Abstract
AIM
To investigate the association between SERPING1 rs2511989 (G>A) polymorphism and age-related macular degeneration (AMD).
METHODS
A number of electronic databases (up to July 15, 2014) were searched independently by two investigators. A Meta-analysis was performed on the association between SERPING1 rs2511989 polymorphism and AMD. Pooled odds ratios (ORs) with 95% confidence intervals (CIs) were estimated.
RESULTS
Eight studies with 16 cohorts consisting of 9163 cases and 6813 controls were included in this Meta-analysis. There was no significant association between rs2511989 polymorphism and AMD under all genetic models in overall estimates (A vs G: OR= 0.938, 95%CI =0.858-1.025; AA vs GG:OR =0.871, 95%CI =0.719-1.056; AG vs GG: OR =0.944, 95%CI =0.845-1.054; AA+AG vs GG: OR =0.927, 95% CI =0.823-1.044; AA vs AG+GG: OR =0.890, 95%CI =0.780-1.034). Cumulative Meta-analyses also showed a trend of no association between rs2511989 polymorphism and AMD as information accumulated by year. Subgroup analysis and Meta-regression analysis indicated that age-matching status was the main source of heterogeneity. Sensitivity analysis found the results in overall comparisons and subgroup comparisons of white subjects under the allele model were found to have significantly statistical differences after studies deviating from Hardy-Weinberg equilibrium (HWE) were excluded (overall: OR=0.918, 95%CI = 0.844-0.999, P =0.049; whites: OR =0.901, 95%CI = 0.817-0.994, P =0.038). However, the results were not sufficiently robust for further sensitivity analysis and statistical differences disappeared on applying Bonferroni correction (with a significance level set at 0.05/25).
CONCLUSION
This Meta-analysis indicates that SERPING1 rs2511989 polymorphism and AMD tend to have no association with each other. Age matching status is a big confounding factor, and more studies with subtle designs are warranted in future.
Keywords: age-related macular degeneration, SERPING1, single nucleotide polymorphism, Meta-analysis
INTRODUCTION
Age-related macular degeneration (AMD) is a leading cause of irreversible blindness in older populations in developed countries, and has become a major public health issue[1],[2]. The prevalence of AMD increases strongly with age, affecting 4% of the population over the age of 60, 10% of individuals older than 75, and 64% of the population after the age of 80[2]–[4]. In late-stage AMD, the disease is characterized by drusen (focal depositions of extracellular material in the Bruch's membrane beneath the retinal pigment epithelium) and usually without clinical symptoms[5],[6]. In late-stage AMD, vision-threatening complications of choroidal neovascularization or atrophy develop, which can lead to severe irreversible central vision loss[7].
Although many previous studies have identified that AMD is a complex disorder caused by the interaction of multiple genetic and environmental risk factors[8]–[12], the specific cause still remains to be further investigated, to improve preclinical prediction. So far, remarkable progress has recently been made in understanding the genetic risk factors AMD. Several complement component and regulator genes have been identified, and these highlight the importance of the complement pathway in the pathogenesis of AMD. At least five complement genes have been found to be associated with the development of this disorder, including complement regulator factor H complement factor I, the complement components C3, the complement components C2, and factor B[13]–[16].
The SERPING1 gene encodes complement 1 inhibitor (C1INH), which is a glycoprotein that inhibits complement activation by interfering with the proteolytic activity of C1r/C1s in the classical pathway and mannose-binding protein-associated serine proteases in the lectin pathway[17]. As a member of the classical and lectin complement pathway, SERPING1 is a plausible candidate gene for AMD. Recently, Ennis et al[18] reported a protective effect on AMD for the minor allele of a single nucleotide polymorphism (SNP rs2511989 G>A)) within intron 6 of the SERPING1 gene encoding for C1INH. Since then, many replication studies have been successively completed.
Although reported studies have focused on the association between the SNP rs 2511989 and AMD, the results are contradictory and inconclusive. Hence, we performed a Meta-analysis based on eight candidate eligible studies consisting of 9163 cases and 6813 controls, which may confirm the association between the SNP rs2511989 in SERPING1 gene and AMD. Additional SNP variants in the SERPING1 gene that have been identified relating to AMD, such as rs2244169, rs2511990, rs2509897, rs1005510, and rs2511988, are not discussed in this article, because of a lack of studies.
MATERIALS AND METHODS
Search Strategy
To identify the studies eligible for system-atic review and Meta-analysis, the following electronic data-bases were searched: PubMed, Embase, Web of Science, Wanfang (Chinese), and the China National Knowledge Infrastructure database (CNKI), up to July 15, 2014. The search algorithm is as follows: (SERPING1 OR complement component 1 inhibitor gene) AND (gene OR polymorphism*OR variant*OR single nucleotide polymorphism OR SNP) AND (age related macular degeneration OR macular degeneration OR AMD) AND (Case-control Studies OR Case-Base Studies OR Case-Comparison Studies OR Case-Referent Studies). Additional studies were identi-fied through a manual search of the references of the original studies and review articles. We retrieved through electronic searches to identify studies not yet included in the computerized databases. No language restriction in the search process and all studies performed on human subjects were included in our search.
Inclusion/Exclusion Criteria
Studies included in our Meta-analysis had to meet the following inclusion criteria: 1) the original major objective was to explore the relationship between SERPING1 gene polymorphism and AMD; 2) the studies were designed on the basis of independent case-control study with available data of genotype distributions and sufficient data for estimating odds ratios (ORs) with 95% confidence intervals (CIs); 3) as for the duplicated articles, the latest or the largest one was adopted. If several different cohorts were reported in the same article, they were considered as independent studies.
Data Extraction
Data ware collected carefully and independently by two independent investigators (Fang XY and Dong Y). The characteristics of the selected articles were shown in Table 1, including first author's name, cohorts, publication year, ethnicity, genotyping method, number of cases and controls, distribution of genotype frequency, minor allele frequency (MAF), and Hardy-Weinberg equilibrium (HWE) of control group (P value). Besides, we also listed the demographic characteristics of the study population and the age-matching status. When detailed data of mean ages were not available, we identified whether the age was matched or not according to claims on age matching status in the studies. If there was no claim, we treated cohorts for which the difference of the mean age was greater than 5y as age-unmatched, and vice versa[18],[19]. Details are shown in Table 2. In our study, ethnicities were subgrouped as Asians and whites. Disagreements were settled by discussion or consensus involving a third reviewer (Shi XF) when required.
Table 1. Principle characteristics of the studies included in the Meta-analysis.
| Frist author | Cohorts | Year | Ethnicity | Genotyping | Case |
Control |
Control HWE(p) | ||||||||
| Size | GG | GA | AA | MAF | Size | GG | GA | AA | MAF | ||||||
| Ennis et al[18] | UK | 2008 | Caucasians | Illumina | 479 | 191 | 215 | 70 | 0.37 | 479 | 132 | 236 | 109 | 0.48 | 0.858 |
| Ennis et al[18] | U (Lowa) | 2008 | Caucasians | Taqman | 248 | 100 | 122 | 26 | 0.35 | 252 | 79 | 124 | 49 | 0.44 | 0.978 |
| Park et al[19] | Mayo | 2009 | Caucasians | Taqman | 476 | 179 | 211 | 80 | 0.39 | 310 | 103 | 157 | 50 | 0.41 | 0.445 |
| Park et al[19] | Areds | 2009 | Caucasians | Illumina | 1221 | 436 | 563 | 222 | 0.41 | 295 | 115 | 127 | 53 | 0.39 | 0.088 |
| Allikmets et al[20] | Columbia | 2009 | Caucasians | Taqman | 1004 | 449 | 431 | 124 | 0.34 | 363 | 151 | 171 | 41 | 0.35 | 0.476 |
| Allikmets et al[20] | Lowa | 2009 | Caucasians | Combination | 368 | 116 | 178 | 74 | 0.44 | 115 | 37 | 59 | 19 | 0.42 | 0.578 |
| Allikmets et al[20] | Amsterdam | 2009 | Caucasians | Taqman | 338 | 107 | 184 | 47 | 0.41 | 257 | 84 | 131 | 42 | 0.42 | 0.447 |
| Allikmets et al[20] | Rotterdam | 2009 | Caucasians | Illumina | 1017 | 328 | 518 | 171 | 0.42 | 842 | 285 | 407 | 150 | 0.42 | 0.822 |
| Allikmets et al[20] | Australia | 2009 | Caucasians | Combination | 741 | 251 | 367 | 123 | 0.41 | 327 | 105 | 157 | 65 | 0.44 | 0.649 |
| Allikmets et al[20] | Germany | 2009 | Caucasians | Taqman | 998 | 377 | 485 | 136 | 0.38 | 725 | 284 | 341 | 100 | 0.37 | 0.883 |
| Allikmets et al[20] | Areds | 2009 | Caucasians | Taqman | 415 | 133 | 188 | 94 | 0.45 | 213 | 90 | 81 | 42 | 0.39 | 0.004 |
| Carter and Churchill [21] | UK | 2011 | Caucasians | direct sequencing | 94 | 38 | 39 | 17 | 0.39 | 95 | 29 | 52 | 14 | 0.42 | 0.232 |
| Lee et al[22] | US | 2010 | Caucasians | MassARRAY | 556 | 213 | 273 | 70 | 0.37 | 256 | 74 | 135 | 47 | 0.45 | 0.287 |
| Lu et al[23] | China | 2010 | Asians | SNaPshot | 272 | 198 | 57 | 5 | 0.13 | 285 | 215 | 63 | 3 | 0.12 | 0.494 |
| Nakata et al[24] | Japan | 2011 | Asians | Taqman+Illumina | 401 | 293 | 102 | 6 | 0.14 | 1530 | 1107 | 384 | 37 | 0.15 | 0.591 |
| Tian et al[25] | China | 2012 | Asians | MassARRAY | 535 | 422 | 96 | 13 | 0.11 | 469 | 371 | 86 | 7 | 0.11 | 0.436 |
Combination: SSCP+direct sequencing+Affymetrix; MassARRAY: Sequenom MassARRAY technology; NA: Data not available; HWE: Hardy-Weinberg equilibrium.
Table 2. Demographic characteristics of study population.
| Frist author | Cohorts | Age (mean±SD or mean) |
Age matching status | Sex (male%) |
||
| Case | Control | Case | Control | |||
| Ennis et al[18] | UK | 77.85±8.83 | 70.59±9.35 | unmatched | 37.9 | 48.5 |
| Ennis et al[18] | US (Lowa) | 81.18±9.12 | 74±9.04 | unmatched | 39.4 | 46.5 |
| Park et al[19] | Mayo | 76.9±9.6 | 69.5±8.2 | unmatched | 35.5 | 45.4 |
| Park et al[19] | Areds | 79.9±5.1 | 77.6±4.3 | matched | 40.5 | 44.1 |
| Allikmets et al[20] | Columbia | NA | NA | matched | NA | NA |
| Allikmets et al[20] | Lowa | NA | NA | matched | NA | NA |
| Allikmets et al[20] | Amsterdam | NA | NA | matched | NA | NA |
| Allikmets et al[20] | Rotterdam | NA | NA | matched | NA | NA |
| Allikmets et al[20] | Australia | NA | NA | matched | NA | NA |
| Allikmets et al[20] | Germany | NA | NA | matched | NA | NA |
| Allikmets et al[20] | Areds | NA | NA | matched | NA | NA |
| Carter and Churchill [21] | UK | NA | NA | matched | 28 | 33.7 |
| Lu et al[23] | China | 68.2±9.8 | 68.4±7.2 | matched | 46.3 | 46.3 |
| Lee et al[22] | US | 79.3 | 69.5 | unmatched | 31.9 | 45.3 |
| Nakata et al[24] | Japan | 77.38±8.39 | NA | unmatched | 71.6 | 41.5 |
| Tian et al[25] | China | NA | NA | unmatched | 60.6 | 46.3 |
NA: Data not available; SD: Standard deviation.
Statistical Analysis
Pooled ORs with corresponding 95% CIs were calculated to evaluate the strength of relationship between the SERPING1 gene and AMD for the following five genetic models: the allele model (A vs G), the homozygote model (AA vs GG), the heterozygote model (AG vs GG), the dominant model (AA+AG vs GG), and the recessive model (AA vs AG+GG). A Z test was used to assess the significance of the pooled OR, in which P<0.05 was considered statistically significant. The Q test and I2 statistics were employed to evaluate between-study heterogeneity. If PQ≤0.10 or I2>50%, which indicated significant heterogeneity in the comparison models among studies[26], the estimated pooled ORs for each study were calculated using a random-effects model (DerSimonian and Laird method) [27]. Otherwise, the fixed-effects model was considered more suitable (Mantel-Haenszel method) [28]. We also performed a cumulative Meta-analysis to provide a framework for updating a genetic effect from all studies, to measure how much of the genetic effect changed as evidence accumulated, and to find the trend in estimated risk effect[29]. For the cumulative Meta-analysis, studies were sorted chronologically by publication year.
We performed subgroup and meta-regression analyses to explore potential sources of heterogeneity. All studies were classified based on ethnicity and age-matching status into white and Asian subgroups and age-matched and age-unmatched subgroups, respectively. We utilized univariate and multivariate meta-regression models to conduct the meta-regression. To tackle the issue of multiple testing, 10 000 permutations of Monte Carlo simulation were necessary to adjust the result of the multivariate meta-regression model. The reliability of the results was assessed by sensitivity analysis performed by sequentially omitting individual studies or by excluding studies deviating from HWE. HWE was checked using a Chi-square test in each control group of the included studies, with results of P<0.05 considered as significantly deviating from HWE. Furthermore, to reduce the false positive error rate, the Bonferroni method was used to adjust the results of multiple comparisons. Because 25 comparisons were made in this Meta-analysis, the P value, which was less than 0.05/25 (0.002), indicated statistical significance after Bonferroni correction.
RESULTS
Study Characteristics
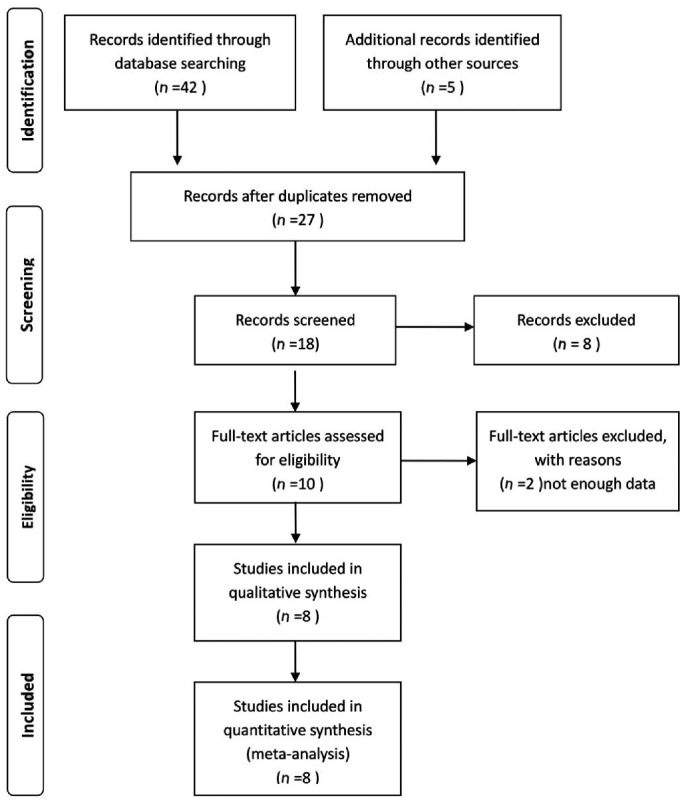
Figure 1 shows the selection process of this study. The initial search strategy identified 47 studies. Of these, 37 were excluded (20 were duplicate studies, 9 were on unrelated topics, and 8 were not case-control studies), leaving 10 studies for full review. Of these, two articles lacked sufficient data to estimate the OR and the 95% CI. Ultimately, eight studies met the inclusion criteria and were included in this Meta-analysis. We treated several different cohorts, which were reported in the same articles as independent studies. Therefore, there were 16 cohort studies consisting of 9163 cases and 6813 controls in our study. The detailed characteristics of the included studies are listed in Tables 1 and 2.
Figure 1. PRISMA flow diagram of studies included in the Meta-analysis.
The genotype distributions in all controls were consistent with HWE, except the Age-related Eye Disease Study (AREDS) cohorts of Allikmets' study[20]. Among the 16 cohorts included, there were 3 studies of Asians[23]–[25] and 13 studies of white subjects[18]–[22]. In terms of age-matching status, there were six age-unmatched studies and ten age-matched studies. The rs2511989 SNP was detected by different assays, as shown in Table 1. The MAF for the rs2511989 SNP varied substantially between studies, from 0.35 to 0.45 in both AMD patients (average 0.40) and control subjects (average 0.42) among the white subgroup. In addition, in the Asian subgroup, the MAF ranged from 0.11 to 0.14 in AMD patients (average 0.13) and from 0.11 to 0.15 in control subjects (average 0.13), respectively.
Overall Comparisons and Cumulative Meta-analysis
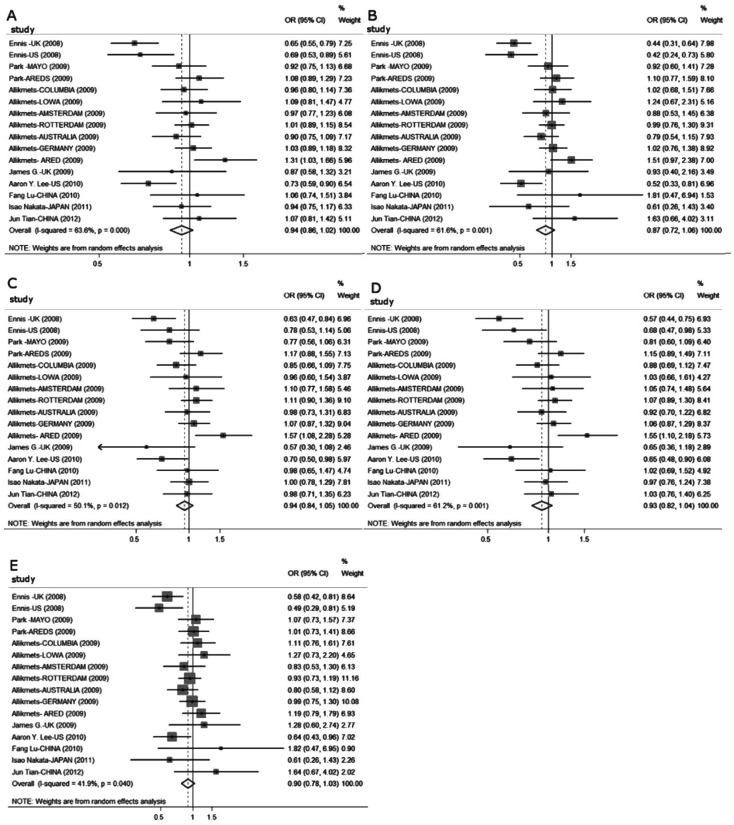
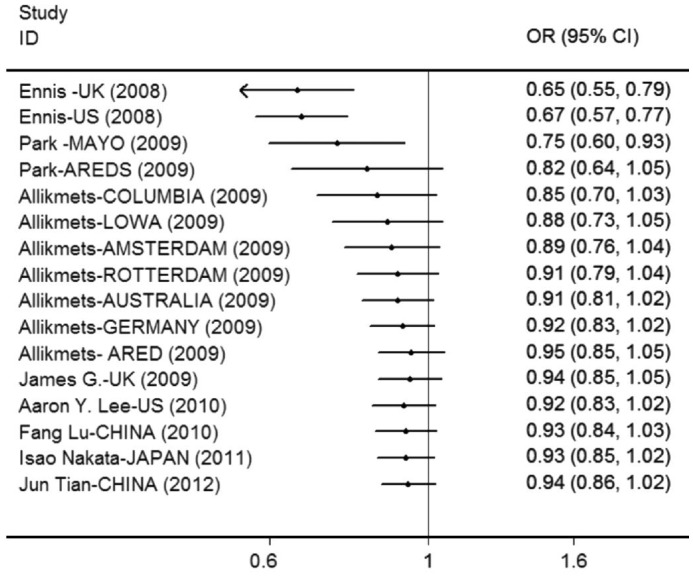
As a result of significant between-study heterogeneity detected in all genetic models for overall analysis (Table 3, Figure 2), a random-effects model was utilized to calculate pooled estimates. Overall, no significant relationship between the SNP rs2511989 and AMD was found in any genetic model (A vs G: OR =0.938, 95%CI =0.858-1.025; AA vs GG: OR =0.871, 95%CI =0.719-1.056; AG vs GG: OR =0.944, 95%CI =0.845-1.054; AA+AG vs GG: OR =0.927, 95%CI =0.823-1.044; AA vs AG+GG: OR =0.890, 95%CI =0.780-1.034). The cumulative Meta-analysis showed a trend of no association between SNP rs2511989 and AMD as information accumulated by year (Figure 3).
Table 3. Summary risk estimates for association between SNP rs2511989 and AMD.
| Stratifications | Studies (n) | Model | Pooled estimate |
Heterogeneity |
Egger's test P |
||
| OR (95%CI) | PZ | I2 (%) | PQ | ||||
| Overall | |||||||
| A vs G | 16 | R | 0.938 (0.858-1.025) | 0.155 | 63.6 | <0.001 | 0.854 |
| AA vs GG | 16 | R | 0.871 (0.719-1.056) | 0.159 | 61.6 | 0.001 | 0.948 |
| AG vs GG | 16 | R | 0.944 (0.845-1.054) | 0.306 | 50.1 | 0.012 | 0.280 |
| AA+AG vs GG | 16 | R | 0.927 (0.823-1.044) | 0.211 | 61.2 | 0.001 | 0.387 |
| AA vs AG+GG | 16 | R | 0.890 (0.780-1.034) | 0.135 | 41.9 | 0.040 | 0.595 |
| Asians | |||||||
| A vs G | 3 | F | 1.002 (0.857-1.171) | 0.984 | 0.0 | 0.725 | 0.615 |
| AA vs GG | 3 | F | 1.042 (0.604-1.800) | 0.882 | 30.6 | 0.237 | 0.407 |
| AG vs GG | 3 | F | 0.993 (0.830-1.187) | 0.935 | 0.0 | 0.993 | 0.352 |
| AA+AG vs GG | 3 | F | 0.998 (0.839-1.187) | 0.980 | 0.0 | 0.948 | 0.435 |
| AA vs AG+GG | 3 | F | 1.043 (0.605-1.797) | 0.880 | 31.6 | 0.232 | 0.614 |
| Caucasians | |||||||
| A vs G | 13 | R | 0.925 (0.835-1.024) | 0.134 | 70.0 | <0.001 | 0.603 |
| AA vs GG | 13 | R | 0.853 (0.697-1.045) | 0.124 | 66.4 | <0.001 | 0.621 |
| AG vs GG | 13 | R | 0.930 (0.811-1.067) | 0.300 | 59.8 | 0.003 | 0.310 |
| AA+AG vs GG | 13 | R | 0.908 (0.785-1.051) | 0.195 | 68.4 | <0.001 | 0.360 |
| AA vs AG+GG | 13 | R | 0.889 (0.769-1.028) | 0.112 | 46.7 | 0.032 | 0.956 |
| Age-matched | |||||||
| A vs G | 10 | F | 1.018 (0.957-1.083) | 0.571 | 0.0 | 0.562 | 0.802 |
| AA vs GG | 10 | F | 1.029 (0.904-1.172) | 0.662 | 0.0 | 0.713 | 0.388 |
| AG vs GG | 10 | F | 1.049 (0.956-1.152) | 0.314 | 25.1 | 0.212 | 0.504 |
| AA+AG vs GG | 10 | F | 1.043 (0.955-1.140) | 0.345 | 17.5 | 0.282 | 0.667 |
| AA vs AG+GG | 10 | F | 0.989 (0.879-1.113) | 0.858 | 0.0 | 0.797 | 0.079 |
| Age-unmatched | |||||||
| A vs G | 6 | R | 0.813 (0.695-0.951) | 0.010 | 66.2 | 0.011 | 0.285 |
| AA vs GG | 6 | R | 0.619 (0.434-0.883) | 0.008 | 60.8 | 0.026 | 0.405 |
| AG vs GG | 6 | F | 0.812 (0.715-0.921) | 0.001 | 36.3 | 0.165 | 0.503 |
| AA+AG vs GG | 6 | R | 0.771 (0.634-0.939) | 0.010 | 61.3 | 0.024 | 0.620 |
| AA vs AG+GG | 6 | R | 0.716 (0.528-0.973) | 0.033 | 55.4 | 0.047 | 0.628 |
R: Random-effects model; F: Fixed-effects mode; PZ: P value for Z test; PQ: P value for Q test.
Figure 2. Forest plots for association between the SERPING1 rs2511989 polymorphism and AMD in different genetic models.
The size of the square indicates the relative weight of each study. Bars: 95% confidence interval (95%CI). A: Allele model (A vs G); B: Homozygote model (AA vs GG); C: Heterozygote model (AG vs GG); D: Dominant model (AA+AG vs GG); E: Recessive model (AA vs AG+GG).
Figure 3. Forest plot of the cumulative Meta-analysis under the allele genetic model.
Cumulative odds ratios are shown for each information stepwise accumulated by year.
Heterogeneity Analysis
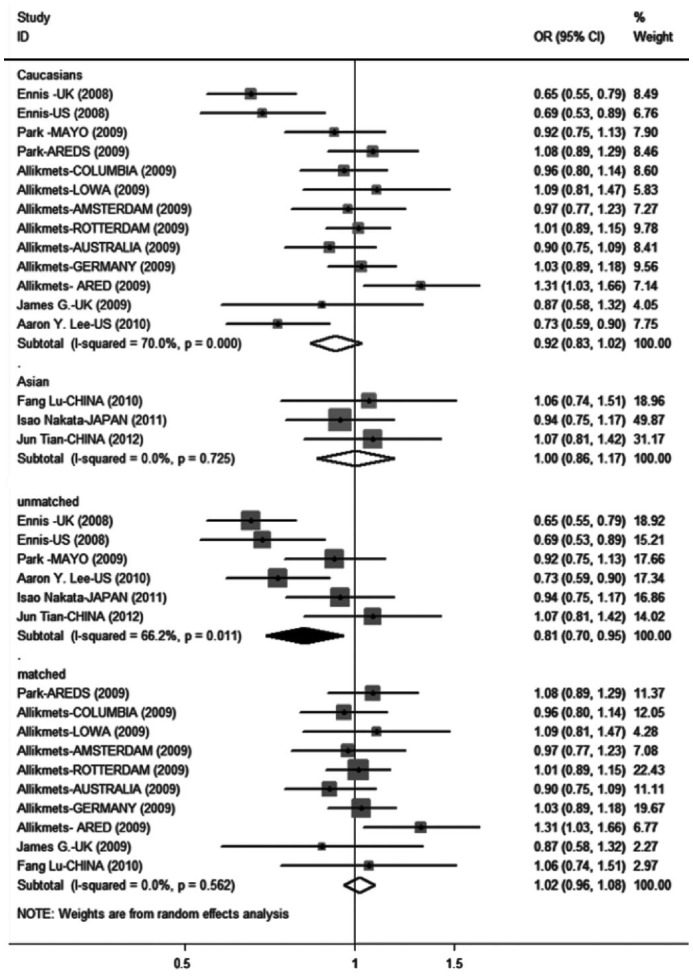
Subgroup analyses were conducted to investigate potential sources of heterogeneity. Among the four subgroups, there was no significant association apart from the age-unmatched subgroup, which showed significant protective associations between SNP rs2511989 and AMD in all genetic models (A vs G: OR =0.813,95%CI =0.695-0.951; AA vs GG:OR =0.619, 95%CI = 0.434-0.883; AG vs GG: OR =0.812, 95%CI =0.715-0.921; AA+AG vs GG: OR =0.771, 95%CI = 0.634-0.939; AA vs AG+GG: OR =0.716, 95%CI =0.528-0.973). We found that the heterogeneity in the Asian subgroup and the age-matched subgroup either disappeared or was moderate. However, there was still significant heterogeneity in the white subgroup and the age-unmatched subgroup. Therefore, ethnicity and age-matching status might be a main source of heterogeneity. Table 3 and Figure 4 showed detailed results.
Figure 4. Subgroup analyses by ethnicity and age matching status for associations between the SERPING1 rs2511989 polymorphism and AMD under the allele contrast.
Univariate and multivariate meta-regression were employed to further explore the influence of ethnicity, age-matching status and genotyping on heterogeneity (Table 4). These results indicated that the majority of the heterogeneity comes from the age-matching status (adjusted P=0.005<0.05), and not ethnicity or genotyping. Moreover, age-matching status could explain 67.0% of the heterogeneity, according to univariate meta-regression.
Table 4. Univariate and multivariate meta-regression analyses of potential source of heterogeneity.
| Factors | Coefficient | Standard error | t | P | 1Adjusted P |
| Ethnicity | |||||
| Univariate | 0.091 | 0.130 | -0.70 | 0.495 | 0.183 |
| Multivariate | -0.288 | 0.154 | -1.88 | 0.085 | |
| Age matching status | |||||
| Univariate | 0.235 | 0.074 | -3.19 | 0.006 | 0.005 |
| Multivariate | 0.285 | 0.071 | -4.02 | 0.002 | |
| Genotyping | |||||
| Univariate | 0.000 | 0.027 | 0.02 | 0.987 | 0.904 |
| Multivariate | 0.016 | 0.030 | -0.53 | 0.605 |
1P values were adjusted by Monte Carlo permutation test (10 000 iterations).
Sensitivity Analysis
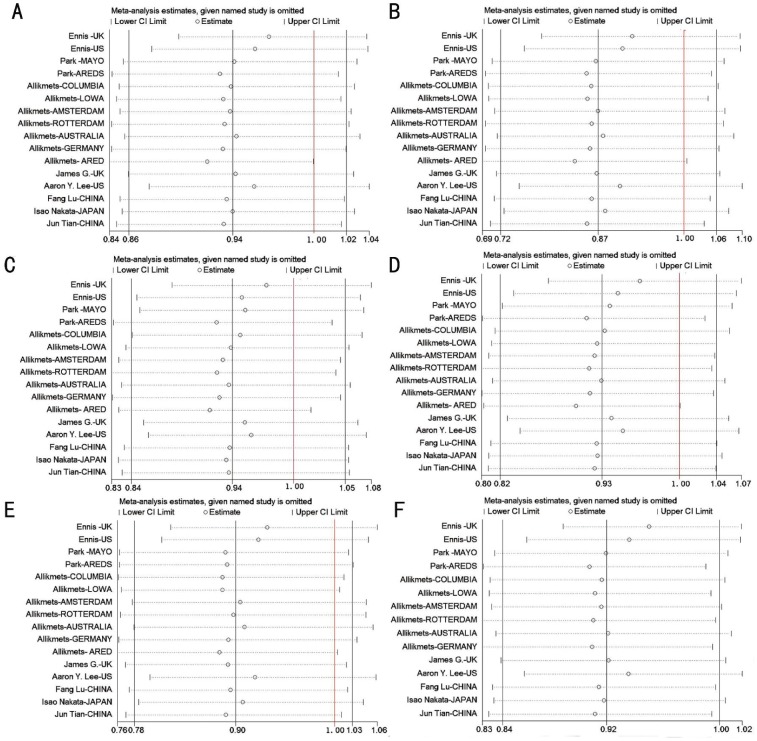
Sensitivity analysis was conducted to evaluate the stability of the results. Using a stepwise process, Meta-analysis was performed repeatedly, with each particular study sequentially excluded. The results of sensitivity analysis suggested that no single study could influence ORs in overall comparisons and subgroup analysis, except the AREDS cohorts of Allikmets' study (Figure 5). Coincidentally, the AREDS cohort was the only study that deviated from HWE in controls. After excluding this cohort, significant associations were observed for overall comparisons and white subgroup comparisons under the allele model (overall: OR = 0.918, 95%CI = 0.844-0.999, P = 0.049; whites: OR = 0.901, 95%CI =0.817-0.994, P =0.038), and the results of the age-matching status subgroup analysis were not changed. However, we found that these associations were not robust after we performed a sensitivity analysis on the remaining 15 studies (Figure 5F). Some of the results were materially altered and suggested different conclusion, as each individual study was sequentially omitted. Moreover, when we applied Bonferroni correction (significance set at 0.05/20) the association did not survive.
Figure 5. Sensitivity analysis of the summary odds ratio (OR) coefficients on the relationships between the SERPING1 rs2511989 polymorphism and AMD under five genetic models.
Results were computed by omitting each study in turn under random-effects model. The red line meant the value of OR is 1.00. The two ends of the dotted lines represented the 95%CI. A: A vs G; B: AA vs GG; C: AG vs GG; D: AA+AG vs GG; E: AA vs AG+GG; F: The result of further sensitivity analysis under allele contrast after exclude the study which deviated from HWE.
Publication Bias
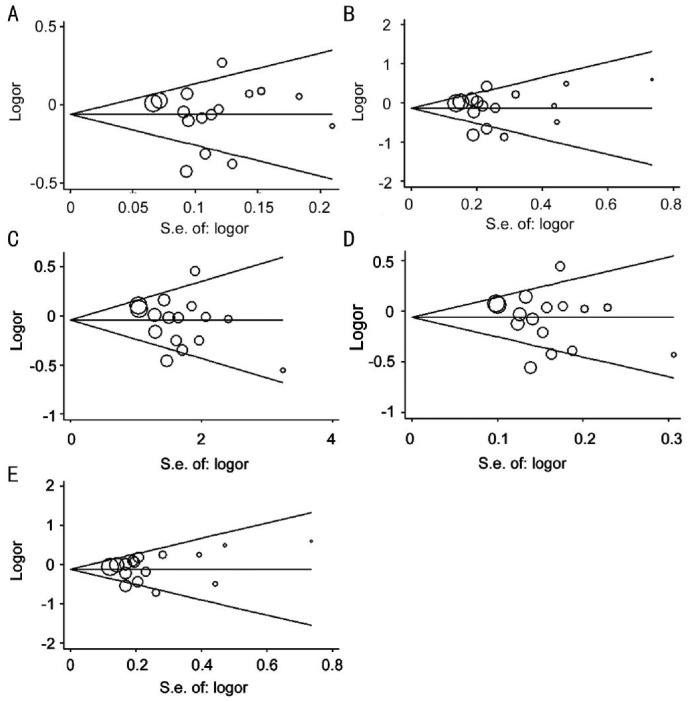
The Begg's funnel plot and Egger's test were performed for the overall comparisons and subgroup analysis. No obvious visual asymmetry was observed in Begg's funnel plots, and all the P values of the Egger's test were greater than 0.05, indicating no statistical evidence for publication bias among studies (Table 3, Figure 6).
Figure 6. Begg's funnel plot for publication bias test under five genetic models.
A: A vs G; B: AA vs GG; C: AG vs GG; D: AA+AG vs GG; E: AA vs AG+GG; Each circle represented a separate study for the indicated association, and its size was proportional to the sample size of each study. The shapes of the funnel plots did not reveal any evidence of obvious asymmetry.
DISCUSSION
As is well-known, the hypothesis that the complement system participates in the pathology of AMD has been supported by many researchers in a number of disciplines, including animal models of neovascularization, histopathology and genetics[30]. As an inhibitor of the classical and lectin pathways of complement activation, SERPING1 expression could plausibly be protective against complement injury in AMD. Recently, several studies have evaluated the association between SERPING1 rs2511989 (G>A) polymorphism and AMD. However, the results were inconsistent. Considering these studies' limitations of small sample size, Meta-analysis is a powerful tool for summarizing the contradicting results from different studies with greater statistical power; we undertook to conduct a Meta-analysis of 16 cohort studies involving 9163 cases and 6813 controls.
Among overall comparisons and subgroup comparisons in our study, we only found a significant positive association between rs2511989 polymorphism and AMD in the age-unmatched subgroup under all genetic models. Moreover, meta-regression analysis showed that age matching status was the main source of heterogeneity; not ethnicity or genotyping. These results led us to attach importance to the influence of age on genetic association. When we reviewed the ages of the patients and control subjects in the age-unmatched subgroups, we found that the control groups were significantly younger than the patient groups (shown in Table 2; some unavailable data can be estimated from the original articles). Ennis et al[18]. thought that this difference in age might reduce the power of detecting genetic association, because some of the controls could develop AMD in later life. Therefore, they considered the significant positive association detected by their study as reliable. However, we think that this explanation is flawed for the following reasons. Firstly, if it were the case, the pooled ORs of the age-matched subgroup should show relatively greater genetic association than the age-unmatched subgroup. In fact, our study obtained the opposite result. Secondly, the control group population might develop AMD, or die, or not return to follow-up in later life, but we are not sure what will happen in the future. Besides, the age of the control groups in the age-unmatched subgroup might be less than the age of the control groups in the age-matched subgroup. Taking this into consideration, we do not know whether the power of detecting genetic association is underestimated or overestimated. So the effect of the difference in age on genetic association is complex and indeterminable.
In fact, many studies have proved that genetic association changes with age[31]–[35]. In particular, the research of Adams et al[34], who studied the associations between four variants in the complement factor H and AMD in different age groups, discovered a change of relationship from protective to risk with increasing age. This finding challenges the conclusion of a strong association between complement factor H and AMD, which has been extensively studied and proved[36],[37]. Adams et al [34]concluded from their study that it is imperative to ensure that cases and controls have the same age distribution when conducting case-control studies of Adams. Therefore, if the design of the age-unmatched studies is not rigorous, it will lead to a bias of genetic association.
If no account is taken of the influence of age, no association of SNP rs2511989 and AMD was found under overall comparisons, or in other subgroup analyses. Our cumulative Meta-analysis also showed a trend of no association between them as information accumulated year by year. However, after excluding the study that deviated from HWE, significant associations were observed for overall comparisons and white subgroup comparisons under the allele model. This association was not robust when further sensitivity analysis was conducted and did not survive after Bonferroni correction. Hence, we cannot draw any definite conclusion from these analyses.
Although a comprehensive analysis was performed, which included overall and subgroup Meta-analysis, cumulative Meta-analysis, meta-regression and sensitivity analysis, we failed to obtain a determinate conclusion of association between SNP rs2511989 and AMD. This may be due to some inevitable limitations of our study. Firstly, 6 of 16 included studies were age-unmatched. The influence of the age-unmatched studies is complex and indeterminable and we do not know whether the power of detecting genetic association is underestimated or overestimated. This is a large confounding factor and must be considered in planning further studies. Secondly, this Meta-analysis was limited by the number of cases and controls, especially in the Asian subgroup analysis, which included only three studies. Moreover, some instability results also indicated the number of studies is insufficient. Thus, additional studies are needed to evaluate the relationship of SNP rs2511989 with AMD. Thirdly, owing to a lack of detailed information, such as sex and smoking status in individual studies, we failed to perform further subgroup analysis to adjust for these possible confounders. Fourthly, there was significant heterogeneity among included studies, which is mainly caused by age matching status. Although we used the random-effects model to calculate pool ORs, the precision of the outcome would be affected.
In view of the large confounding factor of age-unmatched data and unstable results, we are cautious and do not wish to make a final conclusion on the associations between SNP rs2511989 and AMD. However, judging from all of the analyses and the result of the cumulative Meta-analysis, it would appear that SNP rs2511989 tends to have no association with AMD. The reasons are as follows: 1) the overall and ethnicity subgroup Meta-analysis showed no relationship between SNP rs2511989 and AMD; 2) the result of the age-matched subgroup analysis, which showed no association between SNP rs2511989 and AMD, is more valid[38]; 3) the cumulative Meta-analysis also showed a trend of no association between SNP rs2511989 and AMD as information accumulated by year; 4) after excluding the AREDS cohorts of Allikmets et al's[20] study, which deviated from HWE. In controls, we only found an instability and inconspicuous genetic association in one of the five genetic models, which did not survive after Bonferroni correction; 5) there was almost no difference in the mean MAF between cases and controls within all studies, especially in the Asian subgroup.
In summary, our Meta-analysis indicated a tendency of no association between the SNP rs2511989 and AMD. This conclusion is also supported by two genome-wide association studies, which did not identify any SNP of the SERPING1 gene associated with AMD[39],[40]. Be that as it may, the possibility of association cannot be completely ruled out, given current limitations of technology and methodology. The SERPING1 protein is expressed at the macula, as demonstrated in in vitro studies of human donor eyes[18],[41],[42]. Therefore, this gene is worthy of further research in studies with subtler designs, especially where the age of patients and controls is rigorously matched.
Acknowledgments
We thank Qing Liu, who is the director of Tianjin Eye Hospital Library, for the help in the literature search process.
Foundation: Supported by Scientific and Technological Project of Tianjin Health Bureau (No.12KG123).
Conflicts of Interest: Dong Y, None; Li ZD, None; Fang XY, None; Shi XF, None; Chen S, None; Tang X, None.
REFERENCES
- 1.Synek S, Vojnikovic B, Pahor D. Epidemiology and quality of life of patients with age-related macular degeneration. Coll Antropol. 2010;(Suppl. 2):25–28. [PubMed] [Google Scholar]
- 2.Friedman DS, O'Colmain BJ, Muñoz B, Tomany SC, McCarty C, de Jong PT, Nemesure B, Mitchell P, Kempen J, Eye Diseases Prevalence Research Group Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122(4):564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 3.Vingerling JR, Dielemans I, Hofman A, Grobbee DE, Hijmering M, Kramer CF, de Jong PT. The prevalence of age-related maculopathy in the Rotterdam Study. Ophthalmology. 1995;102(2):205–210. doi: 10.1016/s0161-6420(95)31034-2. [DOI] [PubMed] [Google Scholar]
- 4.de Jong PT. Age-related macular degeneration. N Engl J Med. 2006;355(14):1474–1485. doi: 10.1056/NEJMra062326. [DOI] [PubMed] [Google Scholar]
- 5.Booij JC, van Soest S, Swagemakers SM, Essing AH, Verkerk AJ, van der Spek PJ, Gorgels TG, Bergen AA. Functional annotation of the human retinal pigment epithelium transcriptome. BMC Genomics. 2009;10:164. doi: 10.1186/1471-2164-10-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booij JC, Baas DC, Beisekeeva J, Gorgels TG, Bergen AA. The dynamic nature of Bruch's membrane. Prog Retin Eye Res. 2010;29(1):1–18. doi: 10.1016/j.preteyeres.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Bird AC, Bressler NM, Bressler SB, Chisholm IH, Coscas G, Davis MD, de Jong PT, Klaver CC, Klein BE, Klein R, Et A. An international classification and grading system for age-related maculopathy and age-related macular degeneration. The International ARM Epidemiological Study Group. Surv Ophthalmol. 1995;39(5):367–374. doi: 10.1016/s0039-6257(05)80092-x. [DOI] [PubMed] [Google Scholar]
- 8.Swaroop A, Branham KE, Chen W, Abecasis G. Genetic susceptibility to age-related macular degeneration: a paradigm for dissecting complex disease traits. Hum Mol Genet. 2007;16 Spec No. 2(2):R174–182. doi: 10.1093/hmg/ddm212. [DOI] [PubMed] [Google Scholar]
- 9.Haddad S, Chen CA, Santangelo SL, Seddon JM. The genetics of age-related macular degeneration: a review of progress to date. Surv Ophthalmol. 2006;51(4):316–363. doi: 10.1016/j.survophthal.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Seddon JM, George S, Rosner B, Klein ML. CFH gene variant, Y402H, and smoking, body mass index, environmental associations with advanced age-related macular degeneration. Hum Hered. 2006;61(3):157–165. doi: 10.1159/000094141. [DOI] [PubMed] [Google Scholar]
- 11.Francis PJ, George S, Schultz DW, Rosner B, Hamon S, Ott J, et al. The LOC387715 gene, smoking, body mass index, environmental associations with advanced age-related macular degeneration. Hum Hered. 2007;63(3–4):212–218. doi: 10.1159/000100046. [DOI] [PubMed] [Google Scholar]
- 12.Seddon JM, George S, Rosner B. Cigarette smoking, fish consumption, omega-3 fatty acid intake, and associations with age-related macular degeneration: the US Twin Study of Age-Related Macular Degeneration. Arch Ophthalmol. 2006;124(7):995–1001. doi: 10.1001/archopht.124.7.995. [DOI] [PubMed] [Google Scholar]
- 13.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Ven JP, Nilsson SC, Tan PL, Buitendijk GH, Ristau T, Mohlin FC, Nabuurs SB, Schoenmaker-Koller FE, Smailhodzic D, Campochiaro PA, Zack DJ, Duvvari MR, Bakker B, Paun CC, Boon CJ, Uitterlinden AG, Liakopoulos S, Klevering BJ, Fauser S, Daha MR, Katsanis N, Klaver CC, Blom AM, Hoyng CB, den Hollander AI. A functional variant in the CFI gene confers a high risk of age-related macular degeneration. Nat Genet. 2013;45(7):813–817. doi: 10.1038/ng.2640. [DOI] [PubMed] [Google Scholar]
- 15.Yates JR, Sepp T, Matharu BK, Khan JC, Thurlby DA, Shahid H, Clayton DG, Hayward C, Morgan J, Wright AF, Armbrecht AM, Dhillon B, Deary IJ, Redmond E, Bird AC, Moore AT. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357(6):553–61. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 16.Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs K, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38(4):458–62. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagenaar-Bos IG, Hack CE. Structure and function of C1-inhibitor. Immunol Allergy Clin North Am. 2006;26(4):615–32. doi: 10.1016/j.iac.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Ennis S, Jomary C, Mullins R, Cree A, Chen X, Macleod A, Jones S, Collins A, Stone E, Lotery A. Association between the SERPING1 gene and age-related macular degeneration: a two-stage case-control study. Lancet. 2008;372(9652):1828–34. doi: 10.1016/S0140-6736(08)61348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park KH, Ryu E, Tosakulwong N, Wu Y, Edwards AO. Common variation in the SERPING1 gene is not associated with age-related macular degeneration in two independent groups of subjects. Mol Vis. 2009;15:200–207. [PMC free article] [PubMed] [Google Scholar]
- 20.Allikmets R, Dean M, Hageman GS, Baird PN, Klaver CC, Bergen AA, Weber BH. The SERPING1 gene and age-related macular degeneration. Lancet. 2009;374(9693):876–877. doi: 10.1016/S0140-6736(09)61618-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carter JG, Churchill AJ. Analysis of SERPING1 and its association with age-related macular degeneration. Acta Ophthalmol. 2011;89(2):e212–213. doi: 10.1111/j.1755-3768.2009.01788.x. [DOI] [PubMed] [Google Scholar]
- 22.Lee AY, Kulkarni M, Fang AM, Edelstein S, Osborn MP, Brantley MA. The effect of genetic variants in SERPING1 on the risk of neovascular age-related macular degeneration. Br J Ophthalmol. 2010;94(7):915–917. doi: 10.1136/bjo.2009.172007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu F, Zhao P, Fan Y, Tang S, Hu J, Liu X, Yang X, Chen Y, Li T, Lei C, Yang J, Lin Y, Ma S, Li C, Shi Y, Yang Z. An association study of SERPING1 gene and age-related macular degeneration in a Han Chinese population. Mol Vis. 2010;16:1–6. [PMC free article] [PubMed] [Google Scholar]
- 24.Nakata I, Yamashiro K, Yamada R, Gotoh N, Nakanishi H, Hayashi H, Tsujikawa A, Otani A, Saito M, Iida T, Oishi A, Matsuo K, Tajima K, Matsuda F, Yoshimura N. Association between the SERPING1 gene and age-related macular degeneration and polypoidal choroidal vasculopathy in Japanese. PLoS One. 2011;6(4):e19108. doi: 10.1371/journal.pone.0019108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian J, Yu W, Qin X, Fang K, Chen Q, Hou J, Li J, Chen D, Hu Y, Li X. Association of genetic polymorphisms and age-related macular degeneration in Chinese population. Invest Ophthalmol Vis Sci. 2012;53(7):4262–4269. doi: 10.1167/iovs.11-8542. [DOI] [PubMed] [Google Scholar]
- 26.Attia J, Thakkinstian A, D'Este C. Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin Epidemiol. 2003;56(4):297–303. doi: 10.1016/s0895-4356(03)00011-8. [DOI] [PubMed] [Google Scholar]
- 27.DerSimonian R. Meta-analysis in the design and monitoring of clinical trials. Stat Med. 1996;15(12):1237–48; discussion 1249–1252. doi: 10.1002/(SICI)1097-0258(19960630)15:12<1237::AID-SIM301>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 28.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 29.Muellerleile P, Mullen B. Sufficiency and stability of evidence for public health interventions using cumulative Meta-analysis. Am J Public Health. 2006;96(3):515–522. doi: 10.2105/AJPH.2003.036343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson DH, Radeke MJ, Gallo NB, Chapin EA, Johnson PT, Curletti CR, Hancox LS, Hu J, Ebright JN, Malek G, Hauser MA, Rickman CB, Bok D, Hageman GS, Johnson LV. The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog Retin Eye Res. 2010;29(2):95–112. doi: 10.1016/j.preteyeres.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slagboom PE, Beekman M, Passtoors WM, Deelen J, Vaarhorst AA, Boer JM, van den Akker EB, van Heemst D, de Craen AJ, Maier AB, Rozing M, Mooijaart SP, Heijmans BT, Westendorp RG. Genomics of human longevity. Philos Trans R Soc Lond B Biol Sci. 2011;366(1561):35–42. doi: 10.1098/rstb.2010.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams MK, Simpson JA, Aung KZ, Makeyeva GA, Giles GG, English DR. Abdominal obesity and age-related macular degeneration. Abdominal obesity and age-related macular degeneration. Am J Epidemiol. 2011;173(11):1246–1255. doi: 10.1093/aje/kwr005. [DOI] [PubMed] [Google Scholar]
- 33.Middelberg RP, Benyamin B, de Moor MH, Warrington NM, Gordon S, Henders AK, Hopper J, Guymer RH, Baird PN, Robman LD. Loci affecting gamma-glutamyl transferase in adults and adolescents show age x SNP interaction and cardiometabolic disease associations. Hum Mol Genet. 2012;21(2):446–455. doi: 10.1093/hmg/ddr478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adams MK, Simpson JA, Richardson AJ, Guymer RH, Williamson E, Cantsilieris S, English DR, Aung KZ, Makeyeva GA, Giles GG, Hopper J, Robman LD, Baird PN. Can genetic associations change with age? CFH and age-related macular degeneration. Hum Mol Genet. 2012;21(23):5229–5236. doi: 10.1093/hmg/dds364. [DOI] [PubMed] [Google Scholar]
- 35.Ersoy L, Ristau T, Hahn M, Karlstetter M, Langmann T, Droge K, Caramoy A, den Hollander AI, Fauser S. Genetic and environmental risk factors for age-related macular degeneration in persons 90 years and older. Invest Ophthalmol Vis Sci. 2014;55(3):1842–1847. doi: 10.1167/iovs.13-13420. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Geng P, Zhang Y, Zhang M. Association between complement factor H Val62Ile polymorphism and age-related macular degeneration susceptibility: a Meta-analysis. Gene. 2014;538(2):306–312. doi: 10.1016/j.gene.2014.01.032. [DOI] [PubMed] [Google Scholar]
- 37.Sofat R, Casas JP, Webster AR, Bird AC, Mann SS, Yates JR, Moore AT, Sepp T, Cipriani V, Bunce C, Khan JC, Shahid H, Swaroop A, Abecasis G, Branham KE, Zareparsi S, Bergen AA, Klaver CC, Baas DC, Zhang K, Chen Y, Gibbs D, Weber BH, Keilhauer CN, Fritsche LG, Lotery A, Cree AJ, Griffiths HL, Bhattacharya SS, Chen LL, Jenkins SA, Peto T, Lathrop M, Leveillard T, Gorin MB, Weeks DE, Ortube MC, Ferrell RE, Jakobsdottir J, Conley YP, Rahu M, Seland JH, Soubrane G, Topouzis F, Vioque J, Tomazzoli L, Young I, Whittaker J, Chakravarthy U, de Jong PT, Smeeth L, Fletcher A, Hingorani AD. Complement factor H genetic variant and age-related macular degeneration: effect size, modifiers and relationship to disease subtype. Int J Epidemiol. 2012;41(1):250–262. doi: 10.1093/ije/dyr204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falagas ME, Mourtzoukou EG, Ntziora F, Peppas G, Rafailidis PI. Matching criteria in case-control studies on postoperative infections. J Hosp Infect. 2008;69(2):101–113. doi: 10.1016/j.jhin.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Fritsche LG, Chen W, Schu M, et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45(4):433–439, 439e1-2. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheetz TE, Fingert JH, Wang K, Kuehn MH, Knudtson KL, Alward WL, Boldt HC, Russell SR, Folk JC, Casavant TL, Braun TA, Clark AF, Stone EM, Sheffield VC. A genome-wide association study for primary open angle glaucoma and macular degeneration reveals novel Loci. PLoS One. 2013;8(3):e58657. doi: 10.1371/journal.pone.0058657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Soest SS, de Wit GM, Essing AH, Ten BJ, Kamphuis W, de Jong PT, Bergen AA. Comparison of human retinal pigment epithelium gene expression in macula and periphery highlights potential topographic differences in Bruch's membrane. Mol Vis. 2007;13:1608–1617. [PubMed] [Google Scholar]
- 42.Mullins RF, Faidley EA, Daggett HT, Jomary C, Lotery AJ, Stone EM. Localization of complement 1 inhibitor (C1INH/SERPING1) in human eyes with age-related macular degeneration. Exp Eye Res. 2009;89(5):767–773. doi: 10.1016/j.exer.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]