SUMMARY
A decade of genome sequencing has transformed our understanding of how trypanosomatid parasites have evolved and provided fresh impetus to explaining the origins of parasitism in the Kinetoplastida. In this review, I will consider the many ways in which genome sequences have influenced our view of genomic reduction in trypanosomatids; how species-specific genes, and the genomic domains they occupy, have illuminated the innovations in trypanosomatid genomes; and how comparative genomics has exposed the molecular mechanisms responsible for innovation and adaptation to a parasitic lifestyle.
Key words: Trypanosoma, Leishmania, genome, evolution, phylogeny, parasitism
INTRODUCTION
Trypanosomatids are unicellular flagellates and obligate parasites that infect various animals and plants. They include Trypanosoma and Leishmania, species of which cause potent vector-borne diseases in humans, livestock and wildlife; diseases that are responsible for substantial mortality and morbidity across the world. Trypanosoma cruzi causes Chagas disease in South and Central America; Trypanosoma brucei causes Human African Trypanosomiasis in sub-Saharan Africa (and, along with related species, a similar disease in livestock); while Leishmania spp. cause various forms of leishmaniasis in humans. Other species of Trypanosoma and Leishmania infect a wide range of vertebrate hosts, and all are transmitted by invertebrate vectors; predominantly these are biting insects, although some aquatic species are transmitted by leeches (Lom, 1979). Phytomonas spp. are plant parasites transmitted by phloem-sucking insects and are occasionally an agricultural problem in South and Central America (Camargo, 1999). Besides these dixenic (i.e. two-host) parasites that cycle between insect/leech and vertebrate/plant hosts, the trypanosomatids include various other genera, such as Crithidia, Leptomonas, Herpetomonas, Angomonas and Strigomonas that are cosmopolitan, monoxenic (i.e. one host) parasites of insects (Maslov et al. 2013). The diverse associations of trypanosomatids indicate that the origin of parasitism is singular and ancient (Simpson et al. 2006).
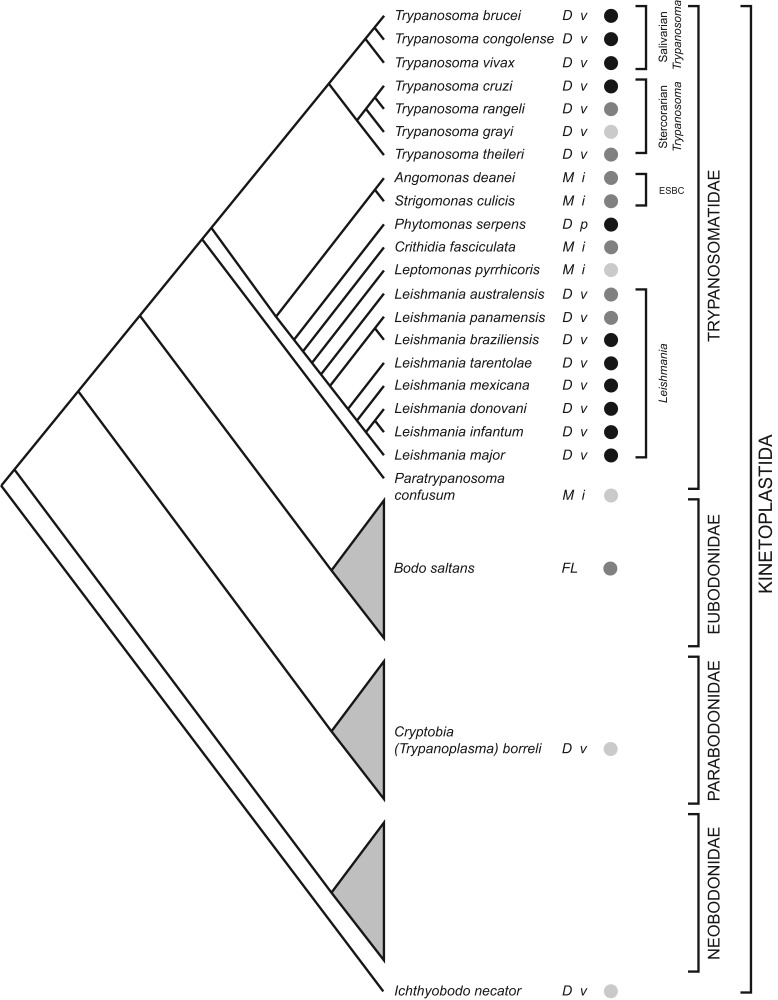
The order Trypanosomatidae is one part of the phylum Kinetoplastida; most other Kinetoplastids live freely or as commensals in marine, terrestrial and aquatic environments. The current consensus on Kinetoplastid phylogeny is summarized in Fig. 1; trypanosomatids are monophyletic and the sister clade to eubodonids (Callahan et al. 2002; Simpson et al. 2004; Moreira et al. 2004; von der Heyden et al. 2004; Deschamps et al. 2011). The closest known relative among eubodonids is Bodo saltans, a free-living bacteriovore of terrestrial and freshwater microbiota. Hence, the phylogeny indicates that parasitism in trypanosomatids had a single origin; although the position of the fish parasites Cryptobia spp. and Ichthyobodo spp. show that parasitism has appeared on other occasions within the Kinetoplastida (Simpson et al. 2006; von der Heyden et al. 2004). This is the context in which I review the contribution of trypanosomatid genome sequences to our understanding of how parasitism evolved and subsequently diversified.
Fig. 1.
Kinetoplastid phylogeny. A cladogram depicting the current consensus on Kinetoplastid phylogenetic relationships (adapted from von der Heyden et al. 2004; Simpson et al. 2004, 2006; Deschamps et al. 2011; Flegontov et al. 2013). Each bodonid order is shown as a grey triangle, representing an indeterminate, but large, number of species. The status of genome sequencing projects for each named species is indicated by filled circles (black: finished; dark grey: unfinished draft; light grey: sequencing in progress). The life cycle of each species is indicated (D: dixenic; M: monoxenic; FL: free-living), as well as the host type(s) (v: vertebrate; i: insect; p: plant). ESBC: ‘Endosymbiont-bearing clade’.
Since the publication of the ‘TriTryp’ genome sequences for T. cruzi, T. brucei and Leishmania major in 2005 (Berriman et al. 2005; El-Sayed et al. 2005a; Ivens et al. 2005), there has been much comparative analysis of these seminal resources. They have been complemented by transcriptomic (Holzer et al. 2006; Leifso et al. 2007; Saxena et al. 2007; Rochette et al. 2008, 2009; Alcolea et al. 2009, 2010; Depledge et al. 2009; Jensen et al. 2009; Kabani et al. 2009; Minning et al. 2009; Veitch et al. 2010; Adaui et al. 2011) and proteomic analyses (Atwood et al. 2005; Rosenzweig et al. 2008a, b ; Alcolea et al. 2011; Eyford et al. 2011; Urbaniak et al. 2012; Butter et al. 2013) of gene expression at various life-cycle stages. Genome sequences for additional species of Trypanosoma (Jackson et al. 2009, 2012), Leishmania (Peacock et al. 2007; Downing et al. 2011; Rogers et al. 2011; Raymond et al. 2012; Real et al. 2013) and Phytomonas (Porcel et al. 2014) have been produced, with several more in progress (see Fig. 1).
Comparison of the Tritryp genomes showed that both gene order and gene repertoire are broadly conserved within chromosomal cores (El-Sayed et al. 2005b). It is generally thought that the considerable co-linearity displayed by trypanosomatid genomes, despite their apparently ancient divergences, reflects strong and fundamental selective constraints on genome structure (Ghedin et al. 2004). Analysis of gene order conservation across Eukaryotic genomes indicates that highly conserved gene pairs are retained for both functional and transcriptional regulation (Dávila-López et al. 2010). While there is little to suggest that the conserved proximity of genes in trypanosomatids reflects their shared or related functions, it has been suggested that their polycistronic organization necessitates the co-directionality of replication and transcription (Ghedin et al. 2004), and that this structural peculiarity of trypanosomatids (the cause of which remains unsolved), is responsible for the strong purifying selection that maintains gene order.
Beyond the chromosomal cores, within sub-telomeric regions for instance, there are numerous species-specific features (El-Sayed et al. 2005b). From the outset it was appreciated that these genes are very often associated with disease mechanisms (El-Sayed et al. 2005b) and are the basis for the distinctive cell surface architectures displayed by each parasite (Acosta-Serrano et al. 2007; Handman et al. 2008). Thus, after 10 years of comparative and experimental analysis of these genomes the principal genomic features that distinguish the stem trypanosomatid lineages, and which are most likely to have been instrumental in the evolution of parasitism, are apparent.
GENOMIC REDUCTION
Parasites were once thought to be ‘degenerate’; while this view is no longer prevalent, it remains intuitive that some characters vital to free-living organisms, but no longer necessary for parasites within a host environment, are lost when the selection pressure to retain them is removed. Hence, we expect phenotypic reduction, which is often observed of parasites, to be reflected in genomic reduction. For example, the genomes of both schistosomes and cestodes, which are phenotypically reduced relative to free-living platyhelminthes, lack elements of canonical metazoan metabolism and developmental regulation (Berriman et al. 2009; Tsai et al. 2013). Genome reduction reaches its apogee in the microsporidian parasites, which in some cases have reduced their genomes to the physiological minimum required for life, and this corresponds with their extreme host dependence (Nakjang et al. 2013). At such extremes, we also observe physical compaction of the genome, in addition to the loss of genes (Keeling and Slamovits, 2005).
Trypanosomatids do not appear to be reduced physically; the size of their genomes (25–35 mb in the haploid state) and the gene density (2·8–4·6 Kb/gene) is comparable with free-living unicellular eukaryotes, for instance Saccharomyces cerevisiae (12·5 mb/2·09 Kb/gene) and Dictyostelium discoideum (33·8 mb/2·72 Kb/gene). However, trypanosomatid genomes might still be functionally reduced, having lost genes essential to free-living Kinetoplastids.
Before the advent of genome sequences, it was known that trypanosomatids lacked certain common metabolic capabilities. For example, they are auxotrophic for pteridine and folate, which are essential co-factors in macromolecule biosynthesis, because they lack the ability to synthesize tetrahydrobiopterin (Beck and Ullman, 1990, Bello et al. 1994, Nare et al. 1997; Ouellette et al. 2002). Similarly, they must scavenge haem from their hosts (or obtain it from bacterial endosymbionts; Alves et al. 2011), because they lack a native haem biosynthesis pathway (Chang et al. 1975; Korený et al. 2010). Trypanosomatids are also auxotrophic for purines (Marr et al. 1978; Gutteridge and Gaborak, 1979), vital in the biosynthesis of nucleic acids and energy metabolism. Other aspects of model eukaryotic physiology are also absent, for example, a system of redox homoeostasis based on catalase and glutathione reductase. Instead, trypanosomatids rely on a unique thiol-based redox metabolism based on trypanothione for the deactivation of oxidizing agents (Oza et al. 2005; Krauth-Siegel and Comini, 2008; Comini and Flohé, 2013). The initial Tritryp comparison showed that trypanosomatids do not possess receptor-linked tyrosine kinases (Parsons et al. 2005), canonical mitochondrial import systems (Pusnik et al. 2009), known telomere end binding proteins such as POT1 (Lira et al. 2007), certain genes that regulate autophagy (Herman et al. 2006) and others controlling apoptosis (i.e. TNF-related family receptors, Bcl-2 family members and caspases; Smirlis et al. 2010).
The question relating to these and any other missing features is whether they represent evolutionary losses, or instead, reflect the branching position of the Kinetoplastida in the eukaryotic phylogeny. It may be that certain widely conserved genes are absent from trypanosomatids because Kinetoplastids separated from other eukaryotic lineages early in evolutionary history and before those genes evolved. Furthermore, it could be that we have systematically underestimated genomic and physiological diversity in eukaryotes, and the apparent deficiencies of trypanosomatids reflect a biased perception based on a narrow sampling of animal and plant genomes. In short, the absence of ‘typical’ features from trypanosomatids need not represent evolutionary loss. In fact, detailed comparisons in the years following publication of the Tritryp genome sequences showed that, while trypanosomatids often lack some conserved features and have numerous clade-specific derivations, they are nevertheless comparable to free-living protists in the number and diversity of protein kinases (Parsons et al. 2005; Bahia et al. 2009), phosphatases (Brenchley et al. 2007), GTPases and other genes involved in intracellular trafficking (Field, 2005; Field et al. 2007) and DNA helicases (Gargantini et al. 2012).
In summary, these genomes are not reduced in size or substantially reduced in function. While trypanosomatids employ unique solutions in redox homoeostasis, mitochondrial protein import and telomere regulation, they nonetheless have a broadly typical eukaryotic physiology. Where there are disparities, it is not clear whether these genes were lost or never existed and this will only become clear after we have sampled the genomes of free-living Kinetoplastids for comparison. Instead, there is abundant evidence that trypanosomatid genomes have expanded during their evolution both physically, through the evolution of sub-telomeres and accessory chromosomes, and functionally, with the acquisition of new genes through duplication and horizontal gene transfer.
GENOMIC INNOVATION: SPECIES-SPECIFIC GENE FAMILIES
Trypanosomatid cell surfaces include various polymorphic proteins combined with diverse glycolipid conjugates (Ferguson, 1997). These structures are enigmatic and their origins are mysterious because they are not seen in other organisms; indeed, the highly abundant cell-surface glycoproteins of T. brucei, T. cruzi and L. major are mutually exclusive, making it very hard to infer what the ancestral cell surface looked like (El-Sayed et al. 2005b). The Tritryp genomes revealed the genes that encode these surface features and their non-random distribution in the genome, which has been reviewed in detail elsewhere (Acosta-Serrano et al. 2007; Handman et al. 2008; De Pablos and Osuna, 2012). These cell surface proteins attract considerable interest because they are implicated in disease, virulence and mechanisms of pathogenesis (De Pablos and Osuna, 2012). Species-specific genes provide the clearest insight into genomic innovations associated with parasitism and the multi-copy gene families that encode these cell surface proteins dominate such species-specific genes in comparative analyses (El-Sayed et al. 2005b).
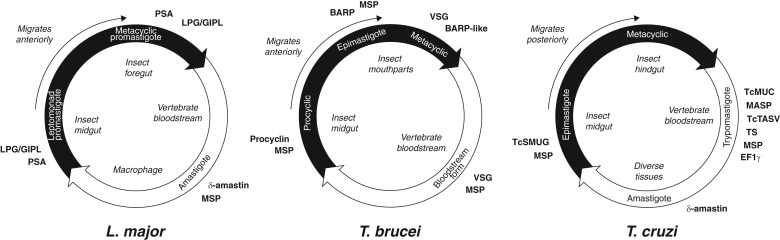
The life cycles of the Tritryp species and the points at which species-specific cell surface proteins are expressed are shown in Fig. 2. Species-specific genes in T. cruzi are dominated by gene families that encode the mucin-based surface coat during its trypomastigote stage (Cerqueira et al. 2008; Nakayasu et al. 2009; De Pablos and Osuna, 2012); primarily mucins (TcMUC; Acosta-Serrano et al. 2001; Buscaglia et al. 2006) and trans-sialidases (TS; Kim et al. 2005; Montagna et al. 2006; Freitas et al. 2011; Oppezzo et al. 2011; Ammar et al. 2013; Oliveira et al. 2014), but also a ‘dispersed gene family’ (DGF-1; El-Sayed et al. 2005b; Kawashita et al. 2009; Lander et al. 2010), the mucin-associated surface protein family (MASP; El-Sayed et al. 2005b; Bartholomeu et al. 2009; dos Santos et al. 2012), and the T. cruzi Trypomastigote Alanine, Serine and Valine-rich proteins (TcTASV; García et al. 2010; Bernabó et al. 2013). Gene families specifically expressed in the other life stages include amastin in the intracellular amastigote stage, and T. cruzi Small MUcin-like Genes (TcSMUG; Urban et al. 2011) in the replicative epimastigote. In addition to these developmentally regulated, surface-expressed gene families, expansions of Retrotransposon Hotspot (RHS) genes and Elongation Factor 1 gamma (EF1γ) genes are prominent innovations of the T. cruzi genome.
Fig. 2.
Trypanosomatid life cycles. Each circle represents the movement of parasites between insect (black) and vertebrate (white) hosts, showing the transition of parasite life stages, the position of each stage within the hosts and the timing of expression of cell surface-expressed protein families mentioned in the text. Note that for T. brucei and L. major, the parasites move anteriorly from the insect gut as they develop, while T. cruzi migrates posteriorly as it prepares for transmission into the vertebrate.
In T. brucei, species-specific genes are dominated by those encoding the Variant Surface Glycoproteins (VSG) that form the surface glycocalyx of all salivarian trypanosomes during their bloodstream stage in the mammal host (Hutchinson et al. 2007; Marcello and Barry, 2007; Jackson et al. 2012; Weirather et al. 2012; Hall et al. 2013). Other species-specific genes like the Invariant Surface Glycoprotein (ISG) genes (Jackson et al. 1993; Ziegelbauer and Overath, 1993) and Expression-Site Associated Genes (ESAGs; Pays et al. 2001; see below) are also preferentially expressed in the bloodstream stage. In the insect host, species-specific genes are dominated by procyclin, encoding the major surface glycoprotein of the procyclic stage while in the insect midgut (Roditi et al. 1998; Berriman et al. 2005), and the Brucei Alanine-Rich Protein (BARP) that, along with related forms, is specifically expressed by the epimastigote and metacyclic stages while in the insect mouthparts (Urwyler et al. 2007; Jackson et al. 2013).
The cell surface of Leishmania is dominated by non-protein lipophosphoglycan (LPG) and glycoinositolphospholipid (GIPL) molecules (de Assis et al. 2012). The LPG/GIPL coat is complemented by species-specific, multi-copy proteins such as δ-amastin, which is specifically expressed during the intracellular amastigote stage (Rochette et al. 2005). While its function is unknown, the evolution of δ-amastin is thought to be an adaptation for infection of, or survival within, macrophages since it is absent from monoxenic species (Crithidia and Leptomonas spp.) lacking a vertebrate stage (Jackson, 2010) and less abundant in Leishmania species that do not routinely infect macrophages (Raymond et al. 2012). Furthermore, a parallel expansion of δ-amastin has occurred in T. cruzi, which also has an amastigote stage, and this is associated with virulence (Kangussu-Marcolino et al. 2013). Another Leishmania-specific family, tuzin (Ivens et al. 2005), is linked to δ-amastin loci physically and phylogenetically (Jackson, 2010); hence, tuzin might be involved in the same adaptation. In the insect life stage, the promastigote surface antigen (PSA or gp46) is preferentially expressed in metacyclic promastigotes (Handman et al. 1995) and is encoded by a diverse gene family in human-infecting species (Devault and Bañuls, 2008). Also specifically expressed in metacyclics are the HASP (Hydrophilic Acylated Surface Protein) and SHERP (Small Hydrophilic ER-associated Protein) gene families (Depledge et al. 2010; Sádlová et al. 2010).
While the precise functions of these enigmatic gene families are unknown, several contribute to parasite fitness. This may be because they initiate infection, for instance, the TcMUC and TS proteins interact to transfer host sialic acid residues to parasite mucins, which is essential for attachment and invasion by T. cruzi trypomastigotes (Acosta-Serrano et al. 2001; Oliveira et al. 2014). Other cell surface protein families are essential for parasite development and transmission through the insect host; for example, HASP and SHERP are required for L. major to form infective metacyclics while in the insect foregut (Sádlová et al. 2010). However, given their prominent roles at the cell surface, most of these species-specific proteins are likely to have immunological roles. These may be in suppressing innate responses, for example by degrading antimicrobial peptides or other effectors of complement-mediated lysis, as has been shown for PSA (Lincoln et al. 2004), or in manipulating cell-mediated immune responses. For instance, TcMUC represses T-cell expansion and cytokine production (Nunes et al. 2013). Salivarian trypanosomes employ VSG in antigenic variation, and have evolved sophisticated mechanisms for regulating VSG expression (see below). The abundance and variety of TcMUC, TS and MASP genes has led some to suggest that a subtler form of antigenic variation operates in T. cruzi as well (Buscaglia et al. 2004, 2006; dos Santos et al. 2012).
GENOMIC INNOVATION: CONTINGENCY ZONES
Trypanosomatids have substantially modified the genome to accommodate these abundant families of cell-surface effectors, by creating genomic sub-domains segregated from the core genome by distance, but also by sequence composition and epigenetic modification (Figueiredo et al. 2009; Rudenko, 2010). We can call these sub-domains ‘contingency zones’ because they provide the environment for flexible expression of what are known as contingency genes (Deitsch et al. 1997). In this trypanosomatids are not alone; diverse parasites possess polymorphic effector protein families that display specialized expression profiles across a wide range of physiological conditions (Deitsch et al. 1997; Kissinger and DeBarry, 2011). It has often been observed that contingency genes aggregate towards the telomeres, a position that promotes both the specific regulation of their expression and their diversification through recombination and gene duplication (Barry et al. 2003; Kissinger and DeBarry, 2011). Thus, both T. brucei and T. cruzi have expanded sub-telomeric regions to contain and regulate their diverse contingency genes (Berriman et al. 2005; El-sayed et al. 2005a, b ; Moraes Barros et al. 2012). It is likely that the strand-switch regions that occur between polycistrons on trypanosomatid chromosomes also serve as incubators of novelty, since they often harbour species-specific genes (Peacock et al. 2007; Jackson et al. 2009).
Perhaps the best example of structural innovation in trypanosomatid genomes is the VSG expression site (ES) in T. brucei. African trypanosomes evade the humoral immune response by periodically switching the VSG monolayer that masks their cell surfaces. This demands that only a single VSG is expressed at a time, while all others are silenced (i.e. monoallelic expression). The function of the ES is to ensure monoallelic expression by providing a dedicated locus for VSG transcription. Thus, the active VSG is transcribed solely from one of several, alternative ESs and antigenic switching occurs when a different VSG from among the many hundreds of silent, sub-telomeric loci, replaces the ES copy through ectopic gene conversion, or by activating an alternative ES (Horn and McCulloch, 2010; Rudenko, 2011). Analysis of ES sequences from several T. brucei strains has identified a canonical ES structure (Graham et al. 1999; Berriman et al. 2002; Hertz-Fowler et al. 2008), which includes not only the VSG and repeat sequences required to promote recombination with sub-telomeric VSG loci, but also the ESAGs (reviewed in Pays et al. 2001; McCulloch and Horn, 2009). The functions of most ESAGs are unclear; however, all are transcribed preferentially in the bloodstream stage (Jensen et al. 2009; Siegel et al. 2010; Veitch et al. 2010) and it is known that they are T. brucei-specific innovations, often derived from conserved gene families with pre-existing cell surface roles (Barker et al. 2008; Barnwell et al. 2010; Salmon et al. 2012; Jackson et al. 2013). Hence, it may be that they support antigenic variation or that the specific regulatory environment of the ES has been exploited secondarily to up-regulate proteins with established and diverse roles during the bloodstream stage.
GENOMIC INNOVATION: THE MAJOR SURFACE PROTEASES
Alongside the many species-specific cell surface proteins, there is one family conserved in all trypansomatid genomes that must have experienced substantial evolution since the origin of parasitism. The Major Surface Protease (MSP) gene family encode a range of metalloproteases that are implicated in various aspects of pathogenesis and virulence in Leishmania (Yao, 2010). MSP subverts the normal host defensive mechanisms by degrading components of immune cell signalling pathways (Gomez et al. 2009; Hallé et al. 2009; Contreras et al. 2010), and suppresses other aspects of innate immunity (Kulkarni et al. 2006; Lieke et al. 2008). In Trypanosoma, MSP is equally abundant in gene copy number and protein abundance but its function is less well understood; it is known to remove the VSG coat from the T. brucei surface during differentiation into the procyclic form (PCF) (Grandgenett et al. 2007) and is thought to have a role in cell invasion by T. cruzi (Cuevas et al. 2003; Kulkarni et al. 2009). As it is present in all trypanosomatids, we can infer the diversification of MSP from its phylogeny, and this too indicates that MSP has been instrumental in parasite adaptation.
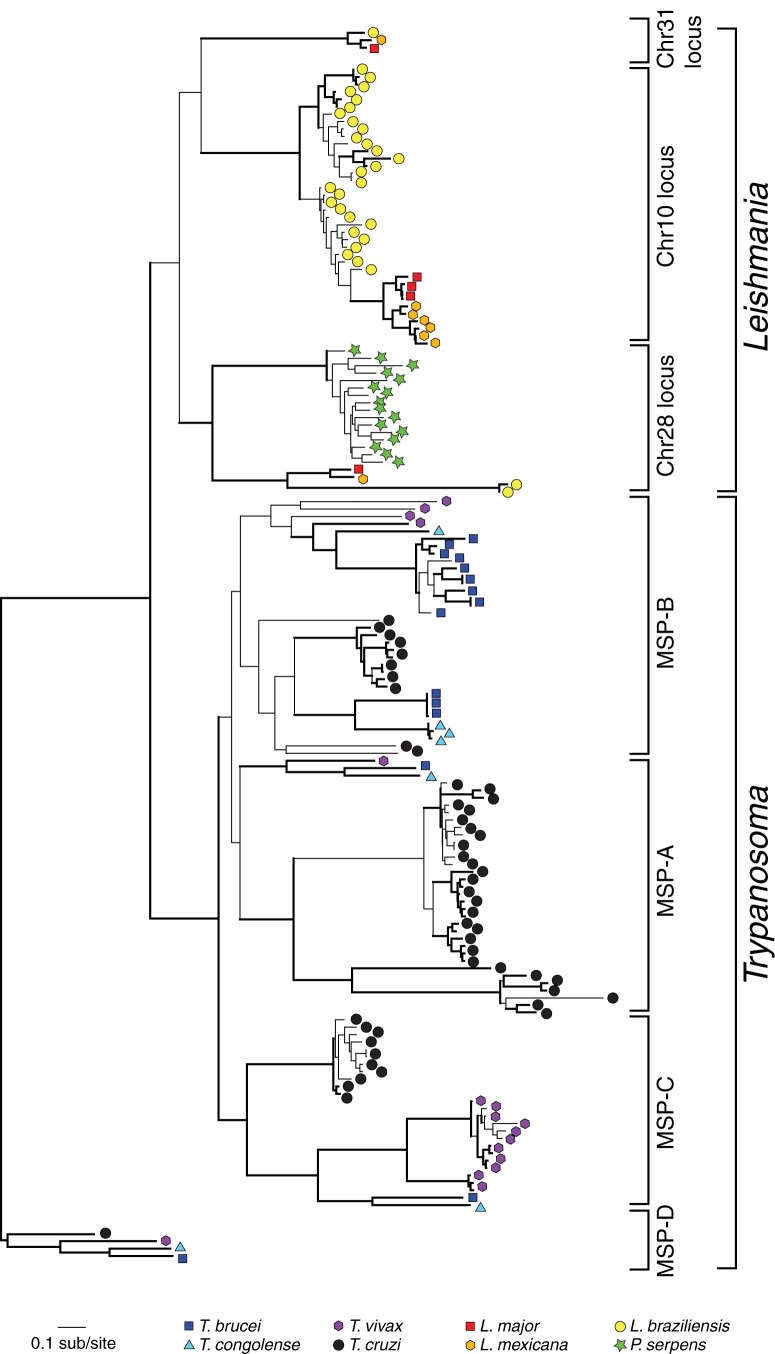
The MSP phylogeny is described in Fig. 3. It shows how, beginning from a much smaller gene repertoire, MSP has differentiated into distinct clades in both Leishmania and Trypanosoma (Victoir et al. 2005; Ma et al. 2011); each clade is associated with a conserved locus, and we know that some of these distinct lineages are developmentally regulated (Yao, 2010). For instance, MSP-A and MSP-C are up-regulated in bloodstream form (BSF) T. brucei, while MSP-B is predominantly seen in the procyclic form (LaCount et al. 2003; Urbaniak et al. 2012). Hence, the trypanosomatids have elaborated their MSP repertoire by creating new loci at least in part to regulate function during the life cycle. Moreover, these different forms have been duplicated to create multiple isoforms, often in species-specific ways; for instance, MSP-C is polymorphic in Trypanosoma vivax while single copy in other salivarian species, and the single-copy MSP gene found on chromosome 28 in Leishmania has been greatly expanded in Phytomonas. However, the phylogeny also demonstrates that MSP in Leishmania and Trypanosoma cluster by genus, and therefore, there is no orthologous MSP shared by all. Thus, MSP repertoires in Leishmania and Trypanosoma have evolved independently, and their similarities in genomic structure, developmental regulation and pathogenesis represent parallel evolution, reflecting a common need for diverse surface proteases throughout trypanosomatid diversification.
Fig. 3.
Major Surface Protease (MSP) gene family phylogeny. This maximum likelihood phylogeny of MSP amino acid sequences sampled from completed genome sequences was estimated using PHYML and a LG+Γ model of amino acid substitution (Guindon et al. 2010). Node robustness was assessed with non-parametric bootstraps; branches with bootstrap support >75 are shown with bold lines. Sequences are labelled with coloured symbols, according to the key. The tree is mid-point rooted, which corresponds with the Trypanosoma-specific MSP-D locus (Marcoux et al. 2010).
DEVELOPMENTAL REGULATION OF GENE EXPRESSION
Trypanosomatids display morphological plasticity that is often associated with developmental transition through a complex life cycle. This is important for the origins of parasitism but not an issue that comparative genomics can illuminate dramatically, without including a comparator lacking developmental complexity. The recent discovery of Paratrypanosoma confusum parasitizing the gut of a Culex pipiens mosquito strengthens the argument that the ancestral trypanosomatid was a monoxenic insect parasite, since P. confusum is a robust outgroup to all other trypanosomatids (Flegontov et al. 2013). As long as P. confusum has no second host, this shows that a dixenic life cycle has evolved on three separate occasions in Trypanosoma, Leishmania and Phytomonas. Trypanosomatids are capable of assuming multiple developmental forms and transition between forms coincides with passing between distinct environments, whether they are in different hosts or a single host, for example from the hindgut to the foregut of an insect. Experimental approaches are beginning to reveal the non-coding sequences (Bringaud et al. 2007; Holzer et al. 2008; Smith et al. 2009; Li et al. 2012; Pastro et al. 2013) and RNA-binding proteins (reviewed in Kolev et al. 2014) that interact to regulate gene expression, as well as genes specifically required for differentiation from one life stage to another (Goldenberg and Avila, 2011; Kolev et al. 2012; Rico et al. 2013). Comparison of life-stage-specific transcriptomes (Holzer et al. 2006; Leifso et al. 2007; Saxena et al. 2007; Rochette et al. 2008, 2009; Alcolea et al. 2009, 2010; Depledge et al. 2009; Jensen et al. 2009; Kabani et al. 2009; Minning et al. 2009; Veitch et al. 2010; Adaui et al. 2011;) and proteomes (Atwood et al. 2005; Rosenzweig et al. 2008a, b ; Alcolea et al. 2011; Urbaniak et al. 2012; Gunasekera et al. 2012; Butter et al. 2013) in various species have estimated the proportion of genes showing preferential expression in the insect or vertebrate stages to be between 2 and 44%; the breadth of these values reflects the diverse conditions and approaches employed. However, it is clear that a significant minority of genes are developmentally regulated. We can predict that this regulation is achieved with layers of interaction between genomic loci, mRNA, non-coding RNA and DNA and RNA-binding proteins. Hence, to understand the origins of complex life cycles we will need to compare the interaction networks of free-living, monoxenic and dixenic Kinetoplastids, and in this P. confusum and the free-living Bodo saltans will be instrumental.
MECHANISMS OF GENOMIC EVOLUTION: GENE DUPLICATION
Besides the genomic innovations themselves, comparative analysis also reveals the molecular mechanisms that create them. These evolutionary events range in size from single amino acid substitutions to chromosomal duplications, and include both coding and non-coding regions, but it is gene duplication above all that creates the raw material for evolutionary novelty (Ohno, 1970). After duplication, paralogs may acquire new functions (neofunctionalization), segregate existing functions (subfunctionalization) or lose function under mutation pressure (pseudogenization) (Lynch and Conery, 2000). Since developmental regulation of gene expression is widespread, it is unsurprising that many gene duplicates are distinguished in the timing or location of their expression. For example, TcMCA5 is an epimastigote-specific metacaspase implicated in programmed cell death of T. cruzi that has evolved from a constitutively expressed metacaspase gene family (Kosec et al. 2006). In Leishmania, Zinoviev et al. (2012) identified two functionally redundant RNA helicases that have evolved purely to perform the same role in insect and vertebrate stages respectively. By contrast, TcPRACA and TcPRACB are two paralogous proline racemases involved in immune-suppression by T. cruzi (Reina-San-Martín et al. 2000); here, function is segregated by location, TcPRACB being expressed intracellularly and TcPRACA secreted (Chamond et al. 2005).
Of course, the derivation of many gene duplicates may be multifactorial; in the example of proline racemases, secretion of TcPRACA may coincide with a new role in the differentiation of infective metacyclics (Chamond et al. 2005). Thus, it is difficult to unambiguously distinguish neofunctionalization from the segregation of the same function by time, space or substrate. However, the transferrin receptor (TFR) in T. brucei, which is required for salvaging haem from the host and is homologous to the VSG (Salmon et al. 1997), is one example. Recently, it was confirmed that the TFR had evolved from an a-type VSG in the ancestor of T. brucei and Trypanosoma congolense, and that, despite their homology, TFR and VSG genes do not recombine, supporting a functionally distinct role from the variant antigen repertoire (Jackson et al. 2012, 2013). As suggested above, the conspicuous abundance and diversity of certain T. cruzi gene families, such as TS, EF1γ and MSP, could indicate that these genes have secondarily evolved a novel role in immune evasion as a consequence of being at the cell surface for their pre-existing functions, i.e. to transfer sialic acid to TcMUC in the case of TS (Oliveira et al. 2014). Furthermore, many TS, EF1γ and MSP genes in T. cruzi are not predicted to encode proteins capable of their putative functions (El-Sayed et al. 2005). At first sight, this would appear to indicate frequent pseudogenization, yet a population of pseudogenes acquiring substitutions under neutral conditions would be expected to display a spectrum of mutational decay that is not seen (El-Sayed et al. 2005). This suggests that these genes may remain under purifying selection for another role, which could represent neofunctionalization.
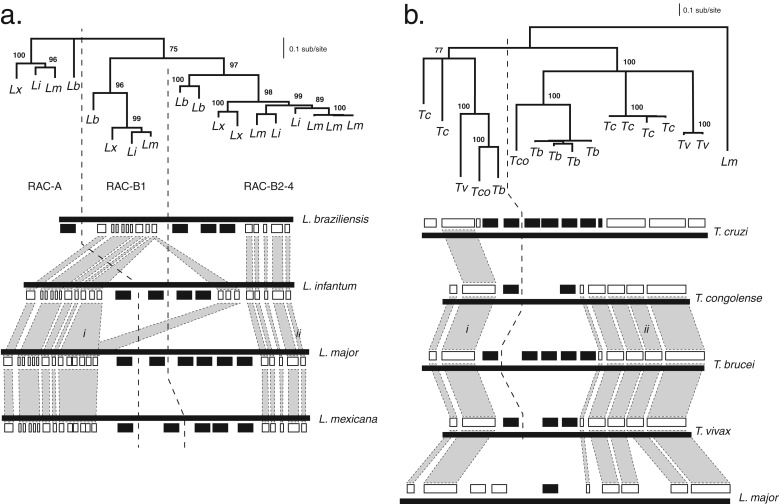
The evolution of gene duplicates is particularly obvious in the abundant tandem gene arrays of trypanosomatid genomes. Tandem duplication is very common in trypanosomatids, perhaps as a means of increasing transcript abundance for highly expressed genes in the presence of polycistronic transcription. Comparative analysis of homologous arrays shows that tandem duplicates can evolve new functions, despite the propensity for concerted evolution of tandemly arrayed genes (Jackson, 2007a), and that this follows a consistent pattern of structural segregation. Figure 4 shows two examples of functional divergence within tandem gene arrays. The expression profiles of adenylate cyclase gene paralogues from the rac array of Leishmania spp. correspond with their position in the array. The 3′-most gene (rac-A) and the gene positioned upstream of rac-A in the array (rac-B1) are expressed specifically in the promastigote (Sanchez et al. 1995; Akopyants et al. 2004), while transcripts for the remaining copies are more abundant in the amastigote (Akopyants et al. 2004). Interestingly, rac-A and rac-B1 may have differentiated in a complementary fashion, since rac-B1 negatively regulates the activity of rac-A in the promastigote (Sanchez et al. 1995). In Trypanosoma, the 5′-most copy of a cation transporter gene array is preferentially expressed in the PCF (Jensen et al. 2009; Urbaniak et al. 2012) (indeed, it is essential to its growth; Alsford et al. 2011), while transcripts for all downstream copies are up-regulated in the bloodstream stage (Jensen et al. 2009; Veitch et al. 2010).
Fig. 4.
Structural differentiation of paralogues in tandem gene arrays. a. Receptor-type adenylate cyclase (rac) genes in four Leishmania species. The rac array (i.e. LmjF.17·0200) is located at the extreme left-end of chromosome 17 in L. major; its conserved position is defined downstream by an EF1α gene array (i) and upstream by a metalloprotease (ii). The structure of the array in four species is depicted, rac genes are shown in black, and other loci are shown in white. Vertical grey shading represents homology between flanking loci, to demonstrate positional conservation. The dashed black line separates array positions that correspond with distinct clades in the phylogeny. A maximum likelihood phylogeny estimated from amino acid sequences using a LG+Γ model is shown, with non-parametric bootstraps applied to nodes where support is >75. Terminal nodes are labelled with species name initials. The tree is midpoint rooted. b. Cation transporter genes in four Trypanosoma species. The transporter array (i.e. Tb927.11.9000) is located on chromosome 11 in T. brucei; its conserved position is defined downstream by a palmitoyl acyltransferase 4 gene (i) and upstream by an EF1γ2 gene (ii). The phylogeny and genome comparison are as depicted in a. except that the tree is rooted with a single-copy orthology from L. major.
The phylogenies of these gene duplicates show that those gene copies that are functionally differentiated retain orthology across species (i.e. they cluster together despite being in different genomes), while undifferentiated copies cluster by species. This shows that gene duplicates that have diverged in their structures and expression for a novel function are preserved by selection over the course of trypanosomatid evolution, despite the pressure exerted by allelic gene conversion in these situations. In fact, when tandem gene duplicates differentiate, this often occurs at either end of the array (Jackson, 2007a), even occurring in otherwise invariant arrays that are exposed to frequent gene conversion; for example, differentiation of the terminal 3′UTR in the β-tubulin array in Leishmania spp. has created a promastigote-specific β-tubulin isoform (Jackson et al. 2006).
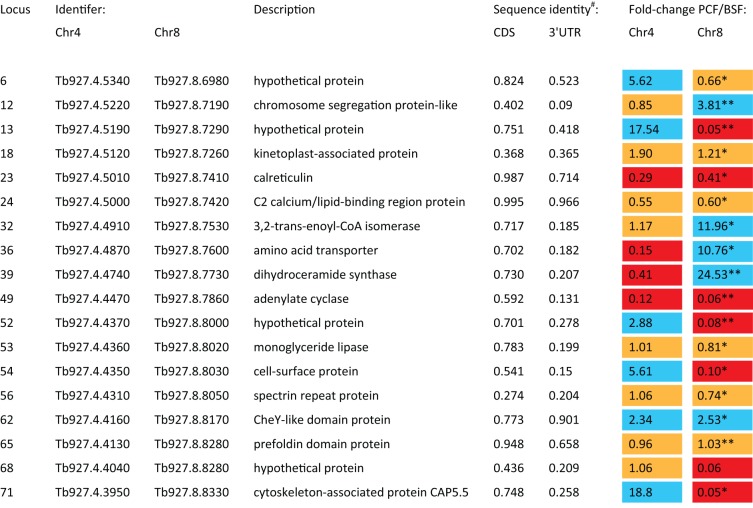
Duplication events do not only affect individual genes. A 0·5 mb segmental duplication in T. brucei was identified that has created duplicons shared by chromosomes 4 and 8 (Jackson, 2007b). Originally, this region contained approximately 158 genes but subsequent deletions from either duplicon have returned many loci to their original copy number. However, 74 loci have been retained as paralogues in both duplicons. Comparison of their coding and flanking sequences indicated that substantial divergence had occurred and this was assumed to reflect functional divergence (Jackson, 2007b). They include CAP5.5, a cysteine peptidase essential for cell morphogenesis, which has been shown to have two paralogues expressed specifically in the insect and vertebrate stages respectively (Hertz-Fowler et al. 2001; Olego-Fernandez et al. 2009). Figure 5 shows how recent proteomic evidence now confirms that several of the paralogues retained after segmental duplication have evolved stage-specific expression profiles, indicating subfunctionalization by life stage. Gene expression in trypanosomatids is largely regulated by sequences within the 3′ untranslated region (UTR) of transcripts (Vanhamme and Pays, 1995; Haile and Papadopoulou, 2007). Accordingly, it is the paralogous pairs with no sequence identity in their 3′ UTRs that have the greatest differences (loci #13, 36, 39, 49 and 71 in Fig. 5), while those paralogues with similar 3′ UTR sequences display similar abundance in both cases (loci #23, 24, 62 and 65 in Fig. 5).
Fig. 5.
Peptide abundance in procyclic form (PCF) and bloodstream form (BSF) T. brucei for selected paralogues resulting from a segmental duplication. 74 loci are present in two forms in T. brucei due to a segmental duplication. The loci listed here are those detected in proteomic analyses. Locus number and sequence identity values refer to the segmental duplication described in Jackson (2007b). Fold change in peptide abundance between PCF and BSF cells taken from * Urbaniak et al. (2012) or ** Butter et al. (2013). Preferential expression in PCF and BSF is indicated by blue and red shading respectively. Constitutive expression is indicated by orange shading.
MECHANISMS OF GENOMIC EVOLUTION: HORIZONTAL GENE TRANSFER
Horizontal gene transfer (HGT) is another mechanism by which many eukaryotic genomes have acquired new functionality. Berriman et al. (2005) identified 49 putative HGT from bacteria and other eukaryotes in trypanosomatid genomes. Confirming HGT rests on sound phylogenetic reconstruction; the most convincing cases are those where the donated gene is closely related to donor genes in unrelated genomes, and nested among these in a phylogeny. Some putative HGT in trypanosomatids achieve this, notably the haem-biosynthesis pathway, absent from Trypanosoma but partially restored in Leishmania and related genera through HGT of three genes (hemF, hemG and hemH encoding coproporphyrinogen oxidase, protoporphyrinogen oxidase and ferrochelatase, respectively) from gamma-proteobacteria. In phylogenies, HemF-H are nested among bacterial homologues and apart from related eukaryotic genes (Ivens et al. 2005, Korený et al. 2010). In salivarian trypanosomes, a phospholipase A1 (PLA1) gene is thought to have been acquired from proteobacteria (Richmond and Smith, 2007). In support of this, the PLA1 gene is absent from all other Kinetoplastids (indeed most other eukaryotes) and it nests among proteobacterial sequences in sequence comparisons. Moreover, the PLA1 locus (Tb927.1.4830) occurs precisely at the boundary between chromosomal core and sub-telomere in African trypanosome genomes, suggesting perhaps that it was recently transposed.
Other good examples of HGT include a cytosolic dihydroorotate dehydrogenase in the pyrimidine biosynthetic pathway, which is unique to Kinetoplastids, and replaces the mitochondrial dihydroorotate dehydrogenase that is typical of euglenids and other eukaryotes. In phylogenies, the cytosolic genes are nested among bacterial taxa, while the mitochondrial genes form a eukaryotic clade (Annoura et al. 2005). Likewise, ornithine decarboxylase genes from salivarian trypanosomes do not cluster with homologues from other trypanosomatids, but instead they are nested among metazoan genes and are the sister taxon to ornithine decarboxylase from vertebrates (Steglich and Schaeffer, 2006). In fact, ornithine decarboxylase is known to be absent from T. cruzi (Carrillo et al. 1999), indicating that this HGT from vertebrates has restored function in African trypanosomes that was lost after the origin of Trypanosoma. However, since the African trypanosome genes are not nested within the vertebrate clade, we can rule out any recent transfer from contemporary hosts and suggest instead a more distant transfer from an ancient chordate.
In other cases of putative HGT the donated gene is not nested among would-be donors, just closest to them in phylogenies. Here, it is possible that the punctate distribution is due to lineage sorting, i.e. patchy inheritance of an ancestral lineage by daughter lineages. When, as is common, eukaryotic diversity is inadequately sampled, it is difficult to distinguish HGT and lineage sorting. For example, trypanosomatid genomes possess four superoxide dismutase genes required for antioxidant defence (soda, sodb1, sodb2 and sodc), which localize to distinct cellular compartments (Dufernez et al. 2006). The four sod genes do not cluster together; soda/sodc cluster most closely to Trichomonas vaginalis, while sodb1/sodb2 cluster with diverse eukaryotes (Dufernez et al. 2006). This suggests sorting of ancestral sod lineages but not necessarily HGT. Similarly, two metallocarboxypeptidases (TcMCP-1 and TcMCP-2) in T. cruzi are found only in Kinetoplastids and prokaryotes, but homologues from the two taxa are sister clades, rather than nested (Niemirowicz et al. 2007). While the original study recognized the possibility of both HGT and lineage sorting, they rejected the latter due to the number of deletions this would require. These losses may not be necessary, however, if eukaryotic diversity were exhaustively sampled. Finally, an uncharacterized protein, META1, is up-regulated in Leishmania metacyclics and is homologous to a bacterial heat-inducible protein, itself similar to a component of the type III secretion system in Shigella (Puri et al. 2011). META1 is hypothesized to have evolved via HGT and may be involved in secretory processes in Leishmania since mutagenesis of select hydrophobic residues in META1 affects the secretion of the secreted acid phosphatase (Puri et al. 2011). However, META1 is not nested among bacterial sequences and, at this stage, the HGT hypothesis rests on it remaining absent from all other eukaryotes.
Although poor sampling continues to limit our ability to distinguish HGT and lineage sorting (Opperdoes and Michels, 2007), HGT has clearly contributed to trypanosomatid genomes; for example, substantial integration of genes from a bacterial endosymbiont has recently been demonstrated in Angomonas deanei (Alves et al. 2011). The role of HGT in the origins of parasitism will be clarified through comparison of trypanosomatids with free-living Kinetoplastids and other neglected unicellular eukaryotes, to reject the lineage sorting hypothesis and to confirm that the HGT is uniquely associated with parasites, such as hemF-H or PLA1, and not Kinetoplastids generally.
CONCLUSION
The genetic content of trypanosomatid genomes indicates that they have been elaborated relative to their common ancestor in terms of both physical structure and physiological capacity. Species-specific gene families, instrumental in cell surface architecture, are central to this history of innovation, and implicitly linked to the origins of complex life cycles and disease. By definition, these unique innovations are mutually exclusive, yet there are themes that cut across species. These gene families are functionally differentiated to perform multiple roles in different host environments through the parasite life cycle. They are positioned in sub-telomeres, tandem gene arrays or other contingency zones that perhaps promote regulatory flexibility and sequence diversity. Their sequences are diverse and often contain low complexity repeats that may promote greater diversity through recombination. In their phylogenies, these gene families display rapid turnover – the gain and loss of lineages – that hint at the importance of host-parasite interactions in genomic evolution. These themes, which would, in fact, apply to parasites of all kinds, suggest how each trypanosomatid lineage has used similar molecular mechanisms to meet the demands of transmission and survival. There are issues in comparative analysis we have not addressed, like protein-protein interactions, the regulatory roles of non-coding regions and regulatory proteins, genomic plasticity or indeed the ~50% of trypanosomatid genes that have no known function. There are also some genes, such as the TcMUC family in T. cruzi, procyclin in T. brucei and T. congolense, and the HASP and SHERP families in L. major, that defy any explanation using a comparative approach, and which may have evolved de novo from non-coding regions. Yet, we have learned enough from the structure and content of trypanosomatid genomes to conclude that becoming parasitic was more an innovative and elaborative process, than one of loss and reduction. With the addition of free-living Kinetoplastids to our comparative analyses, the mechanisms by which these enigmatic genomic adaptations for parasitism came about will be revealed.
FINANCIAL SUPPORT
The author is a Wellcome Trust Tenure-Track Research Fellow, funded by the University of Liverpool and the Wellcome Trust [097826/Z/11/A].
REFERENCES
- Acosta-Serrano A., Almeida I. C., Freitas-Junior L. H., Yoshida N. and Schenkman S. (2001). The mucin-like glycoprotein super-family of Trypanosoma cruzi: structure and biological roles. Molecular and Biochemical Parasitology 114, 143–150. [DOI] [PubMed] [Google Scholar]
- Acosta-Serrano A., Hutchinson C., Nakayasu E. S., Almeida I. and Carrington M. (2007). Comparison and evolution of the surface architecture of trypanosomatid parasites In African Trypanosomes: After the Genome (ed. Barry J. D., McCulloch R., Mottram J. C. and Acosta-Serrano A.), pp. 319–338 Horizon Bioscience, Wymondham, UK. [Google Scholar]
- Adaui V., Castillo D., Zimic M., Gutierrez A., Decuypere S., Vanaerschot M., De Doncker S., Schnorbusch K., Maes I., Van der Auwera G., Maes L., Llanos-Cuentas A., Arevalo J. and Dujardin J. C. (2011). Comparative gene expression analysis throughout the life-cycle of Leishmania braziliensis: diversity of expression profiles among clinical isolates. PLOS Neglected Tropical Diseases 5, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopyants N. S., Matlib R. S., Bukanova E. N., Smeds M. R., Brownstein B. H., Stormo G. D. and Beverley S. M. (2004). Expression profiling using random genomic DNA microarrays identifies differentially expressed genes associated with three major developmental stages of the protozoan parasite Leishmania major. Molecular and Biochemical Parasitology 136, 71–86. [DOI] [PubMed] [Google Scholar]
- Alcolea P. J., Alonso A., Sánchez-Gorostiaga A., Moreno-Paz M., Gómez M. J., Ramos I., Parro V. and Larraga V. (2009). Genome-wide analysis reveals increased levels of transcripts related with infectivity in peanut lectin non-agglutinated promastigotes of Leishmania infantum. Genomics 93, 551–564. [DOI] [PubMed] [Google Scholar]
- Alcolea P. J., Alonso A., Gómez M. J., Moreno I., Domínguez M., Parro V. and Larraga V. (2010). Transcriptomics throughout the life-cycle of Leishmania infantum: high down-regulation rate in the amastigote stage. International Journal for Parasitology 40, 1497–1516. [DOI] [PubMed] [Google Scholar]
- Alcolea P. J., Alonso A. and Larraga V. (2011). Proteome profiling of Leishmania infantum promastigotes. Journal of Eukaryotic Microbiology 58, 352–358. [DOI] [PubMed] [Google Scholar]
- Alsford S., Turner D. J., Obado S. O., Sanchez-Flores A., Glover L., Berriman M., Hertz-Fowler C. and Horn D. (2011). High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Research 21, 915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves J. M., Voegtly L., Matveyev A. V., Lara A. M., da Silva F. M., Serrano M. G., Buck G. A., Teixeira M. M. and Camargo E. P. (2011). Identification and phylogenetic analysis of heme synthesis genes in trypanosomatids and their bacterial endosymbionts. PLOS ONE 6, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammar Z., Plazolles N., Baltz T. and Coustou V. (2013). Identification of trans-sialidases as a common mediator of endothelial cell activation by African trypanosomes. PLOS Pathogens 9, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annoura T., Nara T., Makiuchi T., Hashimoto T. and Aoki T. (2005). The origin of dihydroorotate dehydrogenase genes of kinetoplastids, with special reference to their biological significance and adaptation to anaerobic, parasitic conditions. Journal of Molecular Evolution 60, 113–127. [DOI] [PubMed] [Google Scholar]
- Atwood J. A. 3rd, Weatherly D. B., Minning T. A., Bundy B., Cavola C., Opperdoes F. R., Orlando R. and Tarleton R. L. (2005). The Trypanosoma cruzi proteome. Science 309, 473–476. [DOI] [PubMed] [Google Scholar]
- Bahia D., Oliveira L. M., Lima F. M., Oliveira P., Silveira J. F., Mortara R. A. and Ruiz J. C. (2009). The TryPIKinome of five human pathogenic trypanosomatids: Trypanosoma brucei, Trypanosoma cruzi, Leishmania major, Leishmania braziliensis and Leishmania infantum – new tools for designing specific inhibitors. Biochemical and Biophysical Research Communications 390, 963–970. [DOI] [PubMed] [Google Scholar]
- Barker A. R., Wickstead B., Gluenz E. and Gull K. (2008). Bioinformatic insights to the ESAG5 and GRESAG5 gene families in kinetoplastid parasites. Molecular and Biochemical Parasitology 162, 112–122. [DOI] [PubMed] [Google Scholar]
- Barnwell E. M., van Deursen F. J., Jeacock L., Smith K. A., Maizels R. M., Acosta-Serrano A. and Matthews K. (2010). Developmental regulation and extracellular release of a VSG expression-site-associated gene product from Trypanosoma brucei bloodstream forms. Journal of Cell Science 123, 3401–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry J. D., Ginger M. L., Burton P. and McCulloch R. (2003). Why are parasite contingency genes often associated with telomeres? International Journal of Parasitology 33, 29–45. [DOI] [PubMed] [Google Scholar]
- Bartholomeu D. C., Cerqueira G. C., Leão A. C., daRocha W. D., Pais F. S., Macedo C., Djikeng A., Teixeira S. M. and El-Sayed N. M. (2009). Genomic organization and expression profile of the mucin-associated surface protein (masp) family of the human pathogen Trypanosoma cruzi. Nucleic Acids Research 37, 3407–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J. T. and Ullman B. (1990). Nutritional requirements of wild-type and folate transport-deficient Leishmania donovani for pterins and folates. Molecular and Biochemical Parasitology 43, 221–230. [DOI] [PubMed] [Google Scholar]
- Bello A. R., Nare B., Freedman D., Hardy L. and Beverley S. M. (1994. ). PTR1: a reductase mediating salvage of oxidized pteridines and methotrexate resistance in the protozoan parasite Leishmania major. Proceedings of the National Academy of Sciences USA 91, 11442–11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabó G., Levy G., Ziliani M., Caeiro L. D., Sánchez D. O. and Tekiel V. (2013). TcTASV-C, a protein family in Trypanosoma cruzi that is predominantly trypomastigote-stage specific and secreted to the medium. PLOS ONE 8, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M., Hall N., Sheader K., Bringaud F., Tiwari B., Isobe T., Bowman S., Corton C., Clark L., Cross G. A., Hoek M., Zanders T., Berberof M., Borst P. and Rudenko G. (2002). The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Molecular and Biochemical Parasitology 122, 131–140. [DOI] [PubMed] [Google Scholar]
- Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D. C., Lennard N. J., Caler E., Hamlin N. E., Haas B., Böhme U., Hannick L., Aslett M. A., Shallom J., Marcello L., Hou L., Wickstead B., Alsmark U. C., Arrowsmith C., Atkin R. J., Barron A. J., Bringaud F., Brooks K., Carrington M., Cherevach I., Chillingworth T. J., Churcher C., Clark L. N., Corton C. H., Cronin A., Davies R. M., Doggett J., Djikeng A., Feldblyum T., Field M. C., Fraser A., Goodhead I., Hance Z., Harper D., Harris B. R., Hauser H., Hostetler J., Ivens A., Jagels K., Johnson D., Johnson J., Jones K., Kerhornou A. X., Koo H., Larke N., Landfear S., Larkin C., Leech V., Line A., Lord A., Macleod A., Mooney P. J., Moule S., Martin D. M., Morgan G. W., Mungall K., Norbertczak H., Ormond D., Pai G., Peacock C. S., Peterson J., Quail M. A., Rabbinowitsch E., Rajandream M. A., Reitter C., Salzberg S. L., Sanders M., Schobel S., Sharp S., Simmonds M., Simpson A. J., Tallon L., Turner C. M., Tait A., Tivey A. R., Van Aken S., Walker D., Wanless D., Wang S., White B., White O., Whitehead S., Woodward J., Wortman J., Adams M. D., Embley T. M., Gull K., Ullu E., Barry J. D., Fairlamb A. H., Opperdoes F., Barrell B. G., Donelson J. E., Hall N., Fraser C. M., Melville S. E. and El-Sayed N. M. (2005). The genome of the African trypanosome Trypanosoma brucei. Science 309, 416–422. [DOI] [PubMed] [Google Scholar]
- Berriman M., Haas B. J., LoVerde P. T., Wilson R. A., Dillon G. P., Cerqueira G. C., Mashiyama S. T., Al-Lazikani B., Andrade L. F., Ashton P. D., Aslett M. A., Bartholomeu D. C., Blandin G., Caffrey C. R., Coghlan A., Coulson R., Day T. A., Delcher A., DeMarco R., Djikeng A., Eyre T., Gamble J. A., Ghedin E., Gu Y., Hertz-Fowler C., Hirai H., Hirai Y., Houston R., Ivens A., Johnston D. A., Lacerda D., Macedo C. D., McVeigh P., Ning Z., Oliveira G., Overington J. P., Parkhill J., Pertea M., Pierce R. J., Protasio A. V., Quail M. A., Rajandream M. A., Rogers J., Sajid M., Salzberg S. L., Stanke M., Tivey A. R., White O., Williams D. L., Wortman J., Wu W., Zamanian M., Zerlotini A., Fraser-Liggett C. M., Barrell B. G. and El-Sayed N. M. (2009). The genome of the blood fluke Schistosoma mansoni. Nature 460, 352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley R., Tariq H., McElhinney H., Szöor B., Huxley-Jones J., Stevens R., Matthews K. and Tabernero L. (2007). The TriTryp phosphatome: analysis of the protein phosphatase catalytic domains. BMC Genomics 8, 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringaud F., Müller M., Cerqueira G. C., Smith M., Rochette A., El-Sayed N. M., Papadopoulou B. and Ghedin E. (2007). Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLOS Pathogens 3, 1291–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaglia C. A., Campo V. A., Di Noia J. M., Torrecilhas A. C., De Marchi C. R., Ferguson M. A., Frasch A. C. and Almeida I. C. (2004). The surface coat of the mammal-dwelling infective trypomastigote stage of Trypanosoma cruzi is formed by highly diverse immunogenic mucins. Journal of Biological Chemistry 279, 15860–15869. [DOI] [PubMed] [Google Scholar]
- Buscaglia C. A., Campo V. A., Frasch A. C. and Di Noia J. M. (2006). Trypanosoma cruzi surface mucins: host-dependent coat diversity. Nature Reviews Microbiology 4, 229–236. [DOI] [PubMed] [Google Scholar]
- Butter F., Bucerius F., Michel M., Cicova Z., Mann M. and Janzen C. J. (2013). Comparative proteomics of two life-cycle stages of stable isotope-labeled Trypanosoma brucei reveals novel components of the parasite's host adaptation machinery. Molecular and Cellular Proteomics 12, 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan H. A., Litaker R. W. and Noga E. J. (2002). Molecular taxonomy of the suborder Bodonina (Order Kinetoplastida), including the important fish parasite, Ichthyobodo necator. Journal of Eukaryotic Microbiology 49, 119–128. [DOI] [PubMed] [Google Scholar]
- Camargo E. P. (1999). Phytomonas and other trypanosomatid parasites of plants and fruit. Advances in Parasitology 42, 29–112. [DOI] [PubMed] [Google Scholar]
- Carrillo C., Cejas S., González N. S. and Algranati I. D. (1999). Trypanosoma cruzi epimastigotes lack ornithine decarboxylase but can express a foreign gene encoding this enzyme. FEBS Letters 454, 192–196. [DOI] [PubMed] [Google Scholar]
- Cerqueira G. C., Bartholomeu D. C., DaRocha W. D., Hou L., Freitas-Silva D. M., Machado C. R., El-Sayed N. M. and Teixeira S. M. (2008). Sequence diversity and evolution of multigene families in Trypanosoma cruzi. Molecular and Biochemical Parasitology 157, 65–72. [DOI] [PubMed] [Google Scholar]
- Chamond N., Goytia M., Coatnoan N., Barale J. C., Cosson A., Degrave W. M. and Minoprio P. (2005). Trypanosoma cruzi proline racemases are involved in parasite differentiation and infectivity. Molecular Microbiology 58, 46–60. [DOI] [PubMed] [Google Scholar]
- Chang K. P., Chang C. S. and Sassa S. (1975). Heme biosynthesis in bacterium-protozoon symbioses: enzymic defects in host hemoflagellates and complemental role of their intracellular symbiotes. Proceedings of the National Academy of Sciences USA 72, 2979–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comini M. A. and Flohé L. (2013). The trypanothione-based redox metabolism of trypanosomatids In Trypanosomatid Diseases: Molecular Routes to Drug Discovery (ed. Jäger T., Oliver K. and Flohé L.), pp. 167–200 Wiley-VCH, Weinheim, Germany. [Google Scholar]
- Contreras I., Gómez M. A., Nguyen O., Shio M. T., McMaster R. W. and Olivier M. (2010). Leishmania-induced inactivation of the macrophage transcription factor AP-1 is mediated by the parasite metalloprotease GP63. PLOS Pathogens 6, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas I. C., Cazzulo J. J. and Sánchez D. O. (2003). Gp63 homologues in Trypanosoma cruzi: surface antigens with metalloprotease activity and a possible role in host cell infection. Infection and Immunity 71, 5739–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávila-López M., Martínez-Guerra J. J. and Samuelsson T. (2010). Analysis of gene order conservation in eukaryotes identifies transcriptionally and functionally linked genes. PLOS ONE 5, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch K. W., Moxon E. R. and Wellems T. E. (1997). Shared themes of antigenic variation and virulence in bacterial, protozoal, and fungal infections. Microbiology and Molecular Biology Review 61, 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Assis R. R., Ibraim I. C., Nogueira P. M., Soares R. P. and Turco S. J. (2012). Glycoconjugates in New World species of Leishmania: polymorphisms in lipophosphoglycan and glycoinositolphospholipids and interaction with hosts. Biochimica et Biophysica Acta 1820, 1354–1365. [DOI] [PubMed] [Google Scholar]
- De Pablos L. M. and Osuna A. (2012). Multigene families in Trypanosoma cruzi and their role in infectivity. Infection and Immunity 80, 2258–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depledge D. P., Evans K. J., Ivens A. C., Aziz N., Maroof A., Kaye P. M. and Smith D. F. (2009). Comparative expression profiling of Leishmania: modulation in gene expression between species and in different host genetic backgrounds. PLOS Neglected Tropical Diseases 3, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depledge D. P., MacLean L. M., Hodgkinson M. R., Smith B. A., Jackson A. P., Ma S., Uliana S. R. and Smith D. F. (2010). Leishmania-specific surface antigens show sub-genus sequence variation and immune recognition. PLOS Neglected Tropical Diseases 4, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps P., Lara E., Marande W., López-García P., Ekelund F. and Moreira D. (2011). Phylogenomic analysis of kinetoplastids supports that trypanosomatids arose from within bodonids. Molecular Biology and Evolution 28, 53–58. [DOI] [PubMed] [Google Scholar]
- Devault A. and Bañuls A. L. (2008). The promastigote surface antigen gene family of the Leishmania parasite: differential evolution by positive selection and recombination. BMC Evolutionary Biology 8, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos S. L., Freitas L. M., Lobo F. P., Rodrigues-Luiz G. F., Mendes T. A., Oliveira A. C., Andrade L. O., Chiari E., Gazzinelli R. T., Teixeira S. M., Fujiwara R. T. and Bartholomeu D. C. (2012). The MASP family of Trypanosoma cruzi: changes in gene expression and antigenic profile during the acute phase of experimental infection. PLOS Neglected Tropical Diseases 6, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing T., Imamura H., Decuypere S., Clark T. G., Coombs G. H., Cotton J. A., Hilley J. D., de Doncker S., Maes I., Mottram J. C., Quail M. A., Rijal S., Sanders M., Schönian G., Stark O., Sundar S., Vanaerschot M., Hertz-Fowler C., Dujardin J. C. and Berriman M. (2011). Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Research 21, 2143–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufernez F., Yernaux C., Gerbod D., Noël C., Chauvenet M., Wintjens R., Edgcomb V. P., Capron M., Opperdoes F. R. and Viscogliosi E. (2006). The presence of four iron-containing superoxide dismutase isozymes in trypanosomatidae: characterization, subcellular localization, and phylogenetic origin in Trypanosoma brucei. Free Radical Biology and Medicine 40, 210–225. [DOI] [PubMed] [Google Scholar]
- El-Sayed N. M., Myler P. J., Bartholomeu D. C., Nilsson D., Aggarwal G., Tran A. N., Ghedin E., Worthey E. A., Delcher A. L., Blandin G., Westenberger S. J., Caler E., Cerqueira G. C., Branche C., Haas B., Anupama A., Arner E., Aslund L., Attipoe P., Bontempi E., Bringaud F., Burton P., Cadag E., Campbell D. A., Carrington M., Crabtree J., Darban H., da Silveira J. F., de Jong P., Edwards K., Englund P. T., Fazelina G., Feldblyum T., Ferella M., Frasch A. C., Gull K., Horn D., Hou L., Huang Y., Kindlund E., Klingbeil M., Kluge S., Koo H., Lacerda D., Levin M. J., Lorenzi H., Louie T., Machado C. R., McCulloch R., McKenna A., Mizuno Y., Mottram J. C., Nelson S., Ochaya S., Osoegawa K., Pai G., Parsons M., Pentony M., Pettersson U., Pop M., Ramirez J. L., Rinta J., Robertson L., Salzberg S. L., Sanchez D. O., Seyler A., Sharma R., Shetty J., Simpson A. J., Sisk E., Tammi M. T., Tarleton R., Teixeira S., Van Aken S., Vogt C., Ward P. N., Wickstead B., Wortman J., White O., Fraser C. M., Stuart K. D. and Andersson B. (2005a). The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309, 409–415. [DOI] [PubMed] [Google Scholar]
- El-Sayed N. M., Myler P. J., Blandin G., Berriman M., Crabtree J., Aggarwal G., Caler E., Renauld H., Worthey E. A., Hertz-Fowler C., Ghedin E., Peacock C., Bartholomeu D. C., Haas B. J., Tran A. N., Wortman J. R., Alsmark U. C., Angiuoli S., Anupama A., Badger J., Bringaud F., Cadag E., Carlton J. M., Cerqueira G. C., Creasy T., Delcher A. L., Djikeng A., Embley T. M., Hauser C., Ivens A. C., Kummerfeld S. K., Pereira-Leal J. B., Nilsson D., Peterson J., Salzberg S. L., Shallom J., Silva J. C., Sundaram J., Westenberger S., White O., Melville S. E., Donelson J. E., Andersson B., Stuart K. D. and Hall N. (2005b). Comparative genomics of trypanosomatid parasitic protozoa. Science 309, 404–409. [DOI] [PubMed] [Google Scholar]
- Eyford B. A., Sakurai T., Smith D., Loveless B., Hertz-Fowler C., Donelson J. E., Inoue N. and Pearson T. W. (2011). Differential protein expression throughout the life-cycle of Trypanosoma congolense, a major parasite of cattle in Africa. Molecular and Biochemical Parasitology 177, 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M. A. (1997). The surface glycoconjugates of trypanosomatid parasites. Philosophical Transactions of the Royal Society of London B: Biological Sciences 352, 1295–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field M. C. (2005). Signalling the genome: the Ras-like small GTPase family of trypanosomatids. Trends in Parasitology 21, 447–450. [DOI] [PubMed] [Google Scholar]
- Field M. C., Natesan S. K., Gabernet-Castello C. and Koumandou V. L. (2007). Intracellular trafficking in the trypanosomatids. Traffic 8, 629–639. [DOI] [PubMed] [Google Scholar]
- Figueiredo L. M., Cross G. A. M. and Janzen C. J. (2009). Epigenetic regulation in African trypanosomes: a new kid on the block. Nature Reviews Microbiology 7, 504–513. [DOI] [PubMed] [Google Scholar]
- Flegontov P., Votýpka J., Skalický T., Logacheva M. D., Penin A. A., Tanifuji G., Onodera N. T., Kondrashov A. S., Volf P., Archibald J. M. and Lukeš J. (2013). Paratrypanosoma is a novel early-branching trypanosomatid. Current Biology 23, 1787–1793. [DOI] [PubMed] [Google Scholar]
- Freitas L. M., dos Santos S. L., Rodrigues-Luiz G. F., Mendes T. A., Rodrigues T. S., Gazzinelli R. T., Teixeira S. M., Fujiwara R. T. and Bartholomeu D. C. (2011). Genomic analyses, gene expression and antigenic profile of the trans-sialidase superfamily of Trypanosoma cruzi reveal an undetected level of complexity. PLOS ONE 6, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- García E. A., Ziliani M., Agüero F., Bernabó G., Sánchez D. O. and Tekiel V. (2010). TcTASV: a novel protein family in Trypanosoma cruzi identified from a subtractive trypomastigote cDNA library. PLOS Neglected Tropical Diseases 4, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargantini P. R., Lujan H. D. and Pereira C. A. (2012). In silico analysis of trypanosomatids’ helicases. FEMS Microbiology Letters 335, 123–129. [DOI] [PubMed] [Google Scholar]
- Ghedin E., Bringaud F., Peterson J., Myler P., Berriman M., Ivens A., Andersson B., Bontempi E., Eisen J., Angiuoli S., Wanless D., Von Arxa A., Murphy L., Lennard N., Salzberg S., Adams M. D., White O., Hall N., Stuart K., Fraser C. M. and El-Sayed N. M. A. (2004). Gene synteny and evolution of genome architecture in trypanosomatids. Molecular and Biochemical Parasitology 134, 183–191. [DOI] [PubMed] [Google Scholar]
- Goldenberg S. and Avila A. R. (2011). Aspects of Trypanosoma cruzi stage differentiation. Advances in Parasitology 75, 285–305. [DOI] [PubMed] [Google Scholar]
- Gomez M. A., Contreras I., Hallé M., Tremblay M. L., McMaster R. W. and Olivier M. (2009). Leishmania GP63 alters host signaling through cleavage-activated protein tyrosine phosphatases. Science Signaling 2, ra58. [DOI] [PubMed] [Google Scholar]
- Graham S. V., Terry S. and Barry J. D. (1999). A structural and transcription pattern for variant surface glycoprotein gene expression sites used in metacyclic stage Trypanosoma brucei. Molecular and Biochemical Parasitology 103, 141–154. [DOI] [PubMed] [Google Scholar]
- Grandgenett P. M., Otsu K., Wilson H. R., Wilson M. E. and Donelson J. E. (2007). A function for a specific zinc metalloprotease of African trypanosomes. PLOS Pathogens 3, 1432–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W. and Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Gunasekera K., Wüthrich D., Braga-Lagache S., Heller M. and Ochsenreiter T. (2012) Proteome remodelling during development from blood to insect-form Trypanosoma brucei quantified by SILAC and mass spectrometry. BMC Genomics 13, 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge W. E. and Gaborak M. (1979). A re-examination of purine and pyrimidine synthesis in the three main forms of Trypanosoma cruzi. International Journal of Biochemistry 10, 415–422. [DOI] [PubMed] [Google Scholar]
- Haile S. and Papadopoulou B. (2007). Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Current Opinion in Microbiology 10, 569–577. [DOI] [PubMed] [Google Scholar]
- Hall J. P., Wang H. and Barry J. D. (2013). Mosaic VSGs and the scale of Trypanosoma brucei antigenic variation. PLOS Pathogens 9, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallé M., Gomez M. A., Stuible M., Shimizu H., McMaster W. R., Olivier M. and Tremblay M. L. (2009). The Leishmania surface protease GP63 cleaves multiple intracellular proteins and actively participates in p38 mitogen-activated protein kinase inactivation. Journal of Biological Chemistry 284, 6893–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handman E., Osborn A. H., Symons F., van Driel R. and Cappai R. (1995). The Leishmania promastigote surface antigen 2 complex is differentially expressed during the parasite life-cycle. Molecular and Biochemical Parasitology 74, 189–200. [DOI] [PubMed] [Google Scholar]
- Handman E., Papenfuss A. T., Speed T. P. and Goding J. W. (2008) Leishmania surface proteins In Leishmania: After the Genome (ed. Myler P. J. and Fasel N.), pp. 177–204 Caister Academic Press, Norfolk, UK. [Google Scholar]
- Herman M., Gillies S., Michels P. A. and Rigden D. J. (2006). Autophagy and related processes in trypanosomatids: insights from genomic and bioinformatic analyses. Autophagy 2, 107–118. [DOI] [PubMed] [Google Scholar]
- Hertz-Fowler C., Ersfeld K. and Gull K. (2001). CAP5.5, a life-cycle-regulated, cytoskeleton-associated protein is a member of a novel family of calpain-related proteins in Trypanosoma brucei. Molecular and Biochemical Parasitology 116, 25–34. [DOI] [PubMed] [Google Scholar]
- Hertz-Fowler C., Figueiredo L. M., Quail M. A., Becker M., Jackson A., Bason N., Brooks K., Churcher C., Fahkro S., Goodhead I., Heath P., Kartvelishvili M., Mungall K., Harris D., Hauser H., Sanders M., Saunders D., Seeger K., Sharp S., Taylor J. E., Walker D., White B., Young R., Cross G. A., Rudenko G., Barry J. D., Louis E. J. and Berriman M. (2008). Telomeric expression sites are highly conserved in Trypanosoma brucei. PLOS ONE 3, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer T. R., McMaster W. R. and Forney J. D. (2006). Expression profiling by whole-genome interspecies microarray hybridization reveals differential gene expression in procyclic promastigotes, lesion-derived amastigotes and axenic amastigotes in Leishmania mexicana. Molecular and Biochemical Parasitology 146, 198–218. [DOI] [PubMed] [Google Scholar]
- Holzer T. R., Mishra K. K., LeBowitz J. H. and Forney J. D. (2008). Coordinate regulation of a family of promastigote-enriched mRNAs by the 3′UTR PRE element in Leishmania mexicana. Molecular Biochemical Parasitology 157, 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D. and McCulloch R. (2010). Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Current Opinion in Microbiology 13, 700–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson O. C., Picozzi K., Jones N. G., Mott H., Sharma R., Welburn S. C. and Carrington M. (2007). Variant surface glycoprotein gene repertoires in Trypanosoma brucei have diverged to become strain-specific. BMC Genomics 8, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens A. C., Peacock C. S., Worthey E. A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M. A., Adlem E., Aert R., Anupama A., Apostolou Z., Attipoe P., Bason N., Bauser C., Beck A., Beverley S. M., Bianchettin G., Borzym K., Bothe G., Bruschi C. V., Collins M., Cadag E., Ciarloni L., Clayton C., Coulson R. M., Cronin A., Cruz A. K., Davies R. M., De Gaudenzi J., Dobson D. E., Duesterhoeft A., Fazelina G., Fosker N., Frasch A. C., Fraser A., Fuchs M., Gabel C., Goble A., Goffeau A., Harris D., Hertz-Fowler C., Hilbert H., Horn D., Huang Y., Klages S., Knights A., Kube M., Larke N., Litvin L., Lord A., Louie T., Marra M., Masuy D., Matthews K., Michaeli S., Mottram J. C., Müller-Auer S., Munden H., Nelson S., Norbertczak H., Oliver K., O'neil S., Pentony M., Pohl T. M., Price C., Purnelle B., Quail M. A., Rabbinowitsch E., Reinhardt R., Rieger M., Rinta J., Robben J., Robertson L., Ruiz J. C., Rutter S., Saunders D., Schäfer M., Schein J., Schwartz D. C., Seeger K., Seyler A., Sharp S., Shin H., Sivam D., Squares R., Squares S., Tosato V., Vogt C., Volckaert G., Wambutt R., Warren T., Wedler H., Woodward J., Zhou S., Zimmermann W., Smith D. F., Blackwell J. M., Stuart K. D., Barrell B. and Myler P. J. (2005). The genome of the kinetoplastid parasite, Leishmania major. Science 309, 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P. (2007a). Tandem gene arrays in Trypanosoma brucei: comparative phylogenomic analysis of duplicate sequence variation. BMC Evolutionary Biology 7, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P. (2007b). Evolutionary consequences of a large duplication event in Trypanosoma brucei: chromosomes 4 and 8 are partial duplicons. BMC Genomics 8, 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P. (2010). The evolution of amastin surface glycoproteins in trypanosomatid parasites. Molecular Biology and Evolution 27, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P., Vaughan S. and Gull K. (2006). Comparative genomics and concerted evolution of beta-tubulin paralogs in Leishmania spp. BMC Genomics 7, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P., Sanders M., Berry A., McQuillan J., Aslett M. A., Quail M. A., Chukualim B., Capewell P., MacLeod A., Melville S. E., Gibson W., Barry J. D., Berriman M. and Hertz-Fowler C. (2009). The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human African trypanosomiasis. PLOS Neglected Tropical Diseases 4, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P., Berry A., Aslett M., Allison H. C., Burton P., Vavrova-Anderson J., Brown R., Browne H., Corton N., Hauser H., Gamble J., Gilderthorp R., Marcello L., McQuillan J., Otto T. D., Quail M. A., Sanders M. J., van Tonder A., Ginger M. L., Field M. C., Barry J. D., Hertz-Fowler C. and Berriman M. (2012). Antigenic diversity is generated by distinct evolutionary mechanisms in African trypanosome species. Proceedings of the National Academy of Sciences USA 109, 3416–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P., Allison H. C., Barry J. D., Field M. C., Hertz-Fowler C. and Berriman M. (2013). A cell-surface phylome for African trypanosomes. PLOS Neglected Tropical Diseases 7, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. G., Windle H. J. and Voorheis H. P. (1993). The identification, purification, and characterization of two invariant surface glycoproteins located beneath the surface coat barrier of bloodstream forms of Trypanosoma brucei. Journal of Biological Chemistry 268, 8085–8095. [PubMed] [Google Scholar]
- Jensen B. C., Sivam D., Kifer C. T., Myler P. J. and Parsons M. (2009). Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei. BMC Genomics 10, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani S., Fenn K., Ross A., Ivens A., Smith T. K., Ghazal P. and Matthews K. (2009). Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics 10, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangussu-Marcolino M. M., de Paiva R. M., Araújo P. R., de Mendonça-Neto R. P., Lemos L., Bartholomeu D. C., Mortara R. A., daRocha W. D. and Teixeira S. M. (2013). Distinct genomic organization, mRNA expression and cellular localization of members of two amastin sub-families present in Trypanosoma cruzi. BMC Microbiology 13, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashita S. Y., da Silva C. V., Mortara R. A., Burleigh B. A. and Briones M. R. (2009). Homology, paralogy and function of DGF-1, a highly dispersed Trypanosoma cruzi specific gene family and its implications for information entropy of its encoded proteins. Molecular and Biochemical Parasitology 165, 19–31. [DOI] [PubMed] [Google Scholar]
- Keeling P. J. and Slamovits C. H. (2005). Causes and effects of nuclear genome reduction. Current Opinion in Genetics and Development 15, 601–608. [DOI] [PubMed] [Google Scholar]
- Kim D., Chiurillo M. A., El-Sayed N., Jones K., Santos M. R., Porcile P. E., Andersson B., Myler P., da Silveira J. F. and Ramírez J. L. (2005). Telomere and subtelomere of Trypanosoma cruzi chromosomes are enriched in (pseudo)genes of retrotransposon hot spot and trans-sialidase-like gene families: the origins of T. cruzi telomeres. Gene 346, 153–161. [DOI] [PubMed] [Google Scholar]
- Kissinger J. C. and DeBarry J. (2011). Genome cartography: charting the apicomplexan genome. Trends in Parasitology 27, 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev N. G., Ramey-Butler K., Cross G. A. M., Ullu E. and Tschudi C. (2012). Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science 338, 1352–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev N. G., Ullu E. and Tschudi C. (2014). The emerging role of RNA-binding proteins in the life-cycle of Trypanosoma brucei. Cellular Microbiology 16, 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korený L., Lukes J. and Oborník M. (2010). Evolution of the haem synthetic pathway in kinetoplastid flagellates: an essential pathway that is not essential after all? International Journal for Parasitology 40, 149–156. [DOI] [PubMed] [Google Scholar]
- Kosec G., Alvarez V. E., Agüero F., Sánchez D., Dolinar M., Turk B., Turk V. and Cazzulo J. J. (2006). Metacaspases of Trypanosoma cruzi: possible candidates for programmed cell death mediators. Molecular and Biochemical Parasitology 145, 18–28. [DOI] [PubMed] [Google Scholar]
- Krauth-Siegel R. L. and Comini M. A. (2008). Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochimica et Biophysica Acta 1780, 1236–1248. [DOI] [PubMed] [Google Scholar]
- Kulkarni M. M., McMaster W. R., Kamysz E., Kamysz W., Engman D. M. and McGwire B. S. (2006). The major surface-metalloprotease of the parasitic protozoan, Leishmania, protects against antimicrobial peptide-induced apoptotic killing. Molecular Microbiology 62, 1484–1497. [DOI] [PubMed] [Google Scholar]
- Kulkarni M. M., Olson C. L., Engman D. M. and McGwire B. S. (2009). Trypanosoma cruzi GP63 proteins undergo stage-specific differential posttranslational modification and are important for host cell infection. Infection and Immunity 77, 2193–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount D. J., Gruszynski A. E., Grandgenett P. M., Bangs J. D. and Donelson J. E. (2003). Expression and function of the Trypanosoma brucei major surface protease (GP63) genes. Journal of Biological Chemistry 278, 24658–24664. [DOI] [PubMed] [Google Scholar]
- Lander N., Bernal C., Diez N., Añez N., Docampo R. and Ramírez J. L. (2010). Localization and developmental regulation of a dispersed gene family 1 protein in Trypanosoma cruzi. Infection and Immunity 78, 231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifso K., Cohen-Freue G., Dogra N., Murray A. and McMaster W. R. (2007). Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life-stages: the Leishmania genome is constitutively expressed. Molecular and Biochemical Parasitology 152, 35–46. [DOI] [PubMed] [Google Scholar]
- Li Z. H., De Gaudenzi J. G., Alvarez V. E., Mendiondo N., Wang H., Kissinger J. C., Frasch A. C. and Docampo R. (2012). A 43-nucleotide U-rich element in 3′-untranslated region of large number of Trypanosoma cruzi transcripts is important for mRNA abundance in intracellular amastigotes. Journal of Biological Chemistry 287, 19058–19069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieke T., Nylén S., Eidsmo L., McMaster W. R., Mohammadi A. M., Khamesipour A., Berg L. and Akuffo H. (2008). Leishmania surface protein gp63 binds directly to human natural killer cells and inhibits proliferation. Clinical and Experimental Immunology 153, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln L. M., Ozaki M., Donelson J. E. and Beetham J. K. (2004). Genetic complementation of Leishmania deficient in PSA (GP46) restores their resistance to lysis by complement. Molecular and Biochemical Parasitology 137, 185–189. [DOI] [PubMed] [Google Scholar]
- Lira C. B., Giardini M. A., Neto J. L., Conte F. F. and Cano M. I. (2007). Telomere biology of trypanosomatids: beginning to answer some questions. Trends in Parasitology 23, 357–362. [DOI] [PubMed] [Google Scholar]
- Lom J. (1979). Biology of fish trypanosomes and trypanoplasms In Biology of Kinetoplastida Volume II (ed. Lumsden W. H. R. and Evans D. A.), pp. 269–377 Academic Press, New York, NY, USA. [Google Scholar]
- Lynch M. and Conery J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Ma L., Chen K., Meng Q., Liu Q., Tang P., Hu S. and Yu J. (2011). An evolutionary analysis of trypanosomatid GP63 proteases. Parasitology Research 109, 1075–1084. [DOI] [PubMed] [Google Scholar]
- Marcello L. and Barry J. D. (2007). Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Genome Research 17, 1344–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoux V., Wei G., Tabel H. and Bull H. J. (2010). Characterization of major surface protease homologues of Trypanosoma congolense. Journal of Biomedicine and Biotechnology 2010, 418157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr J. J., Berens R. L. and Nelson D. J. (1978). Purine metabolism in Leishmania donovani and Leishmania braziliensis. Biochimica et Biophysica Acta 544, 360–371. [DOI] [PubMed] [Google Scholar]
- Maslov D. A., Votýpka J., Yurchenko V. and Lukeš J. (2013). Diversity and phylogeny of insect trypanosomatids: all that is hidden shall be revealed. Trends in Parasitology 29, 43–52. [DOI] [PubMed] [Google Scholar]
- McCulloch R. and Horn D. (2009). What has DNA sequencing revealed about the VSG expression sites of African trypanosomes? Trends in Parasitology 25, 359–363. [DOI] [PubMed] [Google Scholar]
- Minning T. A., Weatherly D. B., Atwood J. 3rd, Orlando R. and Tarleton R. L. (2009). The steady-state transcriptome of the four major life-cycle stages of Trypanosoma cruzi. BMC Genomics 10, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagna G. N., Donelson J. E. and Frasch A. C. (2006). Procyclic Trypanosoma brucei expresses separate sialidase and trans-sialidase enzymes on its surface membrane. Journal of Biological Chemistry 281, 33949–33958. [DOI] [PubMed] [Google Scholar]
- Moraes Barros R. R., Marini M. M., Antônio C. R., Cortez D. R., Miyake A. M., Lima F. M., Ruiz J. C., Bartholomeu D. C., Chiurillo M. A., Ramirez J. L. and da Silveira J. F. (2012). Anatomy and evolution of telomeric and subtelomeric regions in the human protozoan parasite Trypanosoma cruzi. BMC Genomics 13, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D., López-García P. and Vickerman K. (2004). An updated view of kinetoplastid phylogeny using environmental sequences and a closer outgroup: proposal for a new classification of the class Kinetoplastea. International Journal of Systematics and Evolutionary Microbiology 54, 1861–1875. [DOI] [PubMed] [Google Scholar]
- Nakayasu E. S., Yashunsky D. V., Nohara L. L., Torrecilhas A. C., Nikolaev A. V. and Almeida I. C. (2009). GPIomics: global analysis of glycosylphosphatidylinositol-anchored molecules of Trypanosoma cruzi. Molecular Systems Biology 5, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakjang S., Williams T. A., Heinz E., Watson A. K., Foster P. G., Sendra K. M., Heaps S. E., Hirt R. P. and Embley T. M. (2013). Reduction and expansion in microsporidian genome evolution: new insights from comparative genomics. Genome Biology and Evolution 5, 2285–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nare B., Hardy L. W. and Beverley S. M. (1997). The roles of pteridine reductase 1 and dihydrofolate reductase-thymidylate synthase in pteridine metabolism in the protozoan parasite Leishmania major. Journal of Biological Chemistry 272, 13883–13891. [DOI] [PubMed] [Google Scholar]
- Niemirowicz G., Parussini F., Agüero F. and Cazzulo J. J. (2007). Two metallocarboxypeptidases from the protozoan Trypanosoma cruzi belong to the M32 family, found so far only in prokaryotes. Biochemistry Journal 401, 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes M. P., Fortes B., Silva-Filho J. L., Terra-Granado E., Santos L., Conde L., de Araújo Oliveira I., Freire-de-Lima L., Martins M. V., Pinheiro A. A., Takyia C. M., Freire-de-Lima C. G., Todeschini A. R., Dosreis G. A. and Morrot A. (2013). Inhibitory effects of Trypanosoma cruzi sialoglycoproteins on CD4(+) T cells are associated with increased susceptibility to infection. PLOS ONE 8, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. (1970). Evolution by Gene Duplication. Springer-Verlag, New York, NY, USA. [Google Scholar]
- Olego-Fernandez S., Vaughan S., Shaw M. K., Gull K. and Ginger M. L. (2009). Cell morphogenesis of Trypanosoma brucei requires the paralogous, differentially expressed calpain-related proteins CAP5.5 and CAP5.5V. Protist 160, 576–590. [DOI] [PubMed] [Google Scholar]
- Oliveira I. A., Freire-de-Lima L., Penha L. L., Dias W. B. and Todeschini A. R. (2014). Trypanosoma cruzi trans-sialidase: structural features and biological implications. Subcellular Biochemistry 74, 181–201. [DOI] [PubMed] [Google Scholar]
- Opperdoes F. R. and Michels P. A. (2007). Horizontal gene transfer in trypanosomatids. Trends in Parasitology 23, 470–476. [DOI] [PubMed] [Google Scholar]
- Oppezzo P., Obal G., Baraibar M. A., Pritsch O., Alzari P. M. and Buschiazzo A. (2011). Crystal structure of an enzymatically inactive trans-sialidase-like lectin from Trypanosoma cruzi: the carbohydrate binding mechanism involves residual sialidase activity. Biochimica et Biophysica Acta 1814, 1154–1161. [DOI] [PubMed] [Google Scholar]
- Ouellette M., Drummelsmith J., El-Fadili A., Kündig C., Richard D. and Roy G. (2002). Pterin transport and metabolism in Leishmania and related trypanosomatid parasites. International Journal for Parasitology 32, 385–398. [DOI] [PubMed] [Google Scholar]
- Oza S. L., Shaw M. P., Wyllie S. and Fairlamb A. H. (2005). Trypanothione biosynthesis in Leishmania major. Molecular and Biochemical Parasitology 139, 107–116. [DOI] [PubMed] [Google Scholar]
- Parsons M., Worthey E. A., Ward P. N. and Mottram J. C. (2005). Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics 6, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastro L., Smircich P., Pérez-Díaz L., Duhagon M. A. and Garat B. (2013). Implication of CA repeated tracts on post-transcriptional regulation in Trypanosoma cruzi. Experimental Parasitology 134, 511–518. [DOI] [PubMed] [Google Scholar]
- Pays E., Lips S., Nolan D., Vanhamme L. and Pérez-Morga D. (2001). The VSG expression sites of Trypanosoma brucei: multipurpose tools for the adaptation of the parasite to mammalian hosts. Molecular and Biochemical Parasitology 114, 1–16. [DOI] [PubMed] [Google Scholar]
- Peacock C. S., Seeger K., Harris D., Murphy L., Ruiz J. C., Quail M. A., Peters N., Adlem E., Tivey A., Aslett M., Kerhornou A., Ivens A., Fraser A., Rajandream M. A., Carver T., Norbertczak H., Chillingworth T., Hance Z., Jagels K., Moule S., Ormond D., Rutter S., Squares R., Whitehead S., Rabbinowitsch E., Arrowsmith C., White B., Thurston S., Bringaud F., Baldauf S. L., Faulconbridge A., Jeffares D., Depledge D. P., Oyola S. O., Hilley J. D., Brito L. O., Tosi L. R., Barrell B., Cruz A. K., Mottram J. C., Smith D. F. and Berriman M. (2007). Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nature Genetics 39, 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcel B. M., Denoeud F., Opperdoes F., Noel B., Madoui M. A., Hammarton T. C., Field M. C., Da Silva C., Couloux A., Poulain J., Katinka M., Jabbari K., Aury J. M., Campbell D. A., Cintron R., Dickens N. J., Docampo R., Sturm N. R., Koumandou V. L., Fabre S., Flegontov P., Lukeš J., Michaeli S., Mottram J. C., Szöőr B., Zilberstein D., Bringaud F., Wincker P. and Dollet M. (2014). The streamlined genome of Phytomonas spp. relative to human pathogenic kinetoplastids reveals a parasite tailored for plants. PLOS Genetics 10, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri V., Goyal A., Sankaranarayanan R., Enright A. J. and Vaidya T. (2011). Evolutionary and functional insights into Leishmania META1: evidence for lateral gene transfer and a role for META1 in secretion. BMC Evolutionary Biology 11, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusnik M., Charrière F., Mäser P., Waller R. F., Dagley M. J., Lithgow T. and Schneider A. (2009). The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Molecular Biology and Evolution 26, 671–680. [DOI] [PubMed] [Google Scholar]
- Raymond F., Boisvert S., Roy G., Ritt J. F., Légaré D., Isnard A., Stanke M., Olivier M., Tremblay M. J., Papadopoulou B., Ouellette M. and Corbeil J. (2012). Genome sequencing of the lizard parasite Leishmania tarentolae reveals loss of genes associated to the intracellular stage of human pathogenic species. Nucleic Acids Research 40, 1131–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real F., Vidal R. O., Carazzolle M. F., Mondego J. M., Costa G. G., Herai R. H., Würtele M., de Carvalho L. M., E Ferreira R. C., Mortara R. A., Barbiéri C. L., Mieczkowski P., da Silveira J. F., Briones M. R., Pereira G. A. and Bahia D. (2013). The genome sequence of Leishmania (Leishmania) amazonensis: functional annotation and extended analysis of gene models. DNA Research 20, 567–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martín B., Degrave W., Rougeot C., Cosson A., Chamond N., Cordeiro-Da-Silva A., Arala-Chaves M., Coutinho A. and Minoprio P. (2000). A B-cell mitogen from a pathogenic trypanosome is a eukaryotic proline racemase. Nature Medicine 6, 890–897. [DOI] [PubMed] [Google Scholar]
- Richmond G. S. and Smith T. K. (2007). A novel phospholipase from Trypanosoma brucei. Molecular Microbiology 63, 1078–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico E., Rojas F., Mony B. M., Szoor B., MacGregor P. and Matthews K. R. (2013). Bloodstream form pre-adaptation to the tsetse fly in Trypanosoma brucei. Frontiers in Cellular and Infection Microbiology 3, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette A., McNicoll F., Girard J., Breton M., Leblanc E., Bergeron M. G. and Papadopoulou B. (2005). Characterization and developmental gene regulation of a large gene family encoding amastin surface proteins in Leishmania spp. Molecular and Biochemical Parasitology 140, 205–220. [DOI] [PubMed] [Google Scholar]
- Rochette A., Raymond F., Ubeda J. M., Smith M., Messier N., Boisvert S., Rigault P., Corbeil J., Ouellette M. and Papadopoulou B. (2008). Genome-wide gene expression profiling analysis of Leishmania major and Leishmania infantum developmental stages reveals substantial differences between the two species. BMC Genomics 9, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette A., Raymond F., Corbeil J., Ouellette M. and Papadopoulou B. (2009). Whole-genome comparative RNA expression profiling of axenic and intracellular amastigote forms of Leishmania infantum. Molecular and Biochemical Parasitology 165, 32–47. [DOI] [PubMed] [Google Scholar]
- Roditi I., Furger A., Ruepp S., Schürch N. and Bütikofer P. (1998). Unravelling the procyclin coat of Trypanosoma brucei. Molecular and Biochemical Parasitology 91, 117–130. [DOI] [PubMed] [Google Scholar]
- Rogers M. B., Hilley J. D., Dickens N. J., Wilkes J., Bates P. A., Depledge D. P., Harris D., Her Y., Herzyk P., Imamura H., Otto T. D., Sanders M., Seeger K., Dujardin J. C., Berriman M., Smith D. F., Hertz-Fowler C. and Mottram J. C. (2011). Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Research 21, 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig D., Smith D., Opperdoes F., Stern S., Olafson R. W. and Zilberstein D. (2008a). Retooling Leishmania metabolism: from sand fly gut to human macrophage. FASEB Journal 22, 590–602. [DOI] [PubMed] [Google Scholar]
- Rosenzweig D., Smith D., Myler P. J., Olafson R. W. and Zilberstein D. (2008b). Post-translational modification of cellular proteins during Leishmania donovani differentiation. Proteomics 8, 1843–1850. [DOI] [PubMed] [Google Scholar]
- Rudenko G. (2010). Epigenetics and transcriptional control in African trypanosomes. Essays in Biochemistry 48, 201–219. [DOI] [PubMed] [Google Scholar]
- Rudenko G. (2011). African trypanosomes: the genome and adaptations for immune evasion. Essays in Biochemistry 51, 47–62. [DOI] [PubMed] [Google Scholar]
- Sádlová J., Price H. P., Smith B. A., Votýpka J., Volf P. and Smith D. F. (2010). The stage-regulated HASPB and SHERP proteins are essential for differentiation of the protozoan parasite Leishmania major in its sand fly vector, Phlebotomus papatasi. Cellular Microbiology 12, 1765–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon D., Hanocq-Quertier J., Paturiaux-Hanocq F., Pays A., Tebabi P., Nolan D. P., Michel A. and Pays E. (1997). Characterization of the ligand-binding site of the transferrin receptor in Trypanosoma brucei demonstrates a structural relationship with the N-terminal domain of the variant surface glycoprotein. EMBO Journal 16, 7272–7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon D., Bachmaier S., Krumbholz C., Kador M., Gossmann J. A., Uzureau P., Pays E. and Boshart M. (2012). Cytokinesis of Trypanosoma brucei bloodstream forms depends on expression of adenylyl cyclases of the ESAG4 or ESAG4-like subfamily. Molecular Microbiology 84, 225–242. [DOI] [PubMed] [Google Scholar]
- Sanchez M. A., Zeoli D., Klamo E. M., Kavanaugh M. P. and Landfear S. M. (1995). A family of putative receptor-adenylate cyclases from Leishmania donovani. Journal of Biological Chemistry 270, 17551–17558. [DOI] [PubMed] [Google Scholar]
- Saxena A., Lahav T., Holland N., Aggarwal G., Anupama A., Huang Y., Volpin H., Myler P. J. and Zilberstein D. (2007). Analysis of the Leishmania donovani transcriptome reveals an ordered progression of transient and permanent changes in gene expression during differentiation. Molecular and Biochemical Parasitology 152, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel T. N., Hekstra D. R., Wang X., Dewell S. and Cross G. A. (2010). Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Research 38, 4946–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson A. G., Gill E. E., Callahan H. A., Litaker R. W. and Roger A. J. (2004). Early evolution within kinetoplastids (euglenozoa), and the late emergence of trypanosomatids. Protist 15, 407–422. [DOI] [PubMed] [Google Scholar]
- Simpson A. G., Stevens J. R. and Lukes J. (2006). The evolution and diversity of kinetoplastid flagellates. Trends in Parasitology 22, 168–174. [DOI] [PubMed] [Google Scholar]
- Smirlis D., Duszenko M., Ruiz A. J., Scoulica E., Bastien P., Fasel N. and Soteriadou K. (2010). Targeting essential pathways in trypanosomatids gives insights into protozoan mechanisms of cell death. Parasites and Vectors 3, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M., Bringaud F. and Papadopoulou B. (2009). Organization and evolution of two SIDER retroposon subfamilies and their impact on the Leishmania genome. BMC Genomics 10, 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich C. and Schaeffer S. W. (2006). The ornithine decarboxylase gene of Trypanosoma brucei: evidence for horizontal gene transfer from a vertebrate source. Infection Genetics and Evolution 6, 205–219. [DOI] [PubMed] [Google Scholar]
- Tsai I. J., Zarowiecki M., Holroyd N., Garciarrubio A., Sanchez-Flores A., Brooks K. L., Tracey A., Bobes R. J., Fragoso G., Sciutto E., Aslett M., Beasley H., Bennett H. M., Cai J., Camicia F., Clark R., Cucher M., De Silva N., Day T. A., Deplazes P., Estrada K., Fernández C., Holland P. W., Hou J., Hu S., Huckvale T., Hung S. S., Kamenetzky L., Keane J. A., Kiss F., Koziol U., Lambert O., Liu K., Luo X., Luo Y., Macchiaroli N., Nichol S., Paps J., Parkinson J., Pouchkina-Stantcheva N., Riddiford N., Rosenzvit M., Salinas G., Wasmuth J. D., Zamanian M., Zheng Y., Taenia solium Genome Consortium, Cai X., Soberón X., Olson P. D., Laclette J. P., Brehm K. and Berriman M. (2013). The genomes of four tapeworm species reveal adaptations to parasitism. Nature 496, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban I., Santurio L. B., Chidichimo A., Yu H., Chen X., Mucci J., Agüero F. and Buscaglia C. A. (2011). Molecular diversity of the Trypanosoma cruzi TcSMUG family of mucin genes and proteins. Biochemistry Journal 438, 303–313. [DOI] [PubMed] [Google Scholar]
- Urbaniak M. D., Guther M. L. and Ferguson M. A. (2012). Comparative SILAC proteomic analysis of Trypanosoma brucei bloodstream and procyclic lifecycle stages. PLOS ONE 7, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwyler S., Studer E., Renggli C. K. and Roditi I. (2007). A family of stage-specific alanine-rich proteins on the surface of epimastigote forms of Trypanosoma brucei. Molecular Microbiology 63, 218–228. [DOI] [PubMed] [Google Scholar]
- Vanhamme L. and Pays E. (1995). Control of gene expression in trypanosomes. Microbiological Review 59, 223–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitch N. J., Johnson P. C., Trivedi U., Terry S., Wildridge D. and MacLeod A. (2010). Digital gene expression analysis of two life-cycle stages of the human-infective parasite, Trypanosoma brucei gambiense reveals differentially expressed clusters of co-regulated genes. BMC Genomics 11, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoir K., Arevalo J., De Doncker S., Barker D. C., Laurent T., Godfroid E., Bollen A., Le Ray D. and Dujardin J. C. (2005). Complexity of the major surface protease (msp) gene organization in Leishmania (Viannia) braziliensis: evolutionary and functional implications. Parasitology 131, 207–214. [DOI] [PubMed] [Google Scholar]
- Von der Heyden S., Chao E. E., Vickerman K. and Cavalier-Smith T. (2004). Ribosomal RNA phylogeny of bodonid and diplonemid flagellates and the evolution of euglenozoa. Journal of Eukaryotic Microbiology 51, 402–416. [DOI] [PubMed] [Google Scholar]
- Weirather J. L., Wilson M. E. and Donelson J. E. (2012). Mapping of VSG similarities in Trypanosoma brucei. Molecular and Biochemical Parasitology 181, 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C. (2010). Major surface protease of trypanosomatids: one size fits all? Infection and Immunity 78, 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer K. and Overath P. (1993). Organization of two invariant surface glycoproteins in the surface coat of Trypanosoma brucei. Infection and Immunity 61, 4540–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A., Akum Y., Yahav T. and Shapira M. (2012). Gene duplication in trypanosomatids – two DED1 paralogs are functionally redundant and differentially expressed during the life-cycle. Molecular and Biochemical Parasitology 185, 127–136. [DOI] [PubMed] [Google Scholar]