirus entry protein B (HveB) mediates entry of pseudorabies virus but not herpes simplex virus types 1 and 2. J Virol. 1999;73:4493–4497. doi: 10.1128/jvi.73.5.4493-4497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tiwari V, et al. A role for herpesvirus entry mediator as the receptor for herpes simplex virus 1 entry into primary human trabecular meshwork cells. J Virol. 2005;79:13173–13179. doi: 10.1128/JVI.79.20.13173-13179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lurain NS, Spafford LE, Thompson KD. Mutation in the UL97 open reading frame of human cytomegalovirus strains resistant to ganciclovir. J Virol. 1994;68:4427–4431. doi: 10.1128/jvi.68.7.4427-4431.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kelly T, et al. High heparanase activity in multiple myeloma is associated with elevated microvessel density. Cancer Res. 2003;63:8749–8756. [PubMed] [Google Scholar]
- 57.Lopez P, Van Sant C, Roizman B. Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1. J Virol. 2001;75:3832–3840. doi: 10.1128/JVI.75.8.3832-3840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith CA, Bates P, Rivera-Gonzalez R, Gu B, DeLuca NA. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J Virol. 1993;67:4676–4687. doi: 10.1128/jvi.67.8.4676-4687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fux L, et al. Structure-function approach identifies a COOH-terminal domain that mediates heparanase signaling. Cancer Res. 2009;69:1758–1767. doi: 10.1158/0008-5472.CAN-08-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang P, et al. Cloning and characterization of the human heparanase-1 (HPR1) gene promoter: role of GA-binding protein and Sp1 in regulating HPR1 basal promoter activity. J Biol Chem. 2002;277:8989–8998. doi: 10.1074/jbc.M105682200. [DOI] [PubMed] [Google Scholar]
- 61.Stechschulte SU, et al. Rapid ocular angiogenic control via naked DNA delivery to cornea. Invest Ophthalmol Vis Sci. 2001;42:1975–1979. [PubMed] [Google Scholar]
- 62.Singh N, et al. Unique homologous siRNA blocks hypoxia-induced VEGF upregulation in human corneal cells and inhibits and regresses murine corneal neovascularization. Cornea. 2007;26:65–72. doi: 10.1097/ICO.0b013e31802b4201. [DOI] [PubMed] [Google Scholar]
- 63.Sarkar J, et al. Corneal neurotoxicity due to topical benzalkonium chloride. Invest Ophthalmol Vis Sci. 2012;53:1792–1802. doi: 10.1167/iovs.11-8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Babu JS, et al. Viral replication is required for induction of ocular immunopathology by herpes simplex virus. J Virol. 1996;70:101–107. doi: 10.1128/jvi.70.1.101-107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karaba AH, Kopp SJ, Longnecker R. Herpesvirus entry mediator is a serotype specific determinant of pathogenesis in ocular herpes. Proc Natl Acad Sci. 2012;109:20649–20654. doi: 10.1073/pnas.1216967109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alekseev O, Tran AH, Azizkhan-Clifford J. Ex vivo organotypic corneal model of acute epithelial herpes simplex virus type I infection. J Vis Exp. 2012;69:e3631. doi: 10.3791/3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.