Abstract
The initial contact point between a recipient’s immune system and a transplanted graft is the vascular endothelium. Clinical studies suggest a pathogenic role for non-HLA antiendothelial cell antibodies (AECAs) in allograft rejection; however, evidence linking AECAs of known specificity to in vivo vascular injury is lacking. Here, we used high-density protein arrays to identify target antigens for AECAs isolated from the sera of recipients of kidney transplants experiencing antibody-mediated rejection in the absence of donor-specific HLA antibodies. Four antigenic targets expressed on endothelial cells were identified: endoglin, Fms-like tyrosine kinase-3 ligand, EGF-like repeats and discoidin I-like domains 3, and intercellular adhesion molecule 4; the first three have been implicated in endothelial cell activation and leukocyte extravasation. To validate these findings, ELISAs were constructed, and sera from an additional 150 renal recipients were tested. All four AECAs were detected in 24% of pretransplant sera, and they were associated with post-transplant donor-specific HLA antibodies, antibody-mediated rejection, and early transplant glomerulopathy. AECA stimulation of endothelial cell cultures increased adhesion molecule expression and production of inflammatory cytokines: regulated on activation, normal T cell expressed and secreted PDGF and RESISTIN. These correlations between in vitro experiments and in vivo histopathology suggest that AECAs activate the vascular endothelium, amplifying the alloimmune response and increasing microvascular damage. Given the growing number of transplant candidates, a better understanding of the antigenic targets, beyond HLA, and mechanisms of immune injury will be essential for improving long-term allograft survival.
Keywords: acute allograft rejection, endothelial cells, immunology and pathology, kidney transplantation
Improvements in the ability to detect alloantibodies in the sera of recipients both before and after renal transplantation and the development of methods to identify antibody-mediated damage have highlighted the role of antibodies in both acute and chronic allograft rejection.1–4 However, the majority of these studies have focused on HLA-specific antibodies and their potential to activate complement. The clinical relevance and mechanisms for how alloantibodies can contribute to allograft rejection in the absence of complement activation, such as in the case of low-level HLA antibodies, are currently under investigation.5,6
Reports of allograft rejection in HLA identical sibling transplants suggest a role for non-HLA antibodies in some rejections.7,8 Non-HLA antigens that have been associated with renal allograft rejection include agrin, vimentin, perlecan, Kα-tubulin, protein kinase Cζ, major histocompatibility complex class I-related chain A, and angiotensin II type 1 receptor.9 In addition, studies combining proteomics and genomics are uncovering tissue-specific non-HLA antigens capable of initiating humoral responses in recipients of renal transplants.10
Non-HLA antigens expressed on endothelial cells are of particular interest given that the vascular endothelium serves as the point of contact between the recipient’s immune system and the transplanted allograft. A prospective, multicenter clinical trial using peripheral blood endothelial cell precursors (ECPs) as targets showed that nonsensitized recipients transplanted across a positive endothelial cell crossmatch experienced increased rejection and higher serum creatinine values early post-transplantation.11 Most of these rejections were C4d-negative and classified as cellular rejections. Subsequently, we reported that these antiendothelial cell antibodies (AECAs) are enriched for IgG2 and IgG4, which are IgG subclasses that activate complement poorly or not at all.12,13 In contrast, identification of AECAs in recipients with broad HLA sensitization or in those undergoing HLA-incompatible transplantation was associated with an increased incidence and severity of antibody-mediated rejection (AMR).
Although the endothelial cell crossmatch has been shown to be clinically useful, it has many technical problems that include the expression of HLAs on ECPs, which limits our ability to test for AECAs in HLA-incompatible recipients and impedes our ability to identify those proceeding to transplant across both barriers. Identification of the antigenic targets expressed on ECPs would provide the potential to test for AECAs in solid-phase immunoassays, which in turn, may help in pretransplant risk assessment and provide an opportunity for therapeutic intervention. Here, we describe a proteomics approach to identify novel antigenic targets for AECAs and the development of an ELISA platform to enable testing for these AECAs in patients broadly sensitized to HLA. We show the effect of these AECAs on endothelial cell cultures in vitro and correlate that with increased in vivo microvascular injury in patients who test positive for AECAs.
Results
Identification of Novel Antigenic Endothelial Cell Targets Using Protein Arrays
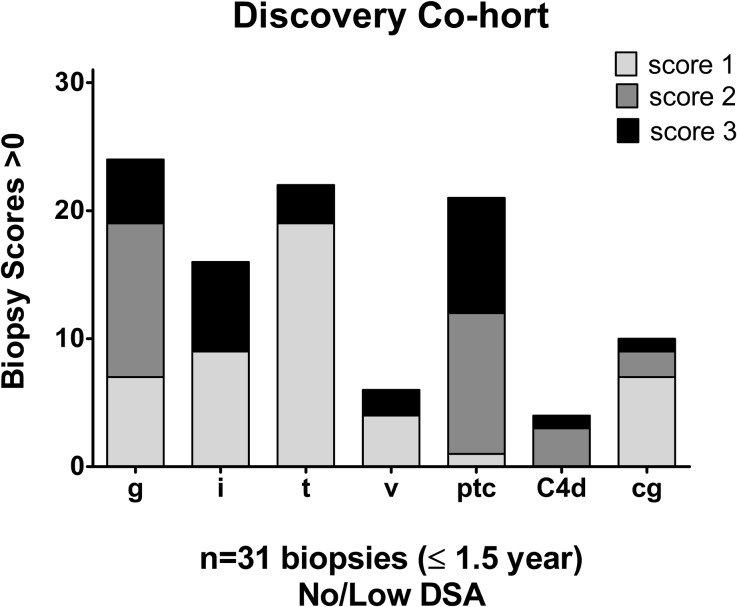
AECAs were isolated from a Discovery Cohort of 10 renal transplant recipients whose demographics are provided in Supplemental Table 1. Most patients (9 of 10) were sensitized to HLA, and all tested positive for AECAs in pretransplant endothelial cell crossmatch tests. Nine patients experienced allograft dysfunction and biopsy-proven rejection with noted glomerulitis and peritubular capillaritis (Figure 1). Only one recipient had low-level antibody, detected by bead assays only, to donor HLA (DR52) at the time of rejection.
Figure 1.
Antibody mediated injury observed in the AECA positive Discovery Cohort. Shown are renal biopsies with positive histologic scores>1 acquired during the 1.5 years post-transplantation according to protocol or at time of dysfunction. Histologic scoring (0–3) was performed using updated Banff 1997–2007 criteria.27–30 Shown are grades for glomerulitis (g), interstitial (i) and tubular (t) inflammation, vasculitis (v), and peritubular capillaritis (ptc). C4d staining was performed on frozen tissue by indirect immunofluorescence. Transplant glomerulopathy (cg) was defined as duplication of the glomerular basement membrane as observed on electron and light microscopy. Low-level DR52 HLA-DSA (median fluorescent intensity<1000) was detected in one recipient at the time of biopsy.
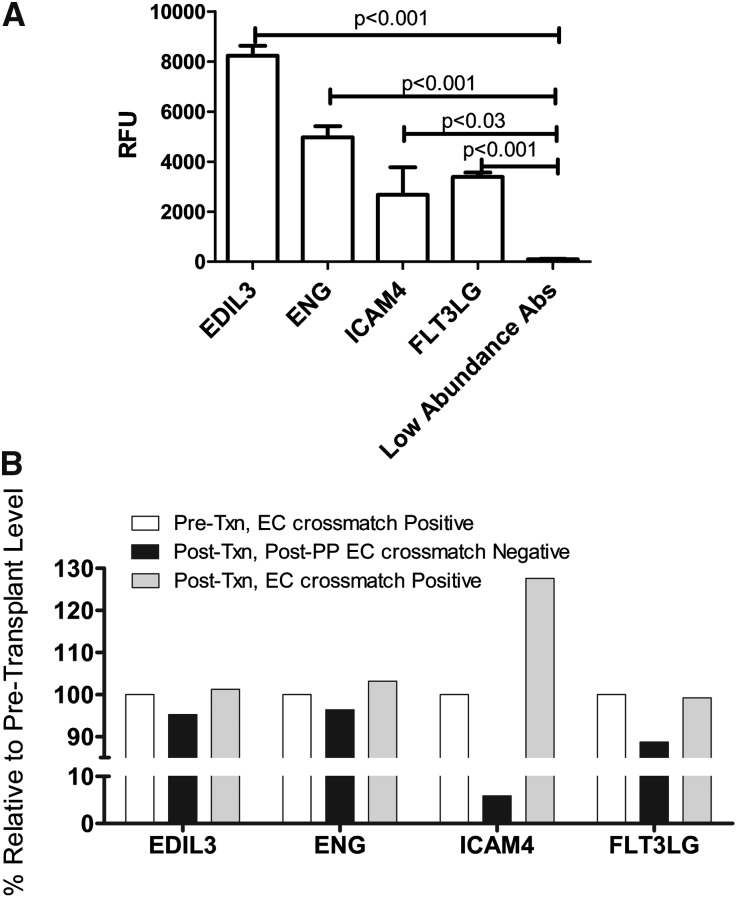
To focus our analyses on AECA target antigens, antibody eluates were generated using ECPs derived from blood. In brief, each serum was incubated with ECPs, and after wash steps, the bound antibodies were eluted. Using a high-density protein platform, we profiled AECA eluates from 10 Discovery Cohort recipients against approximately 9500 human proteins. Four proteins expressed on vascular endothelium, endoglin, EGF-like repeats and discoidin I-like domains 3 (EDIL3), intercellular adhesion molecule 4 (ICAM4), and Fms-like tyrosine kinase-3 (FLT3) ligand, were identified in all eluates. Signal intensities for these four antibodies were significant (endoglin, EDIL3, and FLT3: P<0.001; ICAM4: P<0.03) compared with low-abundance antibodies with signals<2000 relative fluorescent units (Figure 2A, Supplemental Table 2). We also performed analyses of AECA eluates derived from two recipients undergoing therapeutic plasmapheresis for rejection. These AECA eluates were derived from endothelial cell crossmatch-positive sera drawn before transplantation, crossmatch-negative sera obtained at the end of treatment, and sera drawn later post-treatment (3 and 9 months later) that again tested positive in an endothelial cell crossmatch. Protein array data, normalized to total IgG, showed that AECAs for all four protein targets (endoglin, FLT3 ligand, EDIL3, and ICAM4) decreased after plasmapheresis and rebounded again in sera that had AECAs detectable in crossmatch tests (Figure 2B).
Figure 2.
Identification of antigenic targets for AECAs and the effect of desensitization treatment. (A) Protein array analysis of 14 AECA eluates derived from endothelial cell crossmatch-positive sera. Relative fluorescence units (RFUs) for antibodies specific for EDIL3, endoglin (ENG), ICAM4, and FLT3 Ligand (FLT3LG) were significant compared with the mean signal intensity of low-abundance antibodies. We defined a positive threshold of 2000 RFU as a cutoff for abundance. The error bars represent SEM. (B) Effect of plasmapheresis/intravenous Ig (PP) on AECA levels. AECAs specific for ENG, FLT3LG, EDIL3, and ICAM4 decreased after PP treatments and rebounded in a post-transplant serum that yielded a positive endothelial cell crossmatch test. Data shown are from a single renal recipient, and values were normalized to total IgG. EC, endothelial cell; Txn, transplantation.
Expression of Target Antigens on Endothelial Cells
Cell phenotype analysis using flow cytometry confirmed surface expression of endoglin, EDIL3, ICAM4, and FLT3 (receptor for FLT3 ligand) on blood-derived ECPs compared with an isotype control antibody (data not shown). Analysis of two endothelial cell lines, derived from iliac artery and human umbilical vein, yielded positive staining for endoglin but negative staining for EDIL3, ICAM4, and FLT3, even after TNF-α stimulation (data not shown).
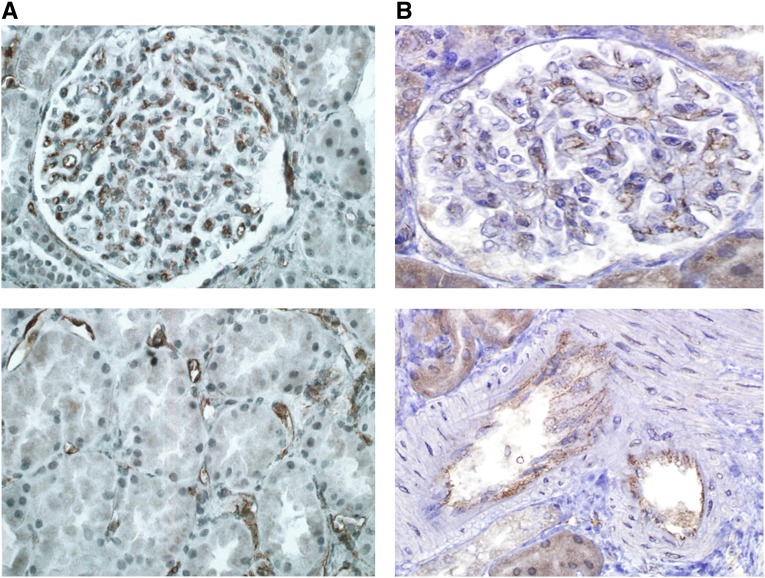
To investigate expression of these antigenic targets in renal tissue, immunohistochemistry was performed on rejection biopsies obtained from nine Discovery Cohort recipients. Figure 3 illustrates representative staining for endoglin and FLT3, which were expressed on arterial endothelium and glomerular and peritubular capillaries. Concomitant staining of biopsy tissue for FLT3 ligand, EDIL3, and ICAM4 yielded negative results.
Figure 3.
Expression of endoglin and FLT3 on renal endothelium. Immunohistochemistry performed on biopsies taken at time of rejection shows expression of (A) endoglin and (B) FLT3 on glomerular and peritubular microvasculature and arteries. Data shown are representative of biopsies tested from nine Discovery Cohort recipients.
Incidence of AECAs Using Antigen-Specific ELISAs
Sera from 150 sequential recipients of renal transplants for whom there were adequate pre- and post-transplant (≤3 months) samples were tested using MSD ELISAs specific for endoglin, EDIL3, ICAM4, and FLT3. This retrospective study cohort was similar to the Discovery Cohort in that it was enriched for recipients sensitized to HLA, with 91% (137 of 150) of recipients testing positive for HLA-specific class I and/or II antibodies (Supplemental Table 1).
We analyzed the most strongly reacting sera in each ELISA with a signal intensity equal to or greater than the trimmed mean. Fifty-six (37%) sera reacted positively with one or more antigenic targets. Within this group, 36 (24%) sera showed strong reactivity with all four antigen targets (Table 1). Pairwise comparisons performed using the top 36 reacting sera yielded highly significant (P<0.001) correlations in antibody production for these four antigens: endoglin and EDIL3 (R2=0.91, r=0.954), endoglin and ICAM4 (R2=0.89, r=0.94), EDIL3 and ICAM4 (R2=0.87, r=0.93), FLT3 and endoglin (R2=0.74, r=0.86), FLT3 and ICAM4 (R2=0.76, r=0.87), and FLT3 and EDIL3 (R2=0.72, r=0.85). AECA levels specific for these four targets decreased in post-transplant sera (≤3 months) in nearly all patients (93%; 140 of 150 patients).
Table 1.
Patient demographics for AECA ELISA categories
| ELISA Categories | Negative (n=40) | Intermediate (n=20) | P Value | Strongly Positive (n=36) | P Value |
|---|---|---|---|---|---|
| ELISA values, mean; median | |||||
| Endoglin | 1058; 992 | 6486; 6357 | <0.001 | 37,432; 26,101 | <0.001 |
| FLT3 | 1311; 1105 | 5499; 5876 | <0.001 | 29,876; 19,104 | <0.001 |
| EDIL3 | 324; 0 | 4437; 4397 | <0.001 | 34,213; 23,290 | <0.001 |
| ICAM4 | 3215; 3272 | 15,378; 13,077 | <0.001 | 56,839; 32,896 | <0.001 |
| Recipient age (yr), mean±SD | 52±16 | 49±13 | 0.47 | 47±15 | 0.22 |
| Race, % not white | 40 | 40 | >0.99 | 50 | 0.49 |
| Men, % | 25 | 50 | 0.08 | 44 | 0.09 |
| Previous transplantation, % | 40 | 50 | 0.58 | 39 | >0.99 |
| HLA sensitization,a % | 90 | 90 | >0.99 | 97 | 0.36 |
| Mean CPRAb (CDC-XM, FCXM), % | 27, 37 | 27, 38 | 0.94 | 35, 44 | 0.48 |
| Original ABO or HLA barrier,c % | |||||
| ABOi | 10 | 15 | 0.68 | 18 | 0.50 |
| CDC-XM+ | 2 | 0 | >0.99 | 6 | 0.60 |
| FCXM+ | 15 | 15 | >0.99 | 19 | 0.76 |
| FCXM−, DSA+ | 48 | 55 | 0.78 | 50 | >0.99 |
| No DSA | 35 | 30 | 0.78 | 25 | 0.45 |
| Donor, mean age (yr)±SD | 38±13 | 43±12 | 0.12 | 42±16 | 0.25 |
| Live donor, % | 45 | 55 | 42 | ||
| Deceased donor, % | 55 | 45 | 0.59 | 58 | 0.82 |
| HLA-A;B;DR;DQ mismatch, mean±SD | 4.7±2.3 | 4.1±2.7 | 0.41 | 5.2±1.5 | 0.21 |
| Plasmapheresis treatments | |||||
| No pre- or post-treatments, % | 50 | 35 | 0.41 | 42 | 0.50 |
| Pretransplant, mean, median | 1.0, 0.0 | 1, 1 | 0.67 | 0.5, 0.0 | 0.17 |
| Post-transplant, mean, median | 3.6, 1.0 | 3, 2 | 0.86 | 4.5, 2.0 | 0.54 |
| Anti-CD25 induction, % | 17 | 20 | >0.99 | 15 | 0.76 |
| Thymoglobulin induction, % | 83 | 80 | >0.99 | 85 | 0.76 |
| Rituximab induction, % | 33 | 45 | 0.40 | 38 | 0.63 |
CPRA, calculated panel reactive antibody; CDC-XM, cytotoxicity crossmatch; ABO, blood group antigen; ABOi, blood group incompatible; DSA, donor HLA-specific antibody; HLA-A;B;DR;DQ, histocompatibility antigens.
HLA-specific antibody detected on Luminex bead immunoassays.
CPRA was determined for HLA antibodies of sufficient strength to yield a positive CDC-XM or FCXM.
Original donor HLA-specific antibody (DSA) strength before desensitization treatment.
AECA Activation of Endothelial Cell Cultures
Primary endothelial cell cultures were established and stimulated with negative control serum, AECA-positive sera, AECA eluates, TNF-α, or a serum containing HLA antibodies. AECA eluates from 5 of the original 10 Discovery Cohort recipients were used as stimulants. The eluates contained a concentration of AECAs compared with the original serum.
Cultured endothelial cells were stimulated for 24 hours, after which time surface phenotype analysis was performed using flow cytometry to assess endothelial cell activation. We observed significant increases in the expression of HLA class I (P=0.04) and adhesion molecules E selectin (P=0.02) and ICAM1 (P=0.005) after stimulation with the AECA eluates compared with negative control serum (data not shown) or the original AECA-positive sera from which the eluates were derived (Figure 4). The increase in PECAM1 after AECA eluate stimulation was not statistically different from the negative controls (P=0.12). Stimulation of endothelial cell cultures using TNF-α or serum containing HLA antibodies increased expression of all markers (HLA class I, PECAM1, E selectin, and ICAM1) compared with negative controls.
Figure 4.
Endothelial cell cultures stimulated with AECA eluates upregulate markers of activation. Primary endothelial cell cultures were stimulated with AECA-positive sera, AECA eluates, TNF-α (10 ng/well), or an HLA antibody-positive serum. Cell surface phenotype analysis was performed 24 hours poststimulation using flow cytometry. Comparisons of median fluorescence values were made between cells stimulated with AECA eluates versus recipient sera (P values shown). Cumulative data were obtained from seven independent experiments, and AECA eluates from five Discovery Cohort recipients were tested. Aby, antibody; PECAM1, platelet/endothelial cell adhesion molecule 1.
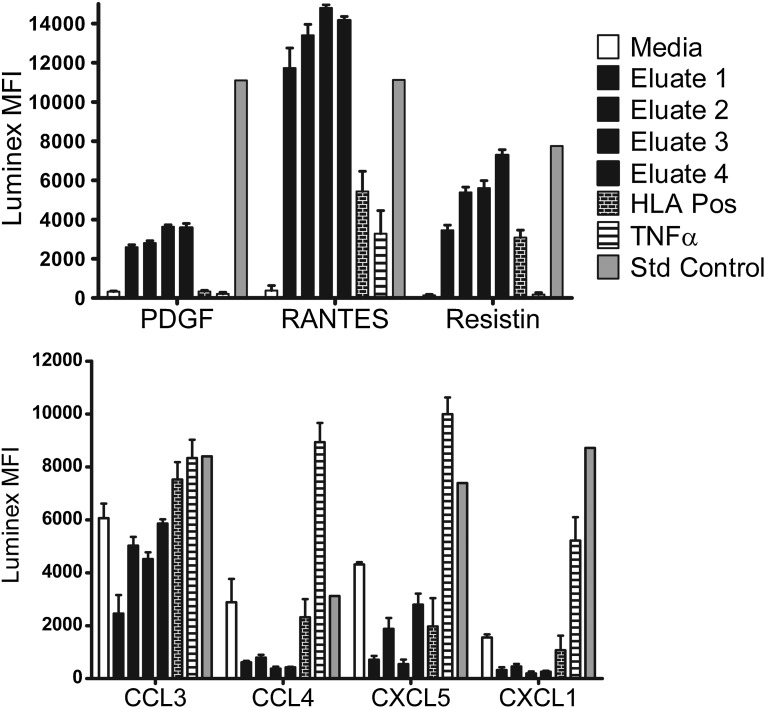
We assessed the production of inflammatory cytokines and chemokines after AECA eluate stimulation using a multiplexed immunoassay run on a Luminex platform. Primary endothelial cell cultures were stimulated with culture medium alone, negative control serum, AECA eluates, TNF-α, or serum containing HLA antibodies specific for the ECP donor. After 72 hours, culture supernatants were harvested and tested in an Affymetrix Procarta human immunoassay; 44 of 54 analytes were negative in all supernatants tested. IL-8, IL-1 receptor antagonist, and Serpin E1 were detected in all test cultures, including those without stimulation (culture medium alone). Inflammatory cytokines PDGF, RANTES (also known as CCL5), and RESISTIN were increased in cultures stimulated with the AECA eluates compared with negative controls or cultures stimulated with TNF-α or HLA antibodies (Figure 5). In contrast, chemokines CCL3, CCL4, CXCL5, and CXCL seemed to be decreased in cultures stimulated with the AECA eluates compared with other stimulants.
Figure 5.
Endothelial cell cultures stimulated with AECA eluates differentially affect production of inflammatory cytokines and chemokines. Primary endothelial cell cultures were stimulated with culture media alone, AECA eluates, an HLA antibody-positive serum, or TNF-α (10 ng/well). Culture supernatant was tested 72 hours poststimulation using a Procarta human 54 analyte immunoassay acquired on a Luminex xMAP multiplex platform. Median fluorescent intensities (MFIs) for differentially expressed analytes and positive control standards are shown. Data are representative of four independent experiments, and testing AECA eluates were derived from five Discovery Cohort recipients. Production of inflammatory cytokines PDGF, regulated on activation normal T cell expressed and presumably secreted (RANTES; also known as CCL5), and RESISTIN were increased after stimulation with the AECA eluates compared with negative controls (P<0.001, P<0.001, and P<0.001, respectively) or cells stimulated with TNF-α or HLA antibodies (P<0.001, P<0.001, and P=0.002, respectively). In contrast, chemokines CCL3, CCL4, CXCL5, and CXCL decreased in cultures stimulated with the AECA eluates compared with stimulation with TNF-α or HLA antibodies (P<0.001, P=0.05, P=0.07, and P=0.04, respectively). Pos, positive; Std, standard.
Clinical Outcomes for Recipients Testing AECA-Positive
To bring this study full circle, we performed a preliminary analysis of the clinical outcomes for 40 recipients with the lowest reacting sera on the ELISAs, an intermediate group of 20 recipients who tested above the trimmed mean for one to three target antigens, and 36 recipients who tested strongly positive for antibodies against all four targets: endoglin, EDIL3, ICAM4, and FLT3 (Table 1). Sera from the remaining recipients tested below the trimmed mean in ELISAs for all four target antigens. The demographics for these three ELISA cohorts were similar for all pretransplantation characteristics, including HLA-DSA strength at the time of transplantation.
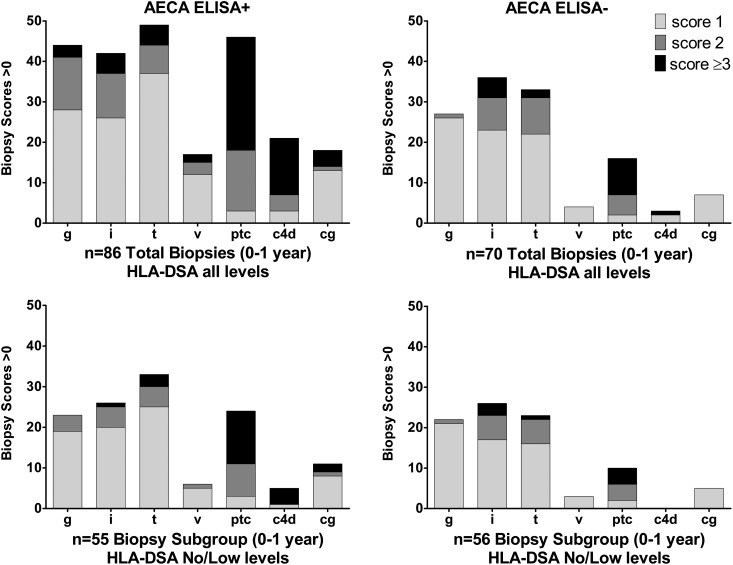
In total, 28% of the ELISA-positive group and 25% of the intermediate group experienced delayed graft function, requiring post-transplant dialysis, compared with 15% in the ELISA-negative cohort (Table 2). The incidence of cellular rejection was not significantly different between the three groups; however, AMR was significantly higher in recipients who tested positive for AECAs (intermediate: 15%, P=0.04; strongly positive: 19%, P=0.007) compared with 4% in recipients who tested ELISA-negative. Analyses of 86 biopsies from 36 recipients with the strongest reacting sera (68 indication and 18 protocol biopsies) revealed significantly more glomerulitis, peritubular capllaritis, C4d positivity, and transplant glomerulopathy than biopsies from 40 ELISA-negative recipients (36 indication and 34 protocol biopsies) (Figure 6, Table 2). The intermediate group (31 indication and 36 protocol biopsies) showed less glomerulitis but more peritubular capllaritis, C4d positivity, and chronic vascular changes than the negative group. The incidence of HLA-DSA at time of biopsy was high, reflecting the high degree of HLA sensitization and the inclusion of incompatible transplants in these patient cohorts (Table 1). HLA-DSA was detected at the time of biopsy in 76% of biopsies from the ELISA-positive recipients, 45% of biopsies from the intermediate positive recipients, and 47% of biopsies from ELISA-negative recipients. Recipients who tested strongly positive for AECAs had a greater number of DSAs at time of biopsy and an increased incidence of antibodies to both HLA classes I and II antigens. From a subsequent analysis, we eliminated biopsies with high-level HLA-DSA sufficient to yield a positive flow cytometric crossmatch (FCXM) or cytotoxicity lymphocyte crossmatch test to examine the effect of low-level HLA-DSA and AECAs. Among remaining biopsies with no or low-level HLA-DSA, which were detected only by bead assays, more microvascular injury was still observed in the strongly positive AECA recipients versus negative recipients (Figure 6, Table 2). Histopathology scores for microvascular injury were not significantly increased in biopsies from recipients with intermediate levels of AECAs with no or low HLA-DSA.
Table 2.
Correlation between AECA ELISAs and clinical outcome
| ELISA Categories | Negative (n=40) | Intermediate (n=20) | P Value | Strongly Positive (n=36) | P Value |
|---|---|---|---|---|---|
| Graft losses,a no. | 0 | 0 | >0.99 | 1 | 0.47 |
| Delayed graft function,b % | 15 | 25 | 0.48 | 28 | 0.26 |
| Acute rejectionc<1 yr, % biopsies | 25 | 32 | 0.76 | 37 | 0.33 |
| CMR, N (%) | 15 (21) | 13 (19) | 0.83 | 16 (19) | 0.69 |
| AMR, N (%) | 3 (4) | 10 (15) | 0.04 | 16 (19) | <0.01 |
| Time of first rejection (d), mean, median | 131, 102 | 76, 17 | 0.63 | 67, 19 | 0.18 |
| Donor-specific HLA antibody (HLA-DSA) | |||||
| All biopsies, % | 44 | 34 | 0.16 | 75 | <0.001 |
| No/low HLA-DSA biopsies, % | 25 | 24 | 0.39 | 38 | <0.01 |
| Number DSA all biopsies, mean±SD | 0.8±1.1 | 1.3±1.5 | 0.04 | 2.5±1.9 | <0.001 |
| Class I, N (% of biopsies) | 11 (14) | 5 (7) | 0.18 | 14 (17) | >0.99 |
| Class II, N (% of biopsies) | 12 (14) | 14 (21) | 0.66 | 24 (28) | 0.13 |
| Classes I and II, N (% of biopsies) | 8 (11) | 5 (7) | 0.56 | 26 (31) | <0.01 |
| Glomerulitis (0–3), mean±SD | |||||
| All biopsies | 0.4±0.5d | 0.2±0.4e | 0.004 | 0.7±0.9f | 0.004 |
| No/low HLA-DSA | 0.4±0.5 | 0.1±0.3 | <0.001 | 0.5±0.6 | 0.47 |
| Inflammation (0–3), mean±SD | |||||
| All biopsies | 0.8±0.9 | 0.8±0.9 | 0.79 | 0.7±0.9 | 0.79 |
| No/low HLA-DSA | 0.7±0.9 | 0.5±0.6 | 0.14 | 0.6±0.7 | 0.53 |
| Tubulitis (0–3), mean±SD | |||||
| All biopsies | 0.7±0.8 | 0.8±0.9 | 0.78 | 0.8±0.8 | 0.41 |
| No/low HLA-DSA | 0.6±0.8 | 0.5±0.6 | 0.43 | 0.8±0.8 | 0.11 |
| Vasculitis (0–3), mean±SD | |||||
| All biopsies | 0.1±0.2 | 0.2±0.5 | 0.18 | 0.3±0.6 | 0.004 |
| No/low HLA-DSA | 0.1±0.2 | 0.0±0.2 | 0.82 | 0.1±0.4 | 0.23 |
| Peritubular capllaritis (0–3), mean±SD | |||||
| All biopsies | 0.6±1.1 | 1.1±1.3 | 0.04 | 1.4±1.4 | <0.001 |
| No/low HLA-DSA | 0.4±0.9 | 0.6±1.1 | 0.33 | 1.1±1.3 | 0.002 |
| C4d (0–3), mean±SD | |||||
| All biopsies | 0.1±0.4 | 0.6±1.1 | 0.004 | 0.7±1.2 | <0.001 |
| No/low HLA-DSA | 0.0±0.0 | 0.0±0.2 | 0.22 | 0.3±0.9 | 0.02 |
| Transplant glomerulopathy (0–3), mean±SD | |||||
| All biopsies | 0.1±0.3 | 0.0±0.3 | 0.85 | 0.3±0.7 | 0.01 |
| No/low HLA-DSA | 0.1±0.3 | 0.0±0.0 | 0.35 | 0.3±0.7 | 0.05 |
| IFTA (0–3), mean±SD | |||||
| All biopsies | 1.4±1.4 | 1.9±1.8 | 0.12 | 1.5±1.5 | 0.43 |
| No/low HLA-DSA | 1.0±1.3 | 1.4±1.5 | 0.07 | 1.7±1.5 | <0.01 |
| Cv (0–3), mean±SD | |||||
| All biopsies | 0.7±0.8 | 1.1±1.1 | 0.02 | 0.9±1.0 | 0.11 |
| No/low HLA-DSA | 0.6±0.8 | 0.9±1.0 | 0.08 | 0.9±0.9 | 0.08 |
| Serum creatinine, mean±SD | |||||
| 7 d | 2.39±2.25 | 3.27±2.52 | 0.19 | 4.69±4.27 | 0.004 |
| 1 mo | 1.38±1.02 | 1.77±0.97 | 0.16 | 1.80±1.07 | 0.09 |
| 3 mo | 1.33±0.64 | 1.90±1.62 | 0.16 | 1.63±1.40 | 0.28 |
| 12 mo | 1.40±0.56 | 1.73±1.47 | 0.40 | 1.61±1.47 | 0.48 |
| Last available value | 1.73±1.47 | 2.05±2.10 | 0.55 | 2.59±2.88 | 0.11 |
| Post-transplant, mo | 18.8±13.4 | 19±2.1 | 0.99 | 24.5±14.9 | 0.09 |
CMR, cellular-mediated rejection; DSA, donor HLA-specific antibody; IFTA, interstitial fibrosis and tubular atrophy; Cv, chronic vascular changes.
Graft loss because of accelerated vascular rejection.
Post-transplant dialysis.
In total, 70 biopsies (34 protocol) and 56 biopsies (28 protocol) with no or low HLA-DSA (detected by Luminex only).
In total, 67 biopsies (36 protocol) and 46 biopsies (30 protocol) with no or low HLA-DSA (detected by Luminex only).
In total, 86 biopsies (18 protocol) and 55 biopsies (16 protocol) with no or low HLA-DSA (detected by Luminex only).
Figure 6.
Histologic scores for recipients who were AECA ELISA strongly positive (n=28) and negative (n=24). Renal biopsies with positive histologic scores>1 were acquired during the first year post-transplantation according to protocol or at the time of dysfunction. Shown are scores from all biopsies or a subset of biopsies when HLA-DSA was not detected at time of biopsy or detected at a negative FCXM level.21 Histologic scoring (0–3) was performed using updated Banff 1997–2007 criteria.27–30 Shown are grades for glomerulitis (g), interstitial (i) and tubular (t) inflammation, vasculitis (v), and peritubular capillaritis (ptc). C4d staining was performed on frozen tissue by indirect immunofluorescence. Transplant glomerulopathy (cg) was defined by duplication of the glomerular basement membrane as observed on electron and light microscopy. Additional details and statistics are provided in Table 2.
Serum creatinine levels in the immediate post-transplant period (7 days) were significantly higher in the ELISA-positive group (P=0.004), corresponding to the high incidence of delayed graft function. The mean serum creatinine at 1 month was higher in the ELISA-positive group (1.80±1.07) compared with the ELISA-negative group (1.38±1.02), but the difference was not significant (P=0.09). Mean serum creatinine levels at 3 months, 1 year, and ≥1.5 years were also higher the ELISA-positive cohort but not significantly different.
Discussion
In this study, a proteomic approach was used to identify novel antigenic targets for AECAs associated with renal allograft dysfunction. Protein array analysis of AECAs eluted from ECPs identified endoglin, EDIL3, ICAM4, and FLT3 ligand as target antigens. Endoglin is constitutively expressed within the vasculature and has been proposed to play a role in endothelial cell activation and inflammation.14 FLT3 is a tyrosine kinase receptor for the cytokine FLT3 ligand. In mice, Thomson and colleagues15 showed that FLT3 stimulation can increase RANTES (CCL5) production by renal endothelial cells, resulting in the recruitment of CD45+ mononuclear cells.
EDIL3 was not detected by immunohistochemistry in renal biopsies taken at the time of rejection from our Discovery Cohort, but inflammation has been shown to reduce EDIL3 expression on vascular endothelium.16 In mice, EDIL3 (Del-1) functions as an inhibitor of LFA1-dependent cell adhesion, resulting in reduced leukocyte extravasation from blood into underlying tissues. Inflammation acts as a switch to upregulate expression of activation molecules, such as ICAM-1 and vascular cell adhesion molecule 1, and downregulate inhibitory molecules, such as EDIL3. Therefore, the inability to detect EDIL3 at the time of a rejection biopsy may not preclude its presence on endothelial cells during the initial stages of rejection. The final protein identified in the AECA eluates was ICAM4 (previously named the Landsteiner–Wiener blood group). This molecule is part of the ICAM family of adhesion molecules and believed to be red blood cell lineage-specific.17 Expression of ICAM4 on renal endothelium could not be validated using immunohistochemistry; however, vascular expression has been documented in microarray literature.18
In the 2011 Banff Conference Allograft Pathology Report, the heterologous phenotypes associated with AMR in renal transplantation were discussed and recognition of AMR in the absence of detectable complement activation, which was observed in our Discovery Cohort, was included.2 Gene array data from Sis et al.19 have shown that increased endothelial cell transcripts are a more sensitive biomarker for AMR than detection of C4d in acute renal biopsies. We and others have reported hyperacute rejections in patients positive for AECAs with no detectable HLA-DSA.13,20 Thus, there is growing evidence that antibody-induced endothelial cell activation, independent of complement, may be sufficient for immune cell recruitment and microvascular injury. Three of the AECA targets identified in this study (endoglin, EDIL3, and FLT3) have been implicated in endothelial activation and leukocyte extravasation, and these findings are consistent with our in vitro data showing that AECA stimulation of endothelial cells results in the upregulation of adhesion and HLA molecules and the production of inflammatory chemokines and cytokines. Admittedly, the histopathology analysis was confounded by the presence of HLA-DSA in many recipients at time of rejection, thus weakening our ability to prove a causal link for AECAs. However, the AECA ELISA-positive and -negative cohorts used for outcome analyses were matched for all relative criteria at time of transplantation, including breadth of sensitization and strength of HLA-DSA at final crossmatch. The divergence of these groups post-transplantation with regards to the severity of rejection and the presence of HLA-DSA at time of rejection supports our hypothesis that pretransplant AECA detection in recipients sensitized to HLA identifies those at higher risk for allograft rejection. Given the increased incidence of transplant glomerulopathy in the AECA-positive cohort, additional studies evaluating the effect of AECAs in long-term allograft survival are needed.
Routine testing for AECAs has not been realized, in part, because of a paucity of tools for inexpensive high-throughput screening of patient sera. The use of cell-based crossmatch methods has been problematic because of the inefficiency of testing single recipient/donor pairs and the inability to identify non-HLA antibodies in patients with known HLA sensitization. In this study, we developed an ELISA platform to increase the sensitivity and efficiency of AECA detection and provided mechanistic data to investigate the role of AECAs in microvascular injury.
Concise Methods
Study Patients
Clinical data and stored sera from 160 recipients of renal transplants transplanted between 2009 and 2011 were studied retrospectively after institutional review board approval. Sera from a discovery group of 10 recipients who tested positive in endothelial cell crossmatch tests and experienced allograft rejection in the first 3 months post-transplant in the absence of donor HLA-specific antibody were tested on a protein array platform. ELISA tests were performed on 150 sequential recipients of renal transplants for whom there were adequate pre- and post-transplant (≤3 months) sera. Patient demographics are provided in Supplemental Table 1.
Immunosuppression
Maintenance immunosuppression included mycophenolate mofetil (2 g/d), tacrolimus (serum level of 8–10 ng/ml), and prednisone (30 mg/d). Prednisone was reduced to 20 mg/d when tacrolimus reached therapeutic range (8–12 ng/dl) and tapered thereafter to 5 or 10 mg/d. Intraoperative induction therapy was either anti–IL-2 receptor antibody (anti-CD25, daclizumab at 2 mg/kg) or Thymoglobulin (1.5 mg/kg per day for 5 days). HLA and ABO incompatible pairs were included in this study. Desensitization included alternate day plasmapheresis immediately followed by low-dose (100 mg/kg) intravenous Ig (Cytogam-CSL Behring, King of Prussia, PA). Mycophenolate mofetil (2 g/d) and tacrolimus (serum level of 8–10 ng/ml) were initiated with the start of plasmapheresis treatments. The number of treatments was dependent on ABO or donor HLA-specific antibody (HLA-DSA) titer. Biopsy-confirmed acute cellular rejection was treated with pulse solumedrol at cumulative doses of 30–50 mg/kg in three to six doses and increased baseline target levels for tacrolimus and/or mycophenolate mofetil. Biopsy-confirmed acute AMR was treated with plasmapheresis and/or rituximab and/or intravenous Ig and pulse solumedrol.
HLA Antibody Detection and Crossmatch Tests
HLA-specific antibodies were evaluated before transplantation and at time of biopsy using solid-phase immunoassays (Lifecodes classes I and II ID panels; Immucor-Lifecodes, Stamford, CT; Single Antigen Beads; One Lambda, Canoga Park, CA) performed on a Luminex platform. Crossmatch tests with donor T and B cells were performed using standard cytotoxicity and/or FCXM protocols. Calculated panel reactive antibody was determined for antibodies sufficient to yield a positive cytotoxicity or FCXM using virtual crossmatch methods and the online UNOS CPRA calculator.21 Non-HLA AECAs were detected using an FCXM performed using angiopoietin receptor positive (Tie-2+) peripheral blood ECPs22 (XM-ONE; Absorber AB, Stockholm, Sweden) acquired on a BD FACSAria using FACSDiva (version 6.1.1; BD Biosciences, Franklin Lakes, NJ).
Non-HLA AECA Eluates
AECA eluates were derived from adsorbing antibody from endothelial cell crossmatch-positive sera onto Tie-2+ ECPs isolated from surrogate donors for whom there was no HLA-DSA. The serum-to-cell ratio, incubation, and wash steps were consistent with the endothelial cell crossmatch procedure. Antibody was acid-eluted 1:10 of the original serum volume as previously described23 and dialyzed 18 hours against PBS. AECA eluates were retested in endothelial cell crossmatch tests to ensure antibody integrity and specificity.
Protein Microarray Analyses
ProtoArray Human Protein Microarrays v5.0 (Life Technologies, Foster City, CA) were used to profile antibodies present in 14 AECA eluates from 10 recipients who were endothelial cell crossmatch-positive. For two recipients, analysis was performed on AECA eluates derived from sera obtained before and after desensitization treatments. The protein arrays contained approximately 9500 recombinant human proteins expressed as N-terminal GST fusion proteins and spotted on nitrocellulose-coated glass slides. Established protocols (http://www.invitrogen.com) were followed24,25 for sample preparation, blocking, probing, drying by centrifugation, scanning, and data acquisition. Slides were scanned using an Axon GenePix 4000B Scanner (Molecular Devices, Sunnyvale, CA) and GenePix pro 6.0 software (Molecular Devices). We used the data analysis software ProtoArray Prospector 5.2 to analyze any bound IgG antibody detected by the secondary Alexa Fluor 647-conjugated antibody. Binding of the secondary antibody on the microarray was then quantified by measuring the fluorescence intensity of each feature on the slide. Data generation and normalization involved calculating appropriate fluorescent signal values by taking into account corrections for background and negative control features printed on the microarray. Z factors for all of the corrected intensities of the human protein features were calculated, and the features that had signal intensities greater than the cutoff value of 2000 were considered as abundant signal. Pathway analysis was performed using Ingenuity Pathway analysis (Ingenuity Systems; www.ingenuity.com) and the GO-Elite Pathway Analysis Tool (http://www.genmapp.org/go_elite/go_elite.html).
Endothelial Cell Antigen-Specific ELISAs
Endothelial cell-specific antigen targets identified by protein arrays were validated using the MSD ELISA platform (Meso Scale Discovery, Gaithersburg, MD). Recombinant proteins/antigens obtained from Abcam, Inc. (Cambridge, MA) included EDIL3 (catalog no. ab94549), FLT3 (catalog no. 83996), and endoglin (catalog no. ab95043), and recombinant ICAM4 (catalog no. H00003386-P) was purchased from Abnova (Walnut, CA). To increase detection specificity and sensitivity, each antigen was biotin-conjugated and incubated with diluted serum before coating the antigen–antibody complex onto the avidin-coated ELISA plate. Biotin labeling of each antigen (5 µg) was performed using EZ-Link Sulfo-NHS-LC_Biotin (catalog no. PI21335), the EZ BIOTIN QUANTIFICATION KIT (catalog no. PI28005), and Zeba* Spin Desalting Columns (7K MWCO, 5 ml; catalog no. PI89892) following the manufacturer’s protocol. The serum was diluted 1:75 with 2% Blocker A (catalog no. R93AA-1; Meso Scale Discovery) in TPBS after titration studies showed it to be the optimal dilution. Equal parts of diluted serum (50 µl) and biotin-labeled antigen (5 ng) or 2% Blocker A in TPBS were added to a Streptavidin Gold Plate (catalog no. L15SA-1; Meso Scale Discovery) and incubated for 90 min at room temperature with shaking (300 rpm). The plate was blocked with 5% Blocker A (150 µl) for 90 minutes at room temperature with shaking and washed one time with 300 μl TPBS, and 100 μl diluted goat anti-human SULFO-TAG (1:1000) detection antibody was added and incubated in the dark for 60 minutes at room temperature with shaking. After three washes with 300 μl TPBS, 150 µl reading buffer was added to each well, and the plate was immediately read with an SECTOR Imager 2400 (Meso Scale Discovery). Reactivity above the trimmed mean was considered positive, and changes between pre- and post-transplant antibody levels were determined by increases or decreases that exceeded a 95% confidence interval for each ELISA.
Endothelial Cell Culture
Primary endothelial cell cultures were established using Tie-2+ ECPs isolated from donors who yielded a positive endothelial cell crossmatch test with sera from the Discovery Cohort as previously described with minor modifications.26 In brief, ECPs were isolated using Tie-2 magnetic beads (XM-ONE Absorber AB), plated at a 1000-cells/mm2 density in fibronectin (1 μg/well)-coated wells, and cultured for 2–3 weeks in supportive media (EGM-2 Bullet Kit; Lonza, Walkersville, MD) to allow for maturation. Endothelial cell lines (CRL-2606; ATCC; HuVEC, gift of Mark K. Halushka) were cultured as above; cultures were passed when 70%–80% confluent and analyzed within the first five passages.
Analyses after Endothelial Cell Culture Stimulation
Primary endothelial cell cultures were washed two times with wash buffer (1× PBS with 5% heat-inactivated FCS). Cells were stimulated using 100 μl negative control AB serum, endothelial cell crossmatch positive sera, AECA eluates, TNF-α (10 ng/well; Abcam, Inc.), or a serum containing antibodies specific for HLA antigens of the ECP donor. Cells were incubated for 1 hour, after which time 1 ml supportive media was added to each well. Cell surface phenotype analysis was performed 24 hours poststimulation. Cells were removed with 0.25% trypsin, washed, and stained according to standard protocols. mAbs included peridinin chlorophyll protein complex-conjugated CD54 (clone HA58), phycoerythrin-conjugated CD62E (clone 68–5H11), fluorescein isothiocyanate-conjugated HLA class I (clone G46–2.6) and CD3 (clone SK7), and Alexa Fluor 647-congugated CD31 (clone WM59; BD Biosciences). Rabbit anti-human polyclonal antibodies included CD105 (endoglin, ab21224), CD242 (ICAM4, ab112554), EDIL3 (ab74775), and CD135 (FLT3, ab37847; Abcam, Inc.). Allophycocyanin-conjugated goat anti-rabbit IgG was purchased from R&D Systems (Minneapolis, MN). Cells were acquired (2000 gated events) and analyzed using BD FACSAria and FACSDiva (version 6.1.1; BD Biosciences) and/or De Novo Software (Los Angeles, CA). After 72 hours of stimulation, culture supernatants were tested using the Procarta human 54 analyte immunoassay (Affymetrix Inc., Santa Clara, CA) according to the manufacturer’s protocol and acquired on a Luminex xMAP multiplex platform. Each assay included a positive control standard for each target protein.
Histopathogy
All biopsies performed during the first year post-transplantation were included in the analysis. Biopsies were performed at the time of graft dysfunction and for HLA-incompatible recipients according to the protocol at 1, 3, 6, and 12 months post-transplantation. Recipients may have been excluded from protocol biopsies because of anticoagulation therapy or if the intra-abdominal transplant could not be safely biopsied. Biopsies were scored using updated Banff 1997–2007 criteria.27–30 Criteria for diagnosis of AMR included detection of C4d by indirect immunofluorescence. C4d staining was considered positive if present in ≥50% of the peritubular capillaries with intensity≥1+ (C4d2–3). Additional staining of biopsy tissue was performed using rabbit anti-human polyclonal antibodies reactive with CD105 (RB9291P1; Thermo Fisher Scientific, Waltham, MA), CD242 (ICAM4, ab112554), EDIL3 (ab74775), and CD135 (FLT3, ab37847; Abcam, Inc.). Detection was performed using a horseradish peroxidase polymer-conjugated secondary antibody (SuperPicture Polymer Detection Kit; Life Technologies, Grand Island, NY).
Statistical Methods
Protein array data were analyzed by Prospector Analyzer (Life Technologies) using robust linear model normalization.31 A minimum relative fluorescent unit>500 and a Z factor of 0.4 were required for positive detection. Summary statistics, including mean, trimmed mean, median, confidence norm distribution, correlation coefficient, coefficient of difference, and SD, were calculated using Microsoft Excel. Statistical significance was determined using chi-squared and t tests (two tailed), and P values<0.05 were considered significant.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank Drs. Andrea A. Zachary and Mary S. Leffell for critical reading of this manuscript.
This work was supported by a grant from the National Kidney Foundation of Maryland (to A.M.J.).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Renal Allograft Rejection: Pieces of the Puzzle,” on pages 1004–1005.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013121277/-/DCSupplemental.
References
- 1.McKenna RM, Takemoto SK, Terasaki PI: Anti-HLA antibodies after solid organ transplantation. Transplantation 69: 319–326, 2000 [DOI] [PubMed] [Google Scholar]
- 2.Mengel M, Sis B, Haas M, Colvin RB, Halloran PF, Racusen LC, Solez K, Cendales L, Demetris AJ, Drachenberg CB, Farver CF, Rodriguez ER, Wallace WD, Glotz D, Banff meeting report writing committee : Banff 2011 Meeting report: New concepts in antibody-mediated rejection. Am J Transplant 12: 563–570, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sis B, Jhangri GS, Riopel J, Chang J, de Freitas DG, Hidalgo L, Mengel M, Matas A, Halloran PF: A new diagnostic algorithm for antibody-mediated microcirculation inflammation in kidney transplants. Am J Transplant 12: 1168–1179, 2012 [DOI] [PubMed] [Google Scholar]
- 4.Loupy A, Hill GS, Jordan SC: The impact of donor-specific anti-HLA antibodies on late kidney allograft failure. Nat Rev Nephrol 8: 348–357, 2012 [DOI] [PubMed] [Google Scholar]
- 5.Morrell CN, Murata K, Swaim AM, Mason E, Martin TV, Thompson LE, Ballard M, Fox-Talbot K, Wasowska B, Baldwin WM, 3rd: In vivo platelet-endothelial cell interactions in response to major histocompatibility complex alloantibody. Circ Res 102: 777–785, 2008 [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Valenzuela NM, Reed EF: HLA class I antibody-mediated endothelial and smooth muscle cell activation. Curr Opin Organ Transplant 17: 446–451, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Opelz G, Collaborative Transplant Study : Non-HLA transplantation immunity revealed by lymphocytotoxic antibodies. Lancet 365: 1570–1576, 2005 [DOI] [PubMed] [Google Scholar]
- 8.Grafft CA, Cornell LD, Gloor JM, Cosio FG, Gandhi MJ, Dean PG, Stegall MD, Amer H: Antibody-mediated rejection following transplantation from an HLA-identical sibling. Nephrol Dial Transplant 25: 307–310, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Sigdel TK, Sarwal MM: Moving beyond HLA: A review of nHLA antibodies in organ transplantation. Hum Immunol 74: 1486–1490, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Sigdel T, Vitalone M, Lee SH, Sarwal M: Differential immunogenicity and clinical relevance of kidney compartment specific antigens after renal transplantation. J Proteome Res 9: 6715–6721, 2010 [DOI] [PubMed] [Google Scholar]
- 11.Breimer ME, Rydberg L, Jackson AM, Lucas DP, Zachary AA, Melancon JK, VonVisger J, Pelletier R, Saidman SL, Williams WW, Jr., Holgersson J, Tyden G, Klintmalm GK, Smith D, Coultrup S, Sumitran-Holgersson S, Grufman P: Multicenter evaluation of a novel endothelial cell crossmatch test in kidney transplantation. Transplantation 87: 549–556, 2009 [DOI] [PubMed] [Google Scholar]
- 12.Jackson AM, Lucas DP, Melancon JK, Desai NM: Clinical relevance and IgG subclass determination of non-HLA antibodies identified using endothelial cell precursors isolated from donor blood. Transplantation 92: 54–60, 2011 [DOI] [PubMed] [Google Scholar]
- 13.Jackson AM, Kuperman MB, Montgomery RA: Multiple hyperacute rejections in the absence of detectable complement activation in a patient with endothelial cell reactive antibody. Am J Transplant 12: 1643–1649, 2012 [DOI] [PubMed] [Google Scholar]
- 14.Docherty NG, López-Novoa JM, Arevalo M, Düwel A, Rodriguez-Peña A, Pérez-Barriocanal F, Bernabeu C, Eleno N: Endoglin regulates renal ischaemia-reperfusion injury. Nephrol Dial Transplant 21: 2106–2119, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Coates PT, Colvin BL, Ranganathan A, Duncan FJ, Lan YY, Shufesky WJ, Zahorchak AF, Morelli AE, Thomson AW: CCR and CC chemokine expression in relation to Flt3 ligand-induced renal dendritic cell mobilization. Kidney Int 66: 1907–1917, 2004 [DOI] [PubMed] [Google Scholar]
- 16.Choi EY, Chavakis E, Czabanka MA, Langer HF, Fraemohs L, Economopoulou M, Kundu RK, Orlandi A, Zheng YY, Prieto DA, Ballantyne CM, Constant SL, Aird WC, Papayannopoulou T, Gahmberg CG, Udey MC, Vajkoczy P, Quertermous T, Dimmeler S, Weber C, Chavakis T: Del-1, an endogenous leukocyte-endothelial adhesion inhibitor, limits inflammatory cell recruitment. Science 322: 1101–1104, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toivanen A, Ihanus E, Mattila M, Lutz HU, Gahmberg CG: Importance of molecular studies on major blood groups—intercellular adhesion molecule-4, a blood group antigen involved in multiple cellular interactions. Biochim Biophys Acta 1780: 456–466, 2008 [DOI] [PubMed] [Google Scholar]
- 18.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB: Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A 99: 4465–4470, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sis B, Jhangri GS, Bunnag S, Allanach K, Kaplan B, Halloran PF: Endothelial gene expression in kidney transplants with alloantibody indicates antibody-mediated damage despite lack of C4d staining. Am J Transplant 9: 2312–2323, 2009 [DOI] [PubMed] [Google Scholar]
- 20.Sumitran-Karuppan S, Tyden G, Reinholt F, Berg U, Moller E: Hyperacute rejections of two consecutive renal allografts and early loss of the third transplant caused by non-HLA antibodies specific for endothelial cells. Transpl Immunol 5: 321–327, 1997 [DOI] [PubMed] [Google Scholar]
- 21.Zachary AA, Sholander JT, Houp JA, Leffell MS: Using real data for a virtual crossmatch. Hum Immunol 70: 574–579, 2009 [DOI] [PubMed] [Google Scholar]
- 22.Jackson AM, Lucas DP, Badders JL: A flow cytometric crossmatch test using endothelial precursor cells isolated from peripheral blood. Methods Mol Biol 1034: 319–329, 2013 [DOI] [PubMed] [Google Scholar]
- 23.Lucchiari N, Panajotopoulos N, Xu C, Rodrigues H, Ianhez LE, Kalil J, Glotz D: Antibodies eluted from acutely rejected renal allografts bind to and activate human endothelial cells. Hum Immunol 61: 518–527, 2000 [DOI] [PubMed] [Google Scholar]
- 24.Sigdel TK, Li L, Tran TQ, Khatri P, Naesens M, Sansanwal P, Dai H, Hsieh SC, Sarwal MM: Non-HLA antibodies to immunogenic epitopes predict the evolution of chronic renal allograft injury. J Am Soc Nephrol 23: 750–763, 2012 [DOI] [PubMed] [Google Scholar]
- 25.Sigdel TK, Shoemaker LD, Chen R, Li L, Butte AJ, Sarwal MM, Steinberg GK: Immune response profiling identifies autoantibodies specific to Moyamoya patients. Orphanet J Rare Dis 8: 45, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asahara T, Murohara T, Sullivan A, Silver M, van der Zee R, Li T, Witzenbichler B, Schatteman G, Isner JM: Isolation of putative progenitor endothelial cells for angiogenesis. Science 275: 964–967, 1997 [DOI] [PubMed] [Google Scholar]
- 27.Racusen LC, Solez K, Colvin RB, Bonsib SM, Castro MC, Cavallo T, Croker BP, Demetris AJ, Drachenberg CB, Fogo AB, Furness P, Gaber LW, Gibson IW, Glotz D, Goldberg JC, Grande J, Halloran PF, Hansen HE, Hartley B, Hayry PJ, Hill CM, Hoffman EO, Hunsicker LG, Lindblad AS, Yamaguchi Y: The Banff 97 working classification of renal allograft pathology. Kidney Int 55: 713–723, 1999 [DOI] [PubMed] [Google Scholar]
- 28.Solez K, Colvin RB, Racusen LC, Sis B, Halloran PF, Birk PE, Campbell PM, Cascalho M, Collins AB, Demetris AJ, Drachenberg CB, Gibson IW, Grimm PC, Haas M, Lerut E, Liapis H, Mannon RB, Marcus PB, Mengel M, Mihatsch MJ, Nankivell BJ, Nickeleit V, Papadimitriou JC, Platt JL, Randhawa P, Roberts I, Salinas-Madriga L, Salomon DR, Seron D, Sheaff M, Weening JJ: Banff ‘05 Meeting Report: Differential diagnosis of chronic allograft injury and elimination of chronic allograft nephropathy (‘CAN’). Am J Transplant 7: 518–526, 2007 [DOI] [PubMed] [Google Scholar]
- 29.Solez K, Colvin RB, Racusen LC, Haas M, Sis B, Mengel M, Halloran PF, Baldwin W, Banfi G, Collins AB, Cosio F, David DS, Drachenberg C, Einecke G, Fogo AB, Gibson IW, Glotz D, Iskandar SS, Kraus E, Lerut E, Mannon RB, Mihatsch M, Nankivell BJ, Nickeleit V, Papadimitriou JC, Randhawa P, Regele H, Renaudin K, Roberts I, Seron D, Smith RN, Valente M: Banff 07 classification of renal allograft pathology: Updates and future directions. Am J Transplant 8: 753–760, 2008 [DOI] [PubMed] [Google Scholar]
- 30.Racusen LC, Colvin RB, Solez K, Mihatsch MJ, Halloran PF, Campbell PM, Cecka MJ, Cosyns JP, Demetris AJ, Fishbein MC, Fogo A, Furness P, Gibson IW, Glotz D, Hayry P, Hunsickern L, Kashgarian M, Kerman R, Magil AJ, Montgomery R, Morozumi K, Nickeleit V, Randhawa P, Regele H, Seron D, Seshan S, Sund S, Trpkov K: Antibody-mediated rejection criteria - an addition to the Banff 97 classification of renal allograft rejection. Am J Transplant 3: 708–714, 2003 [DOI] [PubMed] [Google Scholar]
- 31.Sboner A, Karpikov A, Chen G, Smith M, Mattoon D, Freeman-Cook L, Schweitzer B, Gerstein MB: Robust-linear-model normalization to reduce technical variability in functional protein microarrays. J Proteome Res 8: 5451–5464, 2009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.