Abstract
Environmental factors or socioeconomic status variables used in regression models to explain environmental chemical exposures or health outcomes are often in practice modeled at the same buffer distance or spatial scale. In this paper, we present four model selection algorithms that select the best spatial scale for each buffer-based or area-level covariate. Contamination of drinking water by nitrate is a growing problem in agricultural areas of the United States, as ingested nitrate can lead to the endogenous formation of N-nitroso compounds, which are potent carcinogens. We applied our methods to model nitrate levels in private wells in Iowa. We found that environmental variables were selected at different spatial scales and that a model allowing spatial scale to vary across covariates provided the best goodness of fit. Our methods can be applied to investigate the association between environmental risk factors available at multiple spatial scales or buffer distances and measures of disease, including cancers.
Keywords: model selection, spatial scale, nitrate, environment, cancer risk factors
Introduction
In public health research, it is acknowledged that both compositional (individual-level) and contextual (neighborhood-level) variables are important for explaining variation in health outcomes.1–7 For example, the role of contextual variables continues to be a key focus of investigators studying potential risk factors for obesity,8–10 where neighborhood variables such as fast-food density and green-space presence are thought to be important factors that contribute to obesity status. Neighborhood- or area-level variables can also have an important role in explaining variation in environmental chemical exposures. One example is found with modeling variation in polychlorinated biphenyls (PCBs) measured in carpet dust using percentage of developed land, population density, and number of industrial facilities within 2 km of residences, where total PCB levels are significantly associated with either the percentage of developed land or the population density.11
Through the use of geocoding and geographic information systems (GIS), researchers can link an individual residential address to an external database containing numerous area-level or environmental variables, where the number of variables is denoted by p. Typically, each of the area-level variables is available at multiple geographic scales. Thus, the p number of area-level variables available to consider when explaining variation in a health outcome can quickly multiply. Many socioeconomic status (SES) variables, including race, education, household income, and housing tenure, are available from the U.S. Census at the census block group, census tract, and county level. To clarify, we use the word “level” as a general term for spatial unit. Level is synonymous with region size, which, as will be seen later in our case example, denotes a buffer size. In addition to geopolitical areas, researchers can also create a set of geographic areas of varying size to summarize environmental variables using circular buffers or rings centered at observed data points. As an example, population density can be calculated at distances of 1, 2, and 3 mi from a residential location by intersecting spatial buffers of these sizes with census block group data.
With the abundance of socioeconomic and environmental data available at multiple spatial scales, a natural question arises for researchers who wish to investigate environmental effects: at what spatial scale (geographic areal unit) should each area-level variable be modeled in order to explain a fixed health outcome or environmental exposure of interest? Area-level covariates used as contextual variables in regression models are often in practice modeled at the same spatial scale, where, generally, a smaller spatial unit (ie, census block group versus a county) is thought to better capture heterogeneity in regression relationships. Krieger et al.4 model area-based socioeconomic measures at three spatial scales (census block group, census tract, and ZIP Code) to study mortality outcomes and cancer incidence and find that the effect estimates for the smaller spatial units (census block group and census tract) are similar, while the effect estimates for the larger spatial unit (ZIP Code) differ and are sometimes in the opposite direction. Krieger et al.4 conclude that the level of geography is important and recommend the use of SES variables at a smaller spatial scale, namely census block group or census tract.
The selection of spatial scale for environmental variables is a problem typically encountered in modeling groundwater quality. A variable that is often incorporated into statistical models of groundwater quality is area land use, as it is known to be one of the factors that can affect water quality. Barringer et al.12 find that the use of a circular buffer around a water table well is a simple and effective method for correlating water quality and land use. Regional and national groundwater studies have associated land use near a well with water quality using a fixed circular buffer distance, with 500 m a common choice and 1 km a less common choice.13–17 Some researchers have evaluated the univariate correlation of land use variables with groundwater quality. Ferrari and Ator18 find correlations between agricultural land use and nitrate concentrations using circular buffers of 400 and 800 m. Kolpin19 correlates the concentrations of nitrate, alachlor, and atrazine detected in wells with a variety of land use variables using circular buffers ranging in size from 200 m to 2 km. Johnson and Belitz20 evaluate a range of circular buffer and wedge sizes in a univariate correlation analysis of urban land use and the occurrence of volatile organic compounds (VOCs) in groundwater using Kendall’s tau (τ). They find that the values of τ are within 10% of one another for circles and wedges ranging in size from 500 m to 2 km, with statistically significant correlations for all sizes, and conclude that the popular choice of a 500-m circular buffer is adequate for assigning land use variables to a well.
Other researchers have evaluated the buffer distance to select each type of land use variable to be used in a regression model of groundwater quality. Rupert21 selects the circular buffer size for land use variables to explain the detection of elevated atrazine or desethyl-atrazine (atrazine/DEA) concentrations and elevated concentrations of nitrate using univariate logistic regression. The optimal buffer size is 2 km for agricultural land use variables and 500 m for urban land use variables according to McFadden’s ρ2, which is a transformation of the log-likelihood statistic that is designed to imitate the r2 of linear regression for univariate regression models.21 The buffer distances of 2 km for agricultural land use variables and 500 m for urban land use variables are then used in multiple logistic regression models of the probability of elevated atrazine/DEA detection and the probability of elevated nitrate detection.
An important question is whether all area-level variables should be modeled at the same spatial scale, as recent studies have shown that different area-level covariates are associated with health outcomes at different spatial scales.2,6,7 Root6 finds that the relationship between area-level SES variables and orofacial cleft risk varies when using different spatial scales to define neighborhoods. The study results indicate that poverty has a stronger association with risk for cleft palate at smaller geographic scales, while unemployment has a stronger association at larger scales, thus providing evidence that neighborhood effects operate at different spatial scales. In addition, Flowerdew et al.2 demonstrate in a British study of limiting long-term illness (LLTI) that the correlation strength between area-level variables and LLTI can vary depending on the spatial scale. Stronger correlations are present for LLTI and age group at the smaller enumeration district (ED) scale, while stronger correlations exist for LLTI and unemployment at the larger ward scale. In another study, Block et al.8 examine the relationship between fast-food restaurant density (FFRD) and black and low-income neighborhoods while controlling for various neighborhood variables such as commercial activity and presence of highways, which, in addition to FFRD, are available at two spatial scales (0.5- and 1-mi buffer sizes). They find that, while the results for both buffer analyses are similar, the 1-mi buffer analysis leads to a statistically significant association for median household income, perhaps due to a better capturing of how far people are willing to travel to buy food. In light of these findings, it is important to consider spatial scale for each area-level covariate when studying relationships between environmental variables and a particular outcome variable.
In this paper, we present a novel approach for modeling area-based variables at different spatial scales using four model selection approaches. To demonstrate these methods, we use a nitrate dataset containing numerous geologic and land use variables at different buffer sizes to investigate potential associations with nitrate concentrations in drinking well water. Contamination of drinking water by nitrate is a growing problem in agricultural areas of the United States, as ingested nitrate can lead to the endogenous formation of N-nitroso compounds, which are potent carcinogens. Our methods are not limited to the case example we present, but can be applied to other area-based variables that are related to cancer.
Methods
Statistical methods
We review four established methods used in model selection: forward stepwise regression, incremental forward stagewise regression, least angle regression (LARS), and the lasso.24 Next, we introduce our modified versions of these algorithms to select the spatial scale. Lastly, we present an application of our methods to model groundwater nitrate concentrations in Iowa.
Model selection approaches
Forward stepwise regression
Forward stepwise regression is a common approach used for model selection. A description of the forward stepwise regression algorithm detailed by Berk22 and Wheeler23 is as follows:
Initialize all regression coefficients , and let r = y, where r denotes the residual vector and y denotes the response vector.
Of the candidate variables, find the predictor xj that has the greatest absolute correlation with the residuals r, and add xj to the working design matrix Xin.
Let .
Compute the residuals r = y − ŷ, where ŷ is the vector of fitted values.
Iterate steps (2)–(4) until there is an inadequate improvement in the performance of the model or until all predictors have been added to the model.
For step (5), we consider there to be an inadequate improvement in the model’s performance if the difference in the Akaike information criterion (AIC) between the current model and the proposed model is less than ε, for some ε > 0.
Incremental forward stagewise regression
Incremental forward stagewise regression is another common approach used for model selection. Hastie et al.24 and Hastie et al.25 describe the incremental forward stagewise regression algorithm as follows:
Initialize all regression coefficients , and let r = y, where r denotes the residual vector and y denotes the response vector.
Find the predictor xj that has the greatest absolute correlation with the residuals r.
Let , where δj = τ·sign[corr(r,xj)] for some step size τ>0.
Let r ← r − δjxj.
Iterate steps (2)–(4) until none of the predictors are correlated with the residuals r.
For step (5), we consider none of the predictors to be correlated with the residuals if max|corr(r, X)| is less than a specified tolerance, where the tolerance is some small, positive number.
Least angle regression
Following the notation of Yuan and Lin,26 the LARS algorithm is described as follows:
Initialize all regression coefficients , and let r[0] = y, where r[0] denotes the residual vector at index 0 and y denotes the response vector. Set i = 1, where i is the index for the current iteration count.
Find the predictor xc among the p possible predictors that has the greatest absolute correlation with the residuals r[i−1].
Let the active set be equal to the corresponding column index of X associated with the predictor xc, and add xc to the working design matrix .
- Let γ be a p-dimensional vector where all values are set equal to 0. Calculate the current least squares direction γ by updating
- For every j in X that is not an element of the active set , calculate αj, the minimum distance needed to move the active regression coefficient(s) in direction γ until another predictor xj has as much correlation with the current residuals as the variables in the active set. That is, find αj ∊ (0,1) such that
where j′ is arbitrarily chosen from . If , set , update the current active set , where j* denotes the corresponding column index of X associated with the predictor xj*, and add xj* to the working design matrix ; else, set α = 1.
Let .
Let .
Set i = i + 1, and iterate steps (4)–(8) until α = 1, when all predictors have been added to the model.
For step (5), for ease of computation, we select j′ to always be the first value of the active set . Thus, for each iteration, xj′ corresponds to the first column of the design matrix . After p iterations, the ordinary least squares (OLS) solution is reached.25
Lasso
The lasso, which stands for least absolute shrinkage and selection operator,27 is a shrinkage method that is good for dealing with high-dimensional data and correlated covariates by placing a constraint on the magnitude of the regression coefficients.23,24 Hastie et al.24 define the lasso estimate as follows: , such that , where t is a tuning parameter that determines the shrinkage extent. Efron et al.28 discover that the LARS algorithm can be modified to obtain the lasso solutions. As with LARS, the lasso adds variables to the active set. However, unlike LARS, the lasso does not permit active nonzero coefficients to cross zero, and in cases where a nonzero coefficient reaches zero, that variable is dropped from the active set.24
Following the notation of Efron et al.28, Shi29 describes the lasso algorithm as follows:
Initialize all regression coefficients , and let r[0] = y, where r[0] denotes the residual vector at index 0 and y denotes the response vector. Set i = 1, where i is the index for the current iteration count.
Find the predictor xj among the p possible predictors that has the greatest absolute correlation with the residuals r[i−1], and let the active set be equal to the corresponding column index of X associated with the predictor xj.
Add xj to the working design matrix such that , where sj = sign{corr(xj,r[i−1])} = ±1.
- Compute the following:
- ĉ = corr(X,r[r−1]),
- Ĉ =max{|ĉ|},
- , where is the length of and , and
- , where and .
Find , where “min+” specifies that, for every j not in the active set , the minimum is found over only the positive elements, and ĵ denotes the corresponding column index of X associated with the predictor xĵ.
- Let be a p-dimensional vector where all values are set equal to 0. For every , update by calculating
Find , where denotes the corresponding column index of X associated with the predictor . If ,.let , update , and update the working design matrix ; else, let , update , and update the working design matrix .
Let .
Set i = i + 1, and iterate steps (4)–(8) until all predictors have been added to the model and .
Notice that step (7) is the lasso modification to the LARS algorithm. At the final iteration, the OLS solution is reached.28
Modifications of Model Selection Approaches to Select Spatial Scale
Spatial scale forward stepwise regression
We propose a modified forward stepwise regression algorithm that selects each area-level variable at only one spatial scale in order to build regression models to explain variation in a continuous outcome variable. We use the basic forward stepwise algorithm with adjustments to select the scale for variables available at more than one spatial level. In the algorithm, all variables are considered at all available spatial scales as potential candidates to enter a model. However, due to potentially high correlations present across different scales for a given variable, we constrain the algorithm to select each area-level variable at a single spatial scale.
Our modeling approach uses a 3-D or stacked matrix, where each stack represents a particular level of covariates, including spatial scale. As an example, we might have several individual-level covariates, an area-level covariate available at 1, 2, and 3 mi, and another area-level covariate available at 4 and 6 mi. In this case, the first stack would contain the individual-level variables; the second, third, and fourth stacks would contain the area-level variable at the 1-, 2-, and 3-mi levels; and the fifth and sixth stacks would contain the area-level variable at the 4- and 6-mi levels, respectively. In cases where values are only present for a covariate at certain levels, that covariate is assigned missing values at all other levels. The spatial scale forward stepwise regression algorithm is as follows:
Construct an n × p × S stacked matrix X, where n = the number of observations, p = the number of variables, and S = the number of levels of covariates.
Initialize all regression coefficients , and let r = y, where r denotes the residual vector and y denotes the response vector.
For each stack s, s = 1,…,S: Of the candidate variables, find the predictor xjs that has the greatest absolute correlation with the residuals r.
Of the s correlations, select the predictor xjs that has the maximum correlation, and add that predictor to the working design matrix Xin.
Let .
Compute the residuals r = y – ŷ, where ŷ is the vector of fitted values.
Iterate steps (3)–(6) until there is an inadequate improvement in the performance of the model or until all predictors have been added to the model.
For step (4), it is important to note that, once an overall maximum is determined, we select the corresponding variable at the best spatial scale and remove all other versions (or spatial scales) of that variable from further consideration for model selection. In this way, we constrain the algorithm to select each area-based variable at a single spatial scale. For step (7), we consider there to be an inadequate improvement in the model’s performance if the difference in the AIC between the current model and the proposed model is less than ε, for some ε > 0.
Spatial scale incremental forward stagewise regression
We spatially modify the basic forward stagewise algorithm to select each area-level variable at a single spatial scale. We use the same matrix data structure as with the spatial scale forward stepwise algorithm. The spatial scale incremental forward stagewise regression algorithm is as follows:
Construct an n × p × S stacked matrix X, where n = the number of observations, p = the number of variables, and S = the number of levels of covariates.
Initialize all regression coefficients , and let r = y, where r denotes the residual vector and y denotes the response vector.
For each stack s, s = 1,…,S: Find the predictor xjs that has the greatest absolute correlation with the residuals r.
Of the s correlations, select the predictor xjs that has the maximum correlation.
Let , where δj = τ·sign[corr(r,xjs)] for some step size τ > 0.
Let r ← r − δjxjs.
Iterate steps (3)–(6) until none of the predictors are correlated with the residuals r.
For step (4), once an overall maximum is determined, we select the corresponding variable at the best spatial scale and remove all other versions (or spatial scales) of that variable from further consideration. In this way, we constrain the algorithm to select each area-level variable at a single spatial scale. For step (7), we state that none of the predictors are correlated with the residuals if the overall maximum is less than a specified tolerance, where the tolerance is some small positive number.
Spatial scale least angle regression
Our approach for the spatial modification of the LARS algorithm involves the use of a block diagonal matrix , where each aj,,for j = 1,…, p, is an sj × sj identity matrix such that each column indicates a particular level or spatial scale, where p is the total number of predictor variables disregarding spatial scale and sj is the number of levels of covariates for the jth variable. Acand is a square matrix with rows. Every time a variable enters the active set , Acand is updated by removing all other versions (or spatial scales) of the winning variable. When X is post-multiplied by Acand, we get the candidate design matrix Xcand, which keeps track of variables in the active set as well as candidate variables. Through this updating of Xcand, we modify the basic LARS algorithm to select each area-level variable at a single spatial scale.
Adopting the notation of Yuan and Lin,26 the spatial scale LARS algorithm is as follows:
Initialize all regression coefficients , and let r[0] = y, where r[0] denotes the residual vector at index 0 and y denotes the response vector. Let Acand be a preprocessed p* × p* block diagonal matrix. Set i = 1, where i is the index for the current iteration count.
Find the predictor xc among the p* possible predictors that has the greatest absolute correlation with the residuals r[i−1].
Let the active set be equal to the corresponding column index of X associated with the predictor xc, and add xc to the working design matrix .
Update Acand by removing all other versions of the winning predictor variable xc, and update the candidate design matrix Xcand = X · Acand.
- Let γ be a p*-dimensional vector, where all values are set equal to 0. Calculate the current least squares direction γ by updating
. - For every j in Xcand that is not an element of the active set calculate αj, which is the minimum distance needed to move the active regression coefficient(s) in direction γ until another predictor xj has as much correlation with the current residuals as the variables in the active set. That is, find αj ∊ (0,1) such that
where j′ is arbitrarily chosen from If , set , update the current active set , where j* denotes the corresponding column index of X associated with the predictor xj*, add xj* to the working design matrix , update Acand, and update the candidate design matrix Xcand = X · Acand; else, set α = 1.
Let .
Let .
Set i = i + 1, and iterate steps (5)–(9) until α = 1, when all predictors have been added to the model.
For steps (4) and (7), once a predictor variable is chosen to enter the model, we select that variable at the best spatial scale and remove all other versions (or spatial scales) of that variable from further consideration from the candidate design matrix Xcand. In this way, we constrain the algorithm to select each area-level variable at a single spatial scale. For step (6), for ease of computation, we select j′ to always be the first value of the active set Thus, for each iteration, xj′ corresponds to the first column of the design matrix . After p iterations, the OLS solution is reached.25
Spatial scale lasso
As with the spatial modification of LARS, our modeling approach for the spatial modification of the lasso algorithm involves the use of a p* × p* block diagonal matrix , where each aj, for j = 1,…,p, is an sj × sj identity matrix such that each column indicates a particular level or spatial scale, where p is the total number of predictor variables disregarding spatial scale, sj is the number of levels of covariates for the jth variable, and p* is the total number of predictor variables at all available levels such that . As with the spatially modified LARS method, every time a variable enters the active set , Acand is updated by removing all other versions (or spatial scales) of the winning variable. When X is post-multiplied by Acand, we get the candidate design matrix Xcand, which keeps track of variables in the active set as well as candidate variables. Through this updating of Xcand, we modify the basic lasso algorithm to select each area-level variable at a single spatial scale.
Adopting the notation of Efron et al.28, the spatial scale lasso algorithm is as follows:
Initialize all regression coefficients , and let r[0] = y, where r[0] denotes the residual vector at index 0 and y denotes the response vector. Let Acand be a preprocessed p* × p* block diagonal matrix. Set i = 1, where i is the index for the current iteration count.
Find the predictor xj among the p* possible predictors that has the greatest absolute correlation with the residuals r[i−1], and let the active set be equal to the corresponding column index of X associated with the predictor xj.
Add xj to the working design matrix such that , where sj =sign{corr(xj,r[i−1])} = ±1.
Update Acand by removing all other versions of the winning predictor variable xj, and update the candidate design matrix Xcand = X · Acand.
- Compute the following:
- ĉ = corr(Xcand,r[r−1]),
- Ĉ =max{|ĉ|},
- , where is the length of and , and
- , where and .
Find , where “min+” specifies that, for every j not in the active set the minimum is found over only the positive elements, and ĵ denotes the corresponding column index of X associated with the predictor xĵ.
- Let be a p*-dimensional vector where all values are set equal to 0. For every , update by calculating
Find , where denotes the corresponding column index of X associated with the predictor . If ,.let , update , and update the working design matrix ; else, let , update , update the working design matrix , update Acand, and update the candidate design matrix Xcand = X · Acand.
Let .
Set i = i + 1, and iterate steps (5)–(9) until all predictors have been added to the model and .
Step (8) is the lasso modification. For steps (4) and (8), once a predictor variable is chosen to enter the model, we select that variable at the best spatial scale and remove all other versions (or spatial scales) of that variable from further consideration from the candidate design matrix Xcand. In this way, we constrain the algorithm to select each area-level variable at a single spatial scale. At the final iteration, the OLS solution is reached.28
Application to Groundwater Nitrate
Study data
To model the variation in nitrate in drinking well water in Iowa, we used data for private wells sampled from 1984 to 2011 by the following programs: the Iowa Grants to Counties Water Well Program (GTC), the Iowa Private Well Tracking System, the Iowa Statewide Rural Well Water Survey, the Iowa Community Private Well Study, and the U.S. Geological Survey (USGS). We used only those wells with the most accurate locations as determined by GPS measurements, topographic quad maps, and geocoded residence addresses. Seventy-five percent of the well locations were based on geocoded residential street addresses. Nitrate data were reported either as nitrate or nitrite-plus-nitrate as NO3−, and the latter were converted to nitrate-nitrogen (hereafter referred to as “nitrate”). Values below the detection limit were imputed from a log-normal distribution of uncensored data.30 Same-day samples at the same well location and depth were excluded if their standard deviation was 5 mg/L nitrate-N or more; otherwise the average of such samples was used. Nitrate data were natural-log-transformed prior to modeling. There were 11,931 well measurements in the analysis dataset.
We considered a set of 115 explanatory variables in the statistical analysis (Table 1). Variables were available for characteristics at the individual well location and for characteristics of the surrounding environment over different distance buffers. Variables at the individual well level include longitude, latitude, elevation, well depth, bedrock status, and bedrock depth, among others. A geographic information system was used to calculate the surrounding environmental variables. Most of the environmental variables were calculated using more than one distance buffer to assess the importance of spatial scale. An exception was for counts of animal feeding operations (AFOs) by type (Confined, Open Feedlot, Mixed), which were only calculated at 10 km. The AFO type of the closest AFO was also recorded, along with the number of animals at the nearest AFO (NearAFO_AnimalUnits). Most of the other area-based variables were calculated at distances of 500 m and 1 km. These area-based covariates include average percent sand, average percent clay, average slope length, and mean population density, among others. Only fine-grain thickness (FnGrn_Logs) was calculated at 4- and 6-mi distances. Additional details on the variables are available in Wheeler et al.30 To account for missing data, we excluded 9.3% of the observations that were missing values for any of the covariates and used 10,824 nitrate measurements in our analysis.
Table 1.
Variable definitions for the variables considered in the spatial scale forward stepwise, forward stagewise, LARS, and lasso models. The horizontal dashed line separates the individual-level variables and the area-based variables available at more than one buffer distance. Any variable that falls below the dashed line has a suffix indicating the associated spatial scale.
| VARIABLE NO. | NAME | DESCRIPTION |
|---|---|---|
| 1 | Latitude | Latitude value of well location (degrees) |
| 2 | Longitude | Longitude value of well location (degrees) |
| 3 | SampleYr | Well sample year |
| 4 | Well_Depth | Depth of measurement well (ft) |
| 5 | Elevation | Land-surface elevation at well point (ft) |
| 6 | Bdrk_Dpth | Depth (ft) to bedrock at well point |
| 7 | Bdrk_Flag | Flag indicating if well is within or above bedrock. 0 = Above bedrock; 1 = Within bedrock |
| 8 | NearAFO_Dist | Distance to nearest AFO (Animal Feeding Operation) facility (m) |
| 9 | NearAFO_Type_1 | Type of nearest AFO facility: Open Feedlot |
| 10 | NearAFO_Type_2 | Type of nearest AFO facility: Confined/Open (ie, mixed) |
| 11 | NearAFO_AnimalUnits | Total Animal Units at the nearest AFO facility |
| 12 | Count_10 kmConfmnts | Number of confinement-only AFOs within 10 km of the well point |
| 13 | Count_10 kmFeedlots | Number of feedlot-only AFOs within 10 km of the well point |
| 14 | Count_10 kmMixed | Number of mixed-only AFOs within 10 km of the well point |
| 15 | Count_10 kmHogs | Number of hog facilities within 10 km of the well point |
| 16 | precip | Estimated mean annual precipitation at well point for the time period 1981–2010 (millimeters times 100) |
| 17 | mintemp | Estimated mean annual minimum temperature at well point for the time period 1981–2010 (°C times 100) |
| 18 | maxtemp | Estimated mean annual maximum temperature at well point for the time period 1981–2010 (°C times 100) |
| 19 | SinkholeDist_m | Distance from well point to nearest sinkhole point (m) |
| 20 | K | Average horizontal hydraulic conductivity of all glacial deposits at well point (ft/day) |
| 21 | AvgK | Average horizontal hydraulic conductivity of all glacial deposits within a 4 × 4-mile square around the well point (ft/day) |
| 22 | Kz | Average vertical hydraulic conductivity of all glacial deposits at the well point (ft/day) |
| 23 | AvgKz | Average vertical hydraulic conductivity of all glacial deposits within a 4 × 4-mile square around the well point (ft/day) |
| 24 | Trans | Transmissivity of all glacial deposits at the well point (ft2/day) |
| 25 | AvgTrans | Average transmissivity of all glacial deposits within a 4 × 4-mile square around the well point (ft2/day) |
| 26 | MaxKz | Maximum kz within the 4 × 4-mile square around the well point (ft/day) |
| 27 | KKzT_Logs | Number of USGS water well logs within a 4 × 4-mile square around the well point (count) |
| 28–29 | Sand | Average percent sand within a 500-m/1-km buffer |
| 30–31 | Silt | Average percent silt within a 500-m/1-km buffer |
| 32–33 | Clay | Average percent clay within a 500-m/1-km buffer |
| 34–35 | OM | Average percent organic matter within a 500-m/1-km buffer |
| 36–37 | Db033 | Average bulk density at 1/3 bar within a 500-m/1-km buffer (g/cm3) |
| 38–39 | Dbovendry | Average oven dry bulk density at 1/3 bar within a 500-m/1-km buffer (g/cm3) |
| 40–41 | Ksat | Average saturated hydraulic conductivity within a 500-m/1-km buffer (μm/s) |
| 42–43 | AWC | Average available water capacity within a 500-m/1-km buffer (cm H2O/cm soil) |
| 44–45 | H2O15 | Average water content at 15 bar within a 500-m/1-km buffer (percent by weight) |
| 46–47 | AASHTOGr | Average AASHTO group classification within a 500-m/1-km buffer |
| 48–49 | Kw | Average K factor for whole soil within a 500-m/1-km buffer |
| 50–51 | Kf | Average K factor for rock free soil within a 500-m/1-km buffer |
| 52–53 | CaCO3 | Average calcium carbonate within a 500-m/1-km buffer (percent by weight) |
| 54–55 | CEC7 | Average cation-exchange capacity within a 500-m/1-km buffer (milliequivalents per 100 g) |
| 56–57 | pHH2O | Average pH (1 to 1 water) within a 500-m/1-km buffer |
| 58–59 | Slope | Average percent slope within a 500-m/1-km buffer |
| 60–61 | SlopeLength | Average slope length within a 500-m/1-km buffer (ft) |
| 62–63 | Runoff | Average runoff potential within a 500-m/1-km buffer (Scale: 1–6; negligible to very high) |
| 64–65 | T | Average soil loss tolerance within a 500-m/1-km buffer (tons/acre/year) |
| 66–67 | WEI | Average wind erodibility index within a 500-m/1-km buffer |
| 68–69 | Aspect | Average aspect (direction the surface of the soil faces) within a 500-m/1-km buffer (degrees) |
| 70–71 | MAP | Average mean annual precipitation within a 500-m/1-km buffer (mm) |
| 72–73 | FrostFDays | Average number of frost free days per year within a 500-m/1-km buffer |
| 74–75 | FrostAction | Average degree of frost action within a 500-m/1-km buffer (Scale: 0–3; none to high) |
| 76–77 | CorrosionCon | Average risk of concrete corrosion within a 500-m/1-km buffer (Scale: 1–3; low to high) |
| 78–79 | CorrosionSt | Average risk of steel corrosion within a 500-m/1-km buffer (Scale: 1–3; low to high) |
| 80–81 | IACSR | Average Iowa corn suitability rating within a 500-m/1-km buffer (Scale: 0–100) |
| 82–83 | WaterDepth | Average depth to water within a 500-m/1-km buffer (cm) |
| 84–85 | FloodingFreq | Average flooding frequency within a 500-m/1-km buffer (Scale: 0–4, none to very frequent) |
| 86–87 | PondingFreq | Average ponding frequency within a 500-m/1-km buffer (%) |
| 88–89 | DrainClass | Average drainage classification within a 500-m/1-km buffer (Scale: 1–7, very poorly drained to excessively drained) |
| 90–91 | FarmClass | Percent “not prime farmland” within a 500-m/1-km buffer |
| 92–93 | HELWater | Percent “not highly water erodable land” within a 500-m/1-km buffer |
| 94–95 | HELWind | Percent “not highly wind erodable land” within a 500-m/1-km buffer |
| 96–97 | Basements | Percent “very limited and somewhat limited” basement limitations within a 500-m/1-km buffer |
| 98–99 | SewageLag | Percent “very limited and somewhat limited” sewage lagoon limitations within a 500-m/1-km buffer |
| 100–101 | Trails | Percent “very limited and somewhat limited” path and trail limitations within a 500-m/1-km buffer |
| 102–103 | HydricClas | Percent “all hydric and partially hydric” hydric classifications within a 500-m/1-km buffer |
| 104–105 | TileDrn_USGS | Mean “estimated percent tile drainage on agricultural lands” within a 500-m/1-km buffer |
| 106–107 | TileDrn_IADNR | Mean “estimated percent tile drainage” within a 500-m/1-km buffer |
| 108–109 | PopDen90 | Mean population density within a 500-m/1-km buffer derived from U.S. Census 1990 (persons per km2) |
| 110–111 | PopDen00 | Mean population density within a 500-m/1-km buffer derived from U.S. Census 2000 (persons per km2) |
| 112–113 | Recharge | Estimated mean annual natural ground-water recharge within a 500-m/1-km buffer (millimeters per year) |
| 114–115 | FnGrn_Logs | Number of well logs within a 4 × 4-mile/6 × 6-mile square around the well point used to generate an interpolated total fine-grain thickness grid |
Statistical analysis
We modeled the natural log of nitrate concentrations in well water using our four spatial scale selection algorithms. We built spatial scale forward stepwise regression, spatial scale incremental forward stagewise regression, spatial scale LARS, and spatial scale lasso models to explain variation in log nitrate concentration while allowing any individual-level variable to enter the model and any area-based variable to enter the model at a single spatial scale, considering all available spatial scales. For spatial scale forward stepwise and spatial scale stagewise, we used a stacked data matrix where the first stack contained individual-level variables or area-level variables available at only one spatial scale, the second and third stacks contained area-level variables at the 500-m and 1-km levels, and the fourth and fifth stacks contained area-level variables at the 4- and 6-mi levels, respectively. Given that variables available at multiple spatial scales were limited to enter a model at a single spatial scale, the total number of variables possible for model inclusion was 71 instead of 115. More specifically, the total number of individual-level variables and area-level variables available at one spatial scale possible for model inclusion was 27, and the total number of area-level variables possible was 44.
For spatial scale forward stagewise, spatial scale LARS, and spatial scale lasso, we fitted OLS regression models with the selected covariates to obtain approximate p-values and AIC measures. For ease of computation, approximate significance levels were determined when the covariates selected from the spatial scale stagewise, LARS, and lasso algorithms were plugged into OLS regression models to obtain standard error estimates. Because LARS and lasso yield a sequence of solutions, for each algorithm we selected as the final model the one that had the minimum OLS-based AIC. The outcome and predictor variables were standardized to have a mean of 0 and a standard deviation of 1. We used a significance level of α = 0.05. For the spatial scale forward stepwise algorithm, we set ε = 1 because the rule of thumb for a meaningful difference in AIC is 2 to 3.31 Decreasing the value of ε leads to larger models and better goodness of fit. For the spatial scale forward stagewise algorithm, we used a commonly accepted increment or step size of 0.00122 and set the tolerance = 0.01. Increasing the step size leads to larger coefficient estimates and a decreased number of algorithm iterations, and increasing the tolerance leads to models with fewer selected covariates and decreased goodness of fit. All analyses were performed using R version 3.1.0.32
Evaluation metrics
To evaluate the success of our spatial scale algorithms, we examined our methods using three criteria. First, for each of the four algorithms, we checked to see whether different spatial scales were selected and enumerated the number of selected variables that fell into each spatial scale category. Second, we looked at the agreement in sign and spatial scale for significant variables that were selected across various groupings of the algorithms. Third, in order to evaluate the rationale of including different variables at different spatial scales within the same model, for each algorithm we compared AIC measures across three different scenarios: 1) when limiting all selected area-based variables to be at the smallest available spatial scale, 2) when limiting all selected area-based variables to be at the largest available spatial scale, and 3) when using all selected area-based variables at the spatial scales originally selected by the model.
Results
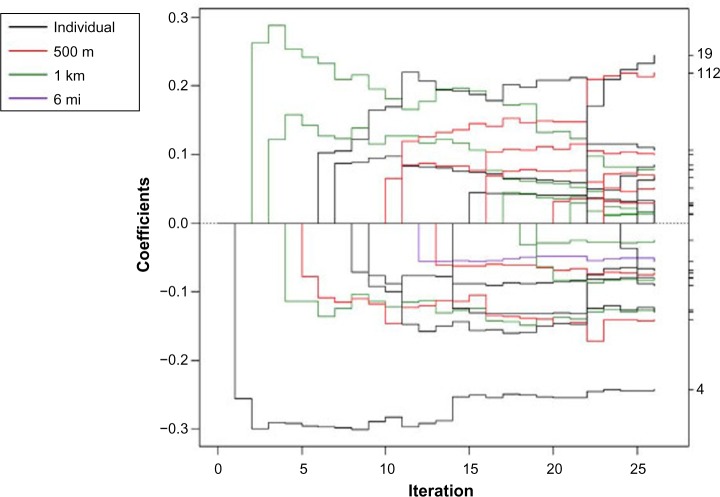
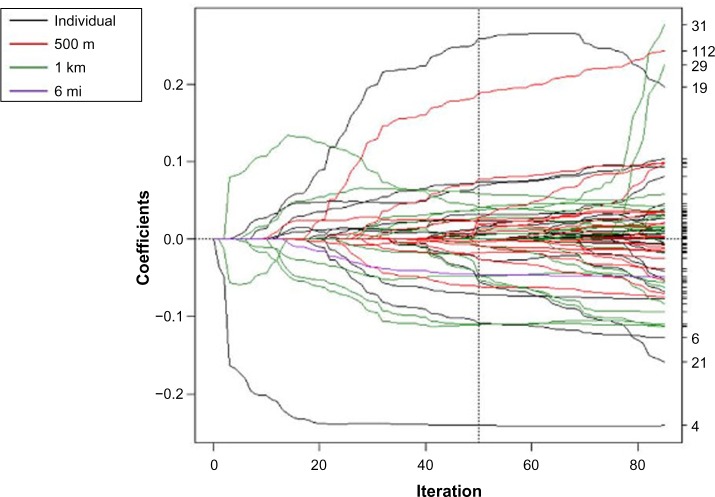
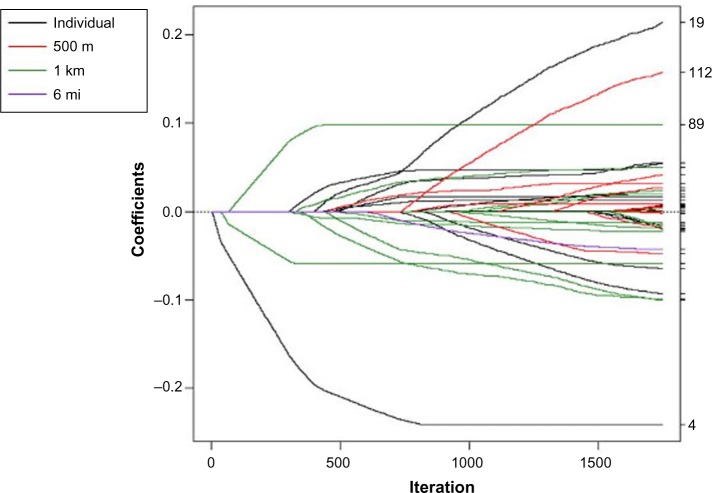
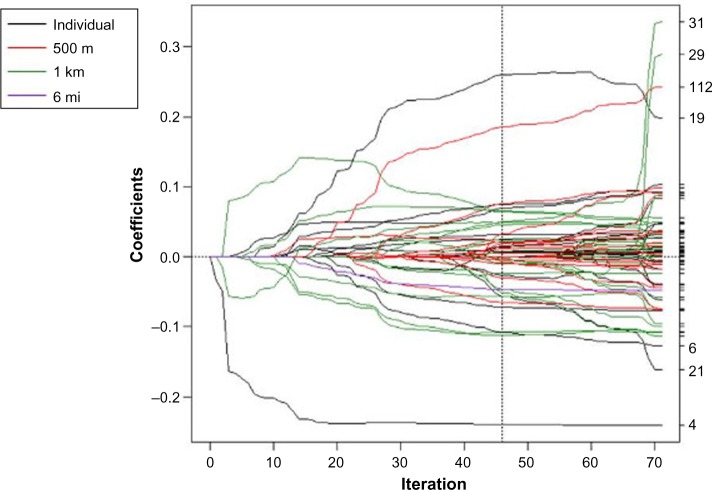
Different variables were selected at different spatial scales using the spatial scale forward stepwise, spatial scale incremental forward stagewise, spatial scale LARS, and spatial scale lasso algorithms (Figs. 1–4). In each coefficient path plot, iterations of the respective algorithm are shown as the model-building progresses, where the coefficient estimates at each iteration change as variables enter or leave a model. Black lines represent individual-level variables, red lines indicate area-based variables at the 500-m level, green lines denote area-based variables at the 1-km level, and purple lines represent area-based variables at the 6-mi level. The forward stepwise algorithm converged after 26 iterations (Fig. 1), and the forward stagewise algorithm converged after 1,747 iterations (Fig. 2). Not surprisingly, it took a large number of iterations before the stagewise algorithm converged because of the incremental updating of the beta coefficient estimates. The LARS algorithm converged to the OLS estimates after 71 iterations (Fig. 3). The dotted vertical line in Figure 3 indicates the chosen model that had the minimum OLS-based AIC. The lasso algorithm converged to the OLS estimates after 85 iterations (Fig. 4). It took more iterations for the lasso algorithm to converge than for the LARS due to lasso’s ability to add and drop variables. The dotted vertical line in Figure 4 indicates the chosen model that had the minimum OLS-based AIC.
Figure 1.
Coefficient paths for spatial scale forward stepwise regression to explain log nitrate concentration in drinking wells in Iowa. The scale the variable entered the model is indicated by the legend.
Figure 4.
Coefficient paths for spatial scale lasso to explain log nitrate concentration in drinking wells in Iowa. The scale the variable entered the model is indicated by the legend. The dotted vertical line indicates the chosen model that had the minimum OLS-based AIC.
Figure 2.
Coefficient paths for spatial scale incremental forward stagewise regression to explain log nitrate concentration in drinking wells in Iowa. The scale the variable entered the model is indicated by the legend.
Figure 3.
Coefficient paths for spatial scale LARS to explain log nitrate concentration in drinking wells in Iowa. The scale the variable entered the model is indicated by the legend. The dotted vertical line indicates the chosen model that had the minimum OLS-based AIC.
The coefficient estimates for each of the covariates selected in each of the algorithms are shown in Table 2, where the horizontal dashed line separates the individual-level variables and the area-based variables that have multiple spatial scales. Across all four algorithms, there were significant positive associations between log nitrate concentration and the following covariates: elevation, number of mixed-only AFOs within a 10-km buffer (Count_10 kmMixed), number of hog facilities within a 10-km buffer (Count_10 kmHogs), distance from well point to nearest sinkhole point (SinkholeDist_m), average transmissivity (AvgTrans), average wind erodibility index within a 500-m buffer (WEI_500 m), and estimated mean annual natural ground-water recharge within a 500-m buffer (Recharge_500 m). There were significant negative associations between log nitrate concentration and the following covariates: latitude, well depth, bedrock depth, bedrock status, average horizontal hydraulic conductivity (AvgK), average soil loss tolerance within a 1-km buffer (T_1 km), percent “not prime farmland” within a 500-m buffer (FarmClass_500 m), mean population density within a 1-km buffer from the U.S. Census 2000 (PopDen00_1 km), and fine-grain thickness at the 6-mi distance (FnGrn_Logs_6 mi).
Table 2.
Estimated coefficients from spatial scale (SS) forward stepwise, forward stagewise, LARS, and lasso models. The blank cells indicate variables not selected for a particular model. The horizontal dashed line separates the individual-level variables and the area-based variables considered at multiple spatial scales.
| VARIABLE NO. | EXPLANATORY VARIABLE | SS-STEPWISE | SS-STAGEWISE | SS-LARS | SS-LASSO |
|---|---|---|---|---|---|
| 1 | Latitude | −0.069 (*) | −0.015 (*) | −0.027 (*) | −0.026 (*) |
| 4 | Well_Depth | −0.243 (*) | −0.242 (*) | −0.240 (*) | −0.241 (*) |
| 5 | Elevation | 0.107 (*) | 0.055 (*) | 0.075 (*) | 0.074 (*) |
| 6 | Bdrk_Dpth | −0.129 (*) | −0.093 (*) | −0.109 (*) | −0.108 (*) |
| 7 | Bdrk_Flag | −0.080 (*) | −0.065 (*) | −0.072 (*) | −0.072 (*) |
| 11 | NearAFO_AnimalUnits | 0.003 | 0.006 | 0.006 | |
| 12 | Count_10 kmConfmnts | 0.001 | |||
| 13 | Count_10 kmFeedlots | 0.013 | 0.008 | 0.007 | |
| 14 | Count_10 kmMixed | 0.026 (*) | 0.020 (*) | 0.024 (*) | 0.024 (*) |
| 15 | Count_10 kmHogs | 0.026 (*) | 0.006 (*) | 0.014 (*) | 0.016 (*) |
| 19 | SinkholeDist_m | 0.244 (*) | 0.214 (*) | 0.260 (*) | 0.259 (*) |
| 21 | AvgK | −0.091 (*) | −0.021 (*) | −0.056 (*) | −0.056 (*) |
| 22 | Kz | 0.067 (*) | 0.008 | 0.032 | 0.032 |
| 23 | AvgKz | 0.017 (+) | 0.016 (*) | 0.015 (*) | |
| 25 | AvgTrans | 0.084 (*) | 0.055 (*) | 0.069 (*) | 0.070 (*) |
| 31 | Silt_1 km | 0.050 (*) | 0.038 (+) | ||
| 32 | Clay_500 m | 0.013 (+) | 0.012 (+) | ||
| 35 | OM_1 km | −0.079 (*) | −0.023 (+) | −0.057 (*) | −0.047 (*) |
| 39 | Dbovendry_1 km | 0.024 (*) | 0.025 (*) | 0.028 (*) | |
| 41 | Ksat_1 km | 0.020 (*) | 0.046 (*) | 0.041 (*) | |
| 42 | AWC_500 m | 0.019 (*) | 0.018 (*) | ||
| 46 | AASHTOGr_500 m | 0.032 | 0.020 | 0.020 | |
| 47 | AASHTOGr_1 km | 0.012 | |||
| 49 | Kw_1 km | −0.004 | |||
| 52 | CaCO3_500 m | −0.141 (*) | |||
| 53 | CaCO3_1 km | −0.099 (*) | −0.108 (*) | −0.110 (*) | |
| 55 | CEC7_1 km | −0.017 (*) | −0.047 (*) | −0.053 (*) | |
| 56 | pHH2O_500 m | −0.002 | −0.030 (*) | −0.026 (*) | |
| 58 | Slope_500 m | −0.019 (*) | −0.007 (+) | −0.009 (+) | |
| 60 | SlopeLength_500 m | 0.009 | −0.004 | ||
| 63 | Runoff_1 km | 0.021 | 0.011 | ||
| 65 | T_1 km | −0.126 (*) | −0.100 (*) | −0.112 (*) | −0.110 (*) |
| 66 | WEI_500 m | 0.100 (*) | 0.042 (*) | 0.076 (*) | 0.077 (*) |
| 71 | MAP_1 km | −0.009 | −0.009 (+) | ||
| 74 | FrostAction_500 m | 0.052 (*) | 0.027 (*) | 0.020 | 0.025 |
| 76 | CorrosionCon_500 m | 0.066 (*) | |||
| 77 | CorrosionCon_1 km | 0.050 (*) | 0.064 (*) | 0.058 (*) | |
| 78 | CorrosionSt_500 m | 0.007 | |||
| 81 | IACSR_1 km | −0.003 | |||
| 82 | WaterDepth_500 m | 0.005 | −0.003 | ||
| 83 | WaterDepth_1 km | −0.013 | |||
| 85 | FloodingFreq_1 km | 0.013 | |||
| 86 | PondingFreq_500 m | 0.029 (*) | 0.005 (+) | 0.011 | 0.009 |
| 89 | DrainClass_1 km | 0.078 (*) | 0.098 (*) | 0.066 (+) | 0.041 |
| 90 | FarmClass_500 m | −0.073 (*) | −0.048 (*) | −0.065 (*) | −0.061 (*) |
| 94 | HELWind_500 m | −0.001 | −0.010 | −0.009 (+) | |
| 97 | Basements_1 km | 0.003 | |||
| 99 | SewageLag_1 km | 0.012 | 0.007 (+) | 0.005 | |
| 100 | Trails_500 m | −0.017 | |||
| 101 | Trails_1 km | −0.024 | |||
| 107 | TileDrn_IADNR_1 km | −0.059 | 0.049 (+) | ||
| 108 | PopDen90_500 m | 0.033 (*) | 0.029 (*) | ||
| 111 | PopDen00_1 km | −0.026 (*) | −0.020 (*) | −0.051 (*) | −0.047 (*) |
| 112 | Recharge_500 m | 0.219 (*) | 0.158 (*) | 0.186 (*) | 0.189 (*) |
| 115 | FnGrn_Logs_6 mi | −0.055 (*) | −0.043 (*) | −0.047 (*) | −0.046 (*) |
Notes: Values marked with (*) have a P-value <0.05, and values marked with (+) have an associated P-value <0.1 (when covariates selected from the SS-Stagewise, SS-LARS, and SS-Lasso algorithms are plugged into OLS regression models).
Multiple spatial scales of 500 m and 1 km were selected for variables by each of the algorithms (Table 3). All four models selected fine-grain thickness (FnGrn_Logs) to enter at the 6-mi level. For the spatial scale forward stepwise model, 26 of the 71 individual- and area-level covariates were selected. With seven variables selected at the 500-m level and seven variables selected at the 1-km level, there was an even split between the number of variables selected at the 500-m level versus the 1-km level. For the spatial scale forward stagewise model, 39 of the 71 individual- and area-level covariates were selected. Again, there was a fairly even split with 11 variables selected at the 500-m level and 12 variables selected at the 1-km level. For the spatial scale LARS model, 46 of the 71 individual- and area-level covariates were selected. More variables were chosen to enter at the 1-km level than the 500-m level, with 14 variables selected for the latter and 17 variables for the former. For the spatial scale lasso model, 42 of the 71 individual- and area-level covariates were selected. There was a fairly even split, with 14 variables selected at the 500-m level and 13 variables selected at the 1-km level.
Table 3.
Number of variables selected at each spatial scale for spatial scale (SS) forward stepwise, forward stagewise, LARS, and lasso models. The last row gives the total number of possible variables at each spatial scale.
| INDIVIDUAL-LEVEL | AREA-LEVEL | NUMBER OF VARIABLES SELECTED | ||||||
|---|---|---|---|---|---|---|---|---|
| 500 m | 1 km | 4 mi | 6 mi | |||||
| SS-Stepwise | 11 | 7 | 7 | 0 | 1 | 26 | ||
| SS-Stagewise | 15 | 11 | 12 | 0 | 1 | 39 | ||
| SS-LARS | 14 | 14 | 17 | 0 | 1 | 46 | ||
| SS-Lasso | 14 | 14 | 13 | 0 | 1 | 42 | ||
| Number of available variables | 27 | 43 | 1 | 71 | ||||
Overall, there was consistency across the spatial scale algorithms in terms of the coefficient signs and spatial scale for the significant selected variables (Table 4). For various groupings of the algorithms, it is evident that, of the commonly selected covariates, the majority of them were significant. There were no instances of significant variables having different signs across algorithms, and only two instances of significant variables being selected at different spatial scales across algorithms. Average calcium carbonate (CaCO3) and average risk of concrete corrosion (CorrosionCon) were selected at the 500-m level by spatial scale stepwise and at the 1-km level by spatial scale stagewise, spatial scale LARS, and spatial scale lasso.
Table 4.
Number of shared significant variables with the same sign and spatial scale and total number of shared variables for spatial scale (SS) forward stepwise, forward stagewise, LARS, and lasso models. The frequency of shared significant variables with the same sign and spatial scale is given along with the total number of shared variables in parentheses.
| INDIVIDUAL-LEVEL | AREA-LEVEL | ||||
|---|---|---|---|---|---|
| 500 m | 1 km | 4 mi | 6 mi | ||
| SS-Stepwise, SS-Stagewise, SS-LARS, SS-Lasso | 10 | 3 | 2 | 0 | 1 |
| No. of shared variables | (11) | (5) | (4) | (0) | (1) |
| SS-Stepwise, SS-Stagewise, SS-LARS | 10 | 3 | 2 | 0 | 1 |
| No. of shared variables | (11) | (5) | (4) | (0) | (1) |
| SS-Stepwise, SS-Stagewise, SS-Lasso | 10 | 3 | 2 | 0 | 1 |
| No. of shared variables | (11) | (5) | (4) | (0) | (1) |
| SS-Stepwise, SS-LARS, SS-Lasso | 10 | 3 | 3 | 0 | 1 |
| No. of shared variables | (11) | (5) | (5) | (0) | (1) |
| SS-Stagewise, SS-LARS, SS-Lasso | 10 | 3 | 7 | 0 | 1 |
| No. of shared variables | (14) | (9) | (9) | (0) | (1) |
| SS-Stepwise, SS-Stagewise | 10 | 4 | 3 | 0 | 1 |
| No. of shared variables | (11) | (5) | (4) | (0) | (1) |
| SS-Stepwise, SS-LARS | 10 | 3 | 3 | 0 | 1 |
| No. of shared variables | (11) | (5) | (5) | (0) | (1) |
| SS-Stepwise, SS-Lasso | 10 | 3 | 3 | 0 | 1 |
| No. of shared variables | (11) | (5) | (5) | (0) | (1) |
| SS-Stagewise, SS-LARS | 10 | 3 | 7 | 0 | 1 |
| No. of shared variables | (14) | (10) | (10) | (0) | (1) |
| SS-Stagewise, SS-Lasso | 10 | 3 | 7 | 0 | 1 |
| No. of shared variables | (14) | (9) | (9) | (0) | (1) |
| SS-LARS, SS-Lasso | 11 | 6 | 8 | 0 | 1 |
| No. of shared variables | (14) | (13) | (13) | (0) | (1) |
| Significant but with different signs | 0 | 0 | 0 | 0 | 0 |
| Significant but with different SS* | – | 2 | 0 | 0 | 0 |
Notes: Variables with a P-value <0.05 are considered significant (when covariates selected from the SS-Stagewise, SS-LARS, and SS-Lasso algorithms are plugged into OLS regression models).
In comparing SS-Stepwise with SS-Stagewise, SS-LARS, and SS-Lasso.
For spatial scale stepwise, spatial scale stagewise, spatial scale LARS, and spatial scale lasso, we fitted OLS regression models based upon the selected covariates to obtain AIC measures for the three different scenarios mentioned previously. The table of AIC measures is shown in Table 5. For all methods, the model using the model-selected spatial scales (Model 3) resulted in the smallest AIC, indicating a better goodness of fit. Thus, we saw a significant improvement in goodness of fit with the spatial scale models in which we used the area-based variables at the spatial scales originally selected by each model. Across all scenarios, the spatial scale lasso had the best goodness of fit.
Table 5.
OLS-based Akaike information criterion (AIC) comparisons across spatial scale (SS) forward stepwise, forward stagewise, LARS, and lasso models.
| SS-STEPWISE | SS-STAGEWISE | SS-LARS | SS-LASSO | |
|---|---|---|---|---|
| Model 1: Smallest SS available | 28,193.57 | 28,196.73 | 28,183.15 | 28,178.17 |
| Model 2: Largest SS available | 28,144.05 | 28,143.04 | 28,133.79 | 28,131.90 |
| Model 3: Model-selected SS | 28,130.90 | 28,135.15 | 28,100.19 | 28,096.65 |
Using the final model provided by the spatial scale lasso, 26 of the 42 selected variables were significant (Table 2). Of the significant variables, several variables had larger magnitudes and stood out as being important for explaining the variation in nitrate. There were significant positive associations between log nitrate concentration and the following covariates: distance from well point to nearest sinkhole point (SinkholeDist_m) and estimated mean annual natural ground-water recharge within a 500-m buffer (Recharge_500 m). In addition, there were significant negative associations between log nitrate concentration and the following covariates: bedrock depth, well depth, average calcium carbonate within a 1-km buffer (CaCO3_1 km), and average soil loss tolerance within a 1-km buffer (T_1 km).
Discussion and Conclusions
To consider the problem of spatial scale selection for area-based variables available at more than one spatial scale in a regression model, we modified the forward stepwise, forward stagewise, LARS, and lasso algorithms to select the best spatial scale for each area-level covariate. Our algorithms allow for any number of spatial scales of covariates to be considered and also enable the inclusion of individual-level covariates or covariates with only one possible spatial scale. We constrained the four algorithms to select each area-based variable to enter the model at a single spatial scale to avoid collinearity effects. When applying the algorithms to model groundwater nitrate exposure in Iowa, we found that not all environmental variables were selected at the same spatial scale. For all four spatial scale algorithms, the regression model that used the model-selected spatial scales had the best model fit. Furthermore, there was an overall agreement in coefficient sign and spatial scale for significant variables that were selected across the algorithms. The selection of area-level variables at different spatial units gives evidence for the environmental effects operating at different spatial scales and demonstrates the importance of considering the spatial scale when modeling environmental exposures.
Other researchers have developed approaches to address the problem of spatial scale selection in regression modeling. For example, rather than choosing the best available scale for each area-level covariate, Root et al.7 use the variance of the outcome variable (eg, disease rates) to select a buffer distance at which to conduct the regression analysis and then use the area-level variables at the selected buffer distance. They propose the Brown–Forsythe (FBF) test of homogeneity of variance to select the optimal neighborhood or buffer size for modeling disease rates. In their approach, Root et al.7 use circular buffers to create a collection of “neighborhoods” of different sizes around each subject and then use the FBF statistical test to select the ideal buffer distance. This approach assumes that small neighborhoods will have high variances (reflective of an individualistic data structure) and large neighborhoods will have low variances (reflective of a global data structure). The goal is to select an “optimal” neighborhood that adequately captures the global characteristics of the neighborhood environment in which a person lives without being so large as to lose applicability to the individual.7
Using the FBF test as a method to choose the optimal neighborhood has its merits. First, it is robust to deviations from the normal distribution in the outcome variables, which can occur when disease rates are modeled as normally distributed outcomes.7 Second, it allows researchers to more specifically define geographic areas that may be more relevant for a particular health outcome, as opposed to using predefined geopolitical spatial scales such as census block groups or counties, which may not adequately capture the proximal environment of an individual.7,33 The FBF approach for selecting spatial scale also has its limitations. First, it may not be suitable for researchers who wish to select neighborhoods other than those defined by using buffers.7 Second, the buffer-based estimates of neighborhood SES variables have measurement error (in addition to the measurement error present in the census data) by assuming that people are equally distributed within a census block group, but this is generally common to buffering approaches. Third, and most importantly, area-level variables are not involved in the selection of the optimal buffer size for calculating disease rates. The buffer distance is selected based on finding a spatial scale with a moderate variance for disease rates, and then the SES variables are modeled at the selected buffer size. Thus, the FBF test does not directly select the spatial scale for area-level covariates.
The strategies for the selection of spatial scale of environmental variables have been primarily univariate, at least in the context of the analysis of groundwater quality. Buffer shape sizes for land use variables have typically been selected independently from one another, as well as from variables measured at the well level.20,21 However, the magnitude of the effect measure and the significance of relationships between area-level variables and the outcome could change when other important variables are considered simultaneously.
Our methods provide a novel approach to the problem of spatial scale selection and have several strengths. First, rather than making an assumption about the appropriate spatial scale at which to model area-based variables, our spatial scale algorithms directly allow the data to drive the selection of spatial scale and permit different spatial scales to be present within a model. Second, our approach to spatial scale selection is multivariate and permits the simultaneous consideration of individual-level variables and area-level variables available at multiple spatial scales to be included in a model. Third, due to the potentially high correlations present across different spatial scales for a given variable, our algorithms constrain each variable to enter the model at a single spatial scale. That is, if a variable is available at two spatial scales, it can enter the model only at one of the two scales. Crowder and South34 permit a variable to enter a regression model at both available spatial scales, and their results show that these variables have opposite signs, suggesting the possibility of collinearity effects. Fourth, to address correlations present across variables, one of our algorithms constrains the regression coefficients in the presence of correlated covariates. Fifth, our methods are scalable and can be extended to accommodate high-dimensional datasets with a large number of covariates at a variety of different spatial scales.
While our initial results when applying our algorithms are encouraging, our analysis of groundwater nitrate has limitations. First, because of limited resources we were unable to consider more buffer distances in our analysis. Second, our analysis of nitrate used fixed buffer sizes for area-level variables across the study area, but adaptive buffer sizes based on population density may be more appropriate. Third, we excluded some observations due to missing values for some of the covariates. Regarding limitations of the algorithms, in the case of the spatial scale lasso, the spatial scale of a variable is fixed once it enters the model. That is, even if a variable is dropped, we constrain that variable to reenter the model at the same spatial scale as was originally selected. This is done to ensure that the correlation between the current residuals and the candidate variables does not exceed the maximum correlation achieved between the current residuals and the variables in the active set. Another limitation is the lack of standard errors for the algorithms, which necessitated our use of OLS models to obtain the P-values. The algorithms we present are for modeling a continuous outcome variable. We are currently working on developing versions of our spatial scale algorithms to model a binary outcome variable. In addition, we plan to run a simulation study to compare model performance across our four algorithms and also aim to evaluate a random effect at different spatial scales.
In the case study of groundwater nitrate exposure, we used our spatial scale algorithms to select area-based variables available at multiple buffer distances in order to explain variation in groundwater nitrate, a known risk factor for cancer. Our methods can be applied to other research problems, where it is of interest to select environmental or area-based risk factors available at multiple spatial scales that are associated with a health outcome of interest such as cancer.
Acknowledgments
We thank Bernard Nolan from the USGS, Abigail Flory from Westat, and Curt DellaValle and Mary Ward from the National Cancer Institute for contributing to the collection and processing of the Iowa nitrate data. We thank Pete Weyer and Jiji Kantamneni for providing nitrate measurement data and Randy Bayless and Les Arihood of the USGS for determining aquifer characteristics at sampled wells. We also thank Mike Giangrande from Westat for performing geocoding of the well locations.
Footnotes
ACADEMIC EDITOR: J.T. Efird, Editor in Chief
FUNDING: The authors (LG) gratefully acknowledge support from the National Institute of Environmental Health Sciences grant (#T32 ES007334). This study was also supported by the Intramural Research Program of the National Cancer Institute. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the methodology: LPG, CG, DCW. Analyzed the data: LPG. Wrote the first draft of the manuscript: LPG, DCW. Contributed to the writing of the manuscript: LPG, DCW. Agree with manuscript results and conclusions: LPG, CG, DCW. Jointly developed the structure and arguments for the paper: LPG, CG, DCW. Made critical revisions and approved final version: LPG, CG, DCW. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Diez Roux AV. The examination of neighbourhood effects on health: Conceptual and methodological issues related to the presence of multiple levels of organization. In: Kawachi I, Berkman L, editors. Neighbourhoods and Health. New York: Oxford University Press; 2003. pp. 45–64. [Google Scholar]
- 2.Flowerdew R, Manley DJ, Sabel CE. Neighborhood effects on health: Does it matter where you draw the boundaries? Soc Sci Med. 2008;66(6):1241–55. doi: 10.1016/j.socscimed.2007.11.042. [DOI] [PubMed] [Google Scholar]
- 3.Kawachi I, Berkman L. Introduction. In: Kawachi I, Berkman L, editors. Neighborhoods and Health. New York: Oxford University Press; 2003. pp. 1–19. [Google Scholar]
- 4.Krieger N, Chen JT, Waterman PD, Soobader M-J, Subramanian SV, Carson R. Geocoding and monitoring of US socioeconomic inequalities in mortality and cancer incidence: Does the choice of area-based measure and geographic level matter? The Public Health Disparities Geocoding Project. Am J Epidemiol. 2002;156(5):471–82. doi: 10.1093/aje/kwf068. [DOI] [PubMed] [Google Scholar]
- 5.Krieger N, Chen JT, Waterman PD, Soobader M-J, Subramanian SV, Carson R. Choosing area based socioeconomic measures to monitor social inequalities in low birth weight and childhood lead poisoning: The Public Health Disparities Geocoding Project (US) J Epidemiol Community Health. 2003;57(3):186–99. doi: 10.1136/jech.57.3.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Root ED. Moving neighborhoods and health research forward: Using geographic methods to examine the role of spatial scale in neighborhood effects on health. Ann Assoc Am Geogr. 2012;102(5):986–95. doi: 10.1080/00045608.2012.659621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Root ED, Meyer RE, Emch M. Socioeconomic context and gastroschisis: Exploring associations at various geographic scales. Soc Sci Med. 2011;72(4):625–33. doi: 10.1016/j.socscimed.2010.11.025. [DOI] [PubMed] [Google Scholar]
- 8.Block JP, Scribner RA, DeSalvo KB. Fast food, race/ethnicity, and income: A geographic analysis. Am J Prev Med. 2004;27(3):211–7. doi: 10.1016/j.amepre.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Galvez MP, Pearl M, Yen IH. Childhood obesity and the built environment: A review of the literature from 2008–2009. Curr Opin Pediatr. 2010;22(2):202–7. doi: 10.1097/MOP.0b013e328336eb6f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prince SA, Kristjansson EA, Russell K, et al. A multilevel analysis of neighborhood built and social environments and adult self-reported physical activity and body mass index in Ottawa, Canada. Int J Environ Res Public Health. 2011;8(10):3953–78. doi: 10.3390/ijerph8103953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DellaValle CT, Wheeler DC, Deziel NC, et al. Environmental determinants of polychlorinated biphenyls concentrations in residential carpet dust. Environ Sci Technol. 2013;47(18):10405–14. doi: 10.1021/es401447w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barringer TH, Dunn D, Battaglin WA, Vowinkel EF. Problems and methods involved in relating land use to ground-water quality. Water Resour Bull. 1990;26(1):1–9. [Google Scholar]
- 13.Gardner KK, Vogel RM. Predicting ground water nitrate concentration from land use. Ground Water. 2005;43(3):343–52. doi: 10.1111/j.1745-6584.2005.0031.x. [DOI] [PubMed] [Google Scholar]
- 14.Moran MJ, Zogorski JS, Squillace PJ. Chlorinated solvents in groundwater of the United States. Environ Sci Technol. 2007;41:74–81. doi: 10.1021/es061553y. [DOI] [PubMed] [Google Scholar]
- 15.Nolan BT, Hitt KJ, Ruddy BC. Probability of nitrate contamination of recently recharged groundwaters in the conterminous United States. Environ Sci Technol. 2002;36(10):2138–45. doi: 10.1021/es0113854. [DOI] [PubMed] [Google Scholar]
- 16.Squillace PJ, Moran MJ. Factors associated with sources, transport, and fate of volatile organic compounds and their mixtures in aquifers of the United States. Environ Sci Technol. 2007;41(7):2123–30. doi: 10.1021/es061079w. [DOI] [PubMed] [Google Scholar]
- 17.Squillace PJ, Scott JC, Moran MJ, Nolan BT, Kolpin DW. VOCs, pesticides, nitrate, and their mixtures in groundwater used for drinking water in the United States. Environ Sci Technol. 2002;36(9):1923–30. doi: 10.1021/es015591n. [DOI] [PubMed] [Google Scholar]
- 18.Ferrari MJ, Ator SW. USGS Water-Resources Investigations Report 1995–4099. USGS; Reston, VA: 1995. Nitrate in ground water in the Great Valley carbonate subunit of the Potomac River Basin. [Google Scholar]
- 19.Kolpin DW. Agricultural chemicals in groundwater of the midwestern United States: Relations to land use. J Environ Qual. 1997;26:1025–37. [Google Scholar]
- 20.Johnson TD, Belitz K. Assigning land use to supply wells for the statistical characterization of regional groundwater quality: Correlating urban land use and VOC occurrence. J Hydrol. 2009;370:100–8. [Google Scholar]
- 21.Rupert MG. USGS Water-Resources Investigations Report 03–4269. USGS; Reston, Virginia: 2003. Probability of detecting atrazine/desethyl-atrazine and elevated concentrations of nitrate in ground water in Colorado. [Google Scholar]
- 22.Berk RA. Statistical Learning from a Regression Perspective. New York: Springer; 2008. [Google Scholar]
- 23.Wheeler DC. Simultaneous coefficient penalization and model selection in geographically weighted regression: The geographically weighted lasso. Environ Plann A. 2009;41(3):722–42. [Google Scholar]
- 24.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd ed. New York: Springer; 2009. [Google Scholar]
- 25.Hastie T, Taylor J, Tibshirani R, Walther G. Forward stagewise regression and the monotone lasso. Electron J Stat. 2007;1:1–29. [Google Scholar]
- 26.Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. J R Stat Soc B. 2006;68(1):49–67. [Google Scholar]
- 27.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc B. 1996;58(1):267–88. [Google Scholar]
- 28.Efron B, Hastie T, Johnstone I, Tibshirani R. Least angle regression. Ann Stat. 2004;32(2):407–51. [Google Scholar]
- 29.Shi F. Available from ProQuest Dissertations and Theses database. (UMI 3556481) 2012. Model Selection in Multivariate Analysis with Missing Data [doctoral dissertation] [Google Scholar]
- 30.Wheeler DC, Nolan BT, Flory AR, DellaValle CT, Ward MH. Modeling groundwater nitrate exposure for an agricultural health study cohort in Iowa. In resubmission. 2015 [Google Scholar]
- 31.Burnham KP, Anderson DR. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. 2nd ed. New York: Springer; 2002. [Google Scholar]
- 32.R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. Available at http://www.R-project.org/ [Google Scholar]
- 33.Diez Roux AV. Investigating neighborhood and area effects on health. Am J Public Health. 2001;91(11):1783–9. doi: 10.2105/ajph.91.11.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crowder K, South SJ. Spatial and temporal dimensions of neighborhood effects on high school graduation. Soc Sci Res. 2011;40:87–106. doi: 10.1016/j.ssresearch.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]