Abstract
T cells critically regulate IBD with T cell dependent experimental colitis models gaining favor in identifying potential pathogenic mechanisms; yet, limited understanding of specific pathogenic molecules or pathways still exists. In this study, we sought to identify changes in whole genome expression profiles using the CD4CD45Rbhi T cell transfer colitis model compared to genome expression differences from Crohn's disease tissue specimens. Colon tissue was used for histopathological and genome expression profiling analysis at 0, 2, 4, or 6 weeks after adoptive T cell transfer. We identified 1,775 genes that were significantly altered during disease progression with 361 being progressively down regulated and 341 progressively up regulated. Gene expression changes were validated by qRT-PCR confirming genome expression analysis data. Differentially expressed genes were clearly related to inflammation/immune responses but also strongly associated with metabolic, chemokine signaling, Jak-STAT signaling, and angiogenesis pathways. Ingenuity network analysis revealed 25 unique network associations that were associated with functions such as antigen presentation, cell morphology, cell-to-cell signaling and interaction, as well as nervous system development and function. Moreover, many of these genes and pathways were similarly identified in Crohn's disease specimens. These findings reveal novel, complex, and dynamic changes in gene expression that may provide useful targets for future therapeutic approaches.
Keywords: inflammation, inflammatory bowel disease, Crohns disease, T cell colitis, angiogenesis
Introduction
Crohn's Disease (CD) is the one of the two major forms of inflammatory bowel disease (IBD) with the incidence of CD being 100–250/100,000 in the United States and the UK (1). As a chronic disease, CD affects many parts of gastroenterological tract including the colon and has the potential to develop into cancer (2). Comprehensive studies have shown that pediatric CD has increased in both developed and developing nations (3). As a multifactor related disease, it is believed that genetic, enteric bacteria, and the immune system components all play a role in the development of CD. Thus, there is a pressing need to better understand molecular mediators involved during disease pathogenesis with the hope of identifying new approaches to treat CD.
Due to the complexity of human inflammatory bowel diseases, no one animal model ideally simulates all aspects of human colitis. However, the adoptive transfer of naïve T cells into recombinase activating gene 1 deficient (Rag1−/−) mice induces both colitis and small bowel inflammation resulting in an experimental model similar to CD pathophysiology (4). It is also known that CD4+ T cells make up a significant portion of cells that infiltrate mucosal tissues during clinical disease and in chronic colitis models, and when CD4+ T cells are experimentally depleted chronic inflammation is attenuated (5). Thus, the murine CD4CD45Rbhi T cell transfer model is a useful model to examine potential disease mechanisms that may be relevant for CD.
Temporal genome transcription studies have been reported using the TNBS colitis model in rats (6) and DSS model in mice (7). However, there are no temporal studies of genome wide expression profile analysis in the T cell transfer model of colitis. In this study, we determined key changes in gene expression throughout the development of CD4CD45Rbhi T cell transfer colitis (week 0, 2, 4, 6) and compared results to clinical CD specimen gene expression changes to identify new molecules involved in chronic colitis and novel targets for future therapy.
Material and methods
T cell transfer of colitis model
Male (Rag1−/−) mice on the C57BL/6 background aged 12–14 weeks were used as recipients for CD4CD45Rbhi T cell transfer as we have previously reported (8). Mice were housed in specific pathogen-free conditions according to Institutional Animal Care and Use Committee (IACUC) regulations and the National Research Council's Guide for the Care and Use of Laboratory Animals and studies approved by the LSU Health Sciences Center IACUC. T cell transfer mice were sacrificed at weeks 0, 2, 4, and 6 (n=4 for each group). Colon length and weight were measured, and the gross colon score was assessed as follows: 1—fairly normal, yellowish, 2—mild bowel wall thickening with no visible hyperemia, 3—severe bowel wall thickening with rigidity and obvious hyperemia, and 4—the colon sample is thick, white, with greater rigidity. Harvested colon tissue was cut in half longitudinally with one half put in RNAlater reagent and stored at −80 °C, and the other half rolled in a Swiss roll fashion and fixed in 10% formaldehyde for histopathological analysis.
Histopathology assessment of T cell transfer colitis
Formalin fixed colon tissue was cut and stained with hematoxylin and eosin (H&E), and scored in a double-blinded manner as we previously reported (8). Tissue sections were scored for the severity of inflammation, goblet cell loss, crypt damage, and neutrophil infiltration, yielding a total histopathology score as we previously reported (8). The degree of inflammation infiltrate in the lamina propria was scored 0 to 3. Goblet cell loss was scored 0 to 2. Neutrophil infiltrates per 40× field were scored 0 to 4. Abnormal crypt structure was scored 0 to 3. The number of crypt abscesses was scored 0 to 2. Mucosal to frank erosion was scored 0 to 1. Submucosal spread to transmural involvement was scored 0 to 3. The histopathological score of each sample is obtained from the summation of these values.
Angiogenic Index assessment of T cell transfer colitis
Angiogenic index assessment was performed as we previously reported (8). Colon tissue from the T cell model at weeks 0, 2, 4, and 6 was cut into five micrometer sections, fixed, washed 3 times with PBS, blocked with 5% horse serum, and stained with rat anti-mouse PECAM-1 (CD31) antibody (BD PharMingen) diluted 1:200 in PBS containing 0.05% horse serum. After washing 3 times with PBS, the slides were counterstained with Cy3-conjugated anti-rat antibody diluted 1:250 in PBS with 0.05% horse serum. Slides were washed three times with PBS and mounted with Vectashield mounting medium containing DAPI nuclear counterstain. Images were taken utilizing a Nikon Eclipse TE2000-S epifluorescence microscope (Melville, NY). The total pixel intensity of CD31 and nuclear staining were quantified for each image using Simple PCI software. All imaging settings for each respective fluorochrome were kept constant between specimens. Total CD31 pixel intensity was divided by total DAPI pixel intensity for each image to calculate the angiogenic index (8).
Microarray expression analysis
Total RNA was isolated using an RNeasy isolation kit (Qiagen). The concentration and integrity of RNA samples were determined by electrophoresis on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Qualified RNA samples were used for gene chip analysis. Sample labelling, hybridization, staining and scanning procedures were carried out as we have described previously (7). GeneChip® Mouse Genome 430 2.0 Arrays, which cover over 39,000 transcripts, were used for colon tissue genome transcription analysis. Sixteen gene chips were used on a total of 16 individual specimens (4 specimens per time point) with no specimens pooled for this study.
Arrays were globally scaled to a target intensity value of 500 in order to compare individual samples. The Genechip Operating Software 1.4 (Affymetrix) was used to indentify the absolute call (present, marginal, absent) of each gene expression in each sample as well as the fold change in expression of individual genes between samples. All microarray data conform to MIAME standards and are uploaded in NCBI Gene Expression Omnibus (GEO) database (accession number GSE 27302) for public access.
The normalized data were log transformed (base 2) and uploaded to the Genesifter program. The minimal threshold value for differences in gene expression was set at 2. One-way ANOVA (analysis of variation) was performed against week 0 samples and corrected by Benjamini and Hochberg posttest to diminish false discovery rates. The differentially expressed gene data was uploaded into Ingenuity software (www.ingenuity.com) to visualize and explore the molecular interaction networks based on Ingenuity Knowledge Database (IKB) that identifies direct and indirect interactions between genes extracted from peer-reviewed publications and databases.
Quantitative real time PCR analysis of T cell induced colitis
Real time PCR validation was carried out by measuring 18 genes that were either progressively up- or down regulated as identified by genome expression profiling. All primers were designed with Beacon Designer software. The primers were designed to avoid primer template structure and cross homology problems, and confirmed by previous reports or sequencing. The qRT-PCR primers used in this study are reported in Table 1. Template cDNA of each sample was synthesized using the iScript cDNA kit from BioRad following the manufacturer's instructions. cDNA was diluted 40 × and used as template for quantitative real time PCR. GAPDH was used as an internal control for all reactions. The threshold cycle (Ct) value formula was used to calculate the relative expression of selected genes as we have previously reported (7). Statistical analysis was performed by one-way ANOVA with Bonferroni post-test to week 0 as control.
Table 1.
Primer sequences used in the T cell microarray qRT-PCR
| Gene Name | Accession No. | Sense primer | Anti-sense primer |
|---|---|---|---|
| C1qa | NM_007572 | AGGGCGTGAAAGGCAATC | CTTGGAAGTTGAAGTAATAGAAGC |
| C3 | K02782 | CCATCAAGATTCCAGCCAGTAAGG | GCTTTCTCCACCACCGTTTCC |
| Ccl8 | NM_021443 | AGAGAATCAACAATATCCAGTG | TCTTAACTCAGGTGTGAAGG |
| Ccr5 | D83648 | GAGGTGAGACATCCGTTCC | GGTGCTGACATACCATAATCG |
| Ecscr | BB736636 | TAGATCCAGAAGAACCTA | GGACAATCTACCTCATTA |
| Emb | BG064842 | GCCACCAAGAGGATGACTTC | GAGAGTGAGGAGCAGAATGAG |
| GAPDH | NM_008084 | GCCTTCCGTGTTCCTACC | CTTCACCACCTTCTTGATGTC |
| Gbp2 | BE197524 | TCAAGACATGTTGTCACAGTGG | ATCACAGAACAGGCAACTTTTAAC |
| Ifitm6 | BB193024 | CCTACATCTACTCGGTGAAG | TCAGAATCTTGGCGGTTG |
| Iigp1 | BM239828 | AATGTAAATATGCTGCTACTTAG | TTTGGGAAATTATTTTGCTTATG |
| Mmp13 | NM_008607 | TATGGTCCAGGCGATGAAG | CCTCGGAGACTGGTAATGG |
| Mmp3 | NM_010809 | AGTTGGAGAACATGGAGAC | AGCAGCAACCAGGAATAG |
| Mpeg1 | L20315 | TGAATGCTTAGGGCTGGTATATG | CCTGGTAGGACCTCCAAGAC |
| Ms4a4b | BB199001 | GCCTTGCTGTGTCCGTCTC | TTGGTCCCTTGGTGTTCTGATG |
| Ptk6 | NM_009184 | AAGCCAGGAGCAGACTATG | CAGACAGATTGGAGAAGGATAC |
| Ptprc | NM_011210 | CCTCCAAGCACAACCATAGC | CACAATCCTCATTTCCACACTTAG |
| Pyhin1 | BM241008 | AAGGAAGCAGGACTGAGTGGTTG | GGATGATGGAAGTGGAGGTCTACG |
| S100a8 | NM_013650 | GGACATCAATAGTGACAATGC | ACCTGAGATATGATGACTTTATTC |
| Wars | BB785450 | GTCTTGGAAAAGCAGAAG | AATGATGCAAGGACAGAA |
Comparison of T cell induced colitis with pediatric CD microarray data
Two sets of CD expression data were downloaded from Subra et al (1) with GEO accession number GSE 10616 consisting of 11 healthy controls and 14 colon-only CD, and from Carey et al (9) with GEO accession number GSE 9686 consisting of 8 control samples and 11 CD samples. Both sets of data used the Affymetrix GeneChip Human Genome U133 plus 2.0 Array, which includes over 38,500 unique transcripts, containing probes for approximately 22,634 genes similarly providing whole genome coverage. These data were normalized using the RMA (Robust Multichip Average) method (10) and uploaded to the Genesifter software. The minimal threshold value for differences in gene expression was set at 2. A t-test was performed against control samples coupled with Benjamini and Hochberg post-test to minimize false discovery rates.
Results
T cell induced colitis parameters progressively increase over time
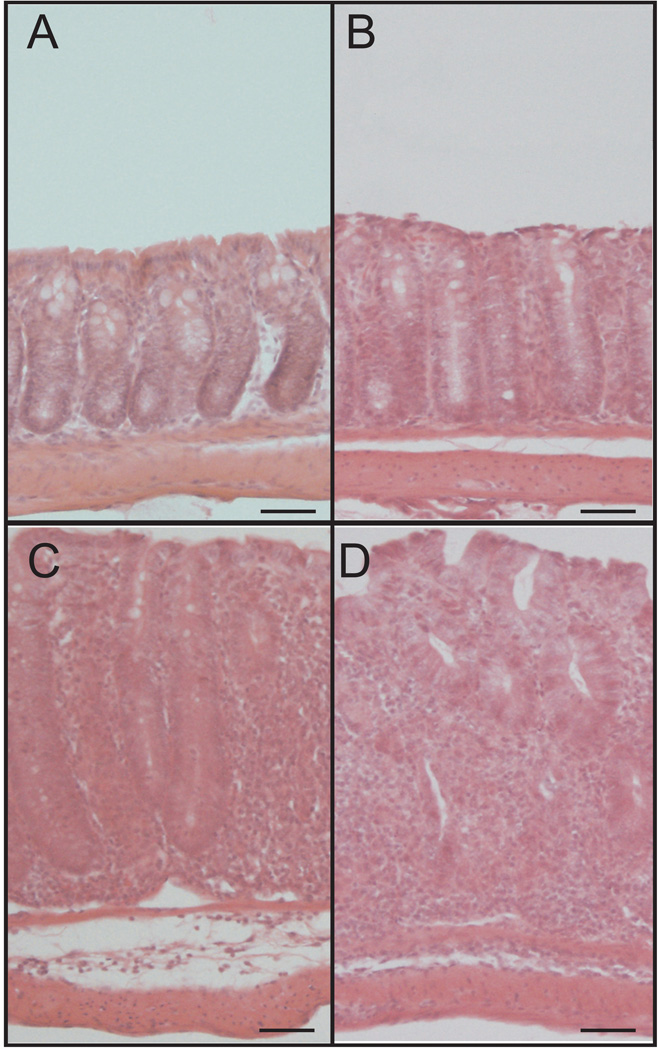
To better understand changes in genome expression profiles, T cell transfer colitis tissue histology was evaluated at weeks 0, 2, 4 and 6. Figure 1 shows representative images of T cell transfer colitis histopathology over time. Panel A shows normal colon histomorphology at week 0, whereas panels B–D demonstrate T cell transfer histological changes at weeks 2, 4 and 6 respectively. These findings are in agreement with our and others' previous results regarding progressive T cell transfer-mediated tissue histopathology including increased neutrophil infiltrates, abnormal crypt architecture, goblet cell loss, and infiltrates within the lamina propria.
Figure 1. T cell transfer colitis-mediated tissue histopathology over time.
A–D: T cell transfer-mediated histological changes illustrated by hematoxylin and eosin staining of tissue harvested at weeks 0, 2, 4 and 6, respectively. Histopathological changes progressively increase over time including neutrophil infiltrate, abnormal crypt architecture, goblet cell loss, and degree of inflammation infiltrate in the lamina propria. Magnification is 200×. Scale bars, 50µm.
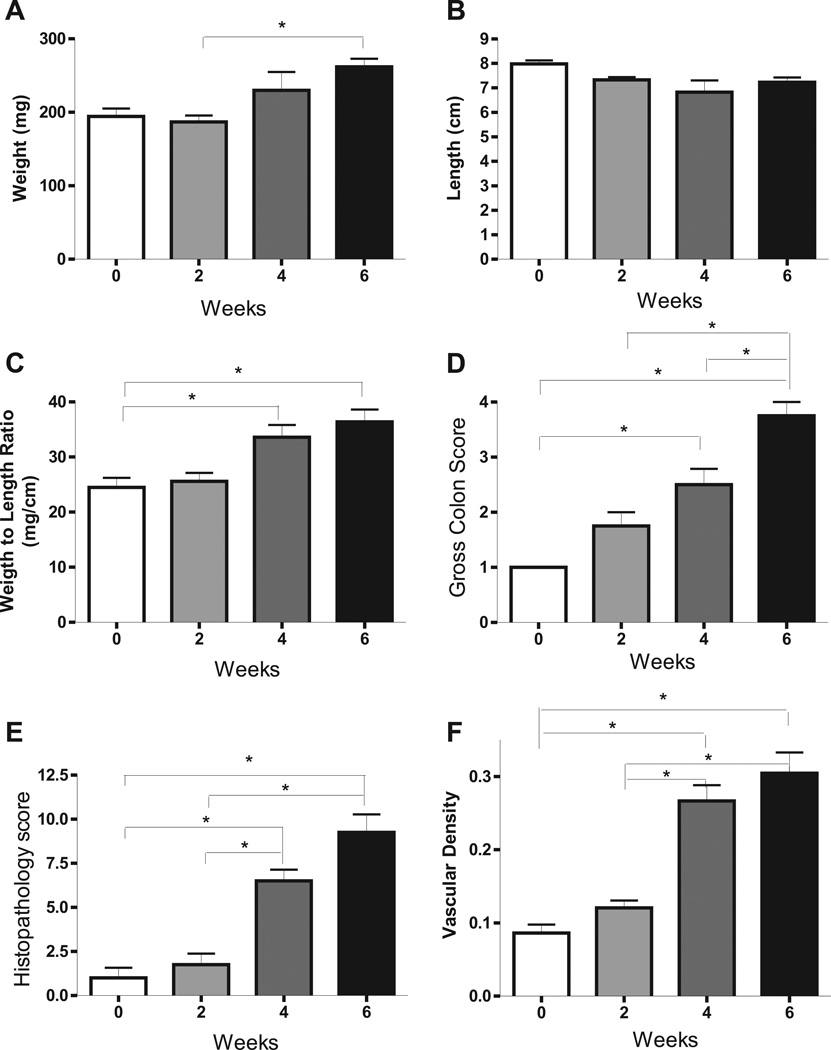
Additional temporal disease measurements are shown in figure 2 during the development of T cell mediated colitis. Panels A, B, and C show colon weight, colon length, and ratio of colon weight to length at each of the 4 time points. Colon weight, colon weight to length ratio at week 0 and week 2 display no difference, but are progressively up at weeks 4 and 6. Colon lengths progressively shorten from week 0 to week 6, which was not statistically different. Colon weight to length ratio was significantly different comparing weeks 4 and 6 with week 0, while gross colon score, histopathology score and angiogenic index are progressively and significantly increased over time as shown in panels D–F.
Figure 2. T cell transfer colitis model progressively increases multiple disease parameters over time.
The tissue histopathology score, colon length, colon weight, and vascular density were measured at weeks 0, 2, 4 and 6. A: colon weight at weeks 0, 2, 4 and 6. B: colon length at weeks 0, 2, 4 and 6. C–F: colon weight to length ration, gross colon score, tissue pathology and vascular density at discrete time points. *P<0.05 vs. week 0 (n=4) by 1-way ANOVA with Bonferroni posttest; n=4 animals per time cohort at weeks 2, 4 and 6.
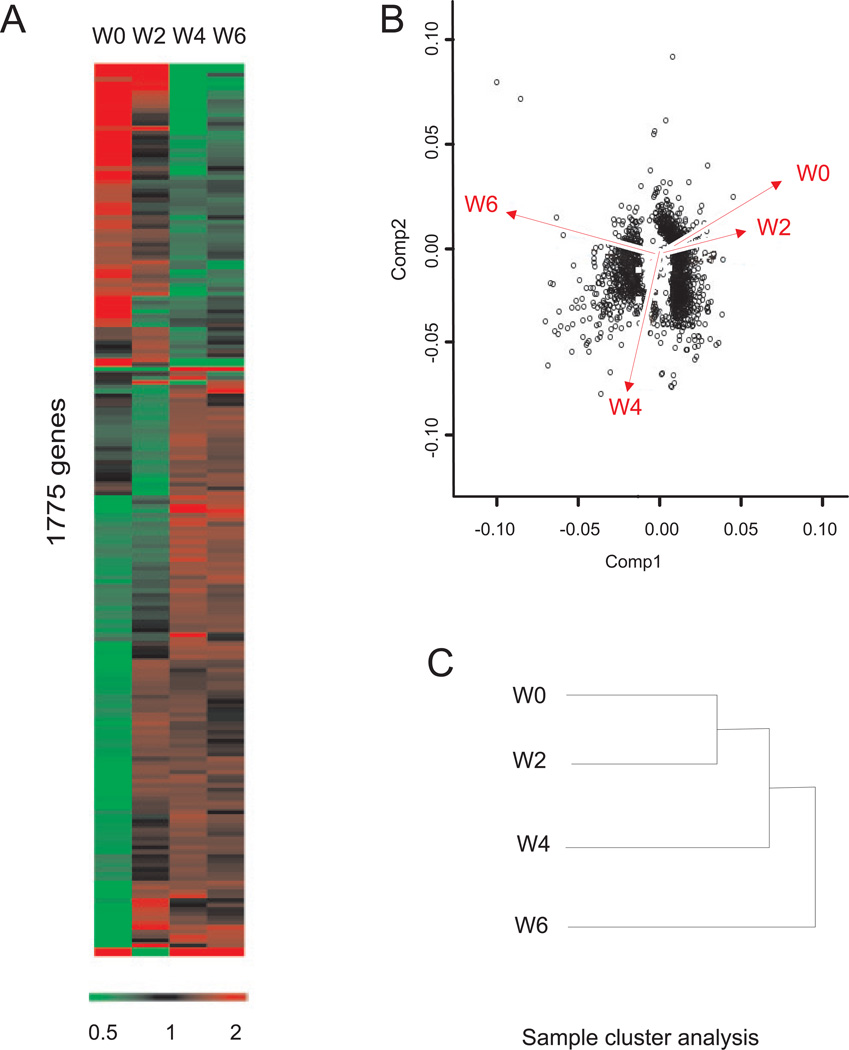
T cell induced temporal change in global gene expression
Global genome transcription change was detected by hybridization of individual colon total RNA with individual Mouse 430 2.0A Affymetrix gene chips. The heat map of Figure 3A shows the expression change of 1,775 genes represented by 2,043 probe sets that were identified as significant by Genesifter software. From the heat map signature, it is clear that sets of genes are progressively up- or down regulated, while other expression profiles fluctuate. Principal component analysis (PCA) was used to visualize the comprehensive response of gene expression changes in the T cell transfer colitis model (panel B). Component 1 (Comp1) represents the actual changes in gene expression profile and component 2 illustrates the trend of change in gene expression. PCA revealed that week 0 and week 2 time points show similar PCA profiles, whereas, week 4 and 6 illustrate large shifts in expression profiles. Similarly, sample cluster analysis showed that gene expression profiles at week 0 and 2 is clustered into one group, whereas week 4 changes are divergent from week 0 and 2, and week 6 is further divergent from all three time points (panel C).
Figure 3. Genesifter analysis of T cell mediated colitis array data.
A: Genesifter program heat map analysis, 1,775 genes were identified from the 39,000 transcripts that significantly changed over the experimental time course at weeks 0, 2, 4 and 6. B: Principal component analysis (PCA) of the gene expression pattern using eigenvalues of covariance. Component 1 (Comp 1) represents the actual change in the gene expression profile, while component 2 (Comp2) illustrates the trend of change in gene expression. X- and y- Axis values are arbitrary units for eigenvalues and gene expression represented by dots. C: sample cluster analysis show that gene expression profiles can be divided into three groups, week 0 and week 2 in one group, which diverge from week 4 (second group) and week 6 (third group).
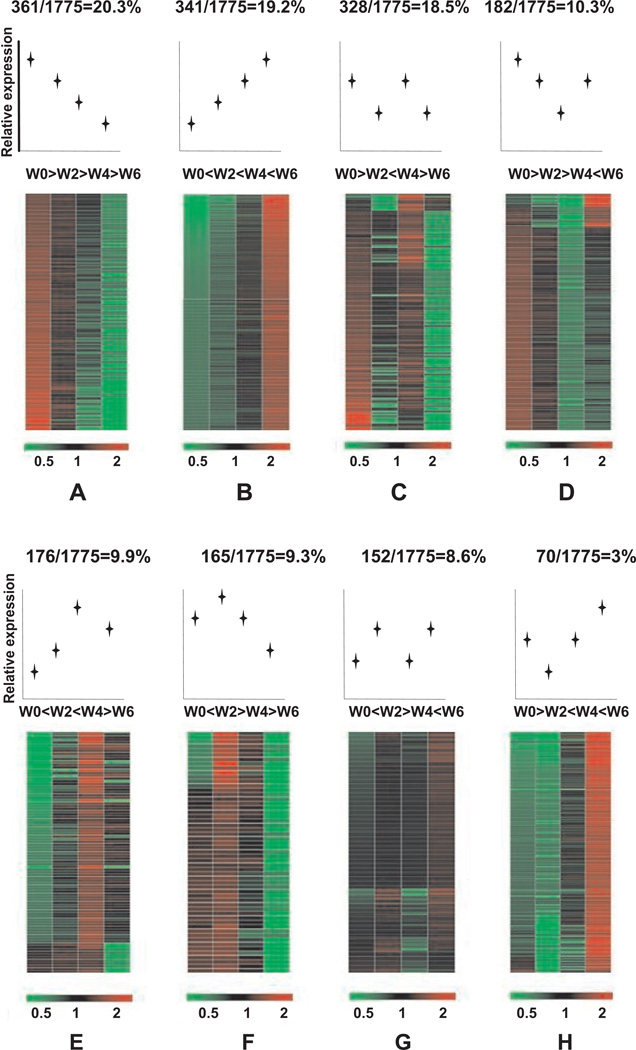
Distribution of gene expression profiles in T cell induced colitis
Having identified 1,775 differentially expressed genes during the progression of T cell mediated colitis; we next sought to better classify these changes with respect to their temporal phenotype. All 1,775 genes were divided into 8 different classes based on expression profile responses between the 4 time points. As shown in figure 4, the majority of genes (group A-20.3%) were progressively down regulated over the 6 weeks. However, the second highest number of genes (group B-19.2%) was progressively upregulated. The other 6 groups of fluctuating expression profiles account for the remaining 60 percent of the identified genes. Supplementary tables A–H provide specific gene names and expression levels over time assigned to each group.
Figure 4. Different gene expression profile classes during the progression of T cell mediated colitis.
1,775 genes for which expression is significantly changed during the time course were classified into 8 types (groups A–H) according to temporal fluctuations in gene expression levels over time. Expression profile classes are ordered from most to least abundant percentage within the total cohort of 1,775 genes.
Quantitative real time PCR validation of T cell induced colitis
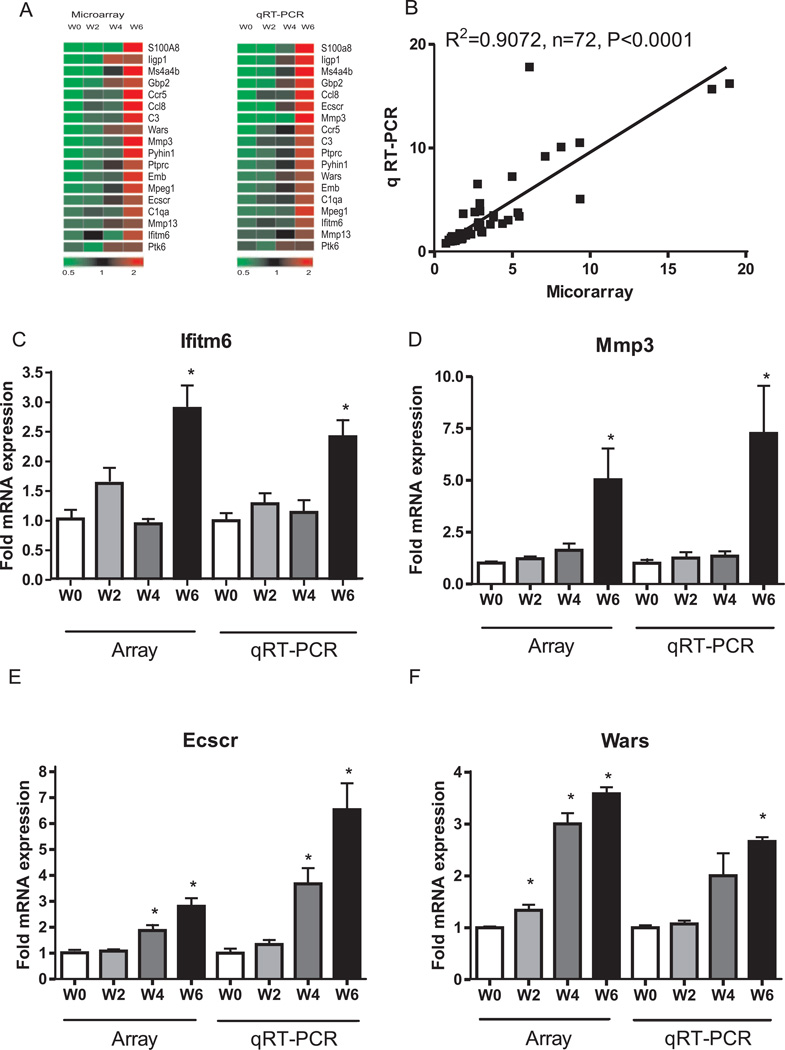
Next we performed qRT-PCR validation on a subset of mRNAs identified in the T cell transfer colitis microarray data. The selected genes represent many functions including inflammation, angiogenesis, and cell adhesion. The changes in gene expression obtained from real time PCR analysis were uploaded to Genesifter to get heat map plots. As shown in figure 5, panel A, the qRT-PCR expression profiles are highly similar to microarray data. Likewise, panel B illustrates the correlation between qRT-PCR and microarray data with an r2 value = 0.9072, p < 0.0001 at the 95% confidence level. Table 2 reports the expression value comparisons between qRT-PCR and microarray data of the selected genes. Importantly, panels C–F shows that the array and qRT-PCR gene expression profiles for Ifitm6, Mmp3, Ecscr, and Wars were all similarly changed over time. Taken together, these results validate our microarray data and verify a diverse array of gene expression changes during T cell mediated colitis.
Figure 5. Quantitative real-time PCR (qRT-PCR) validation of T cell transfer colitis microarray data.
qRT-PCR validation was performed for numerous genes identified by microarray analysis. A: microarray vs. qRT-PCR expression heat map for 18 individual genes. B: correlation in gene expression between microarray (x-axis) and qRT-PCR (y-axis) changes in gene expression with r2=0.9072, P<0.0001 at the 95% confidential level, n=72. C, D, E, F: representative validation between microarray and qRT-PCR gene expression for Ifitm6, Mmp3, Ecscr and Wars respectively. *P<0.01 vs week 0 (n=4), at weeks 2, 4 and 6 (n=4).
Table 2.
Quantitative real-time PCR validation of microarray data
| Gene Symbol |
Accession NO | Affymerix Gene Array | RT-PCR | ||||||
|---|---|---|---|---|---|---|---|---|---|
| W0 | W2 | W4 | W6 | W0 | W2 | W4 | W6 | ||
| C1qa | NM_007572 | 1±0.09 | 1.27±0.05* | 1.13±0.08 | 2.82±0.24* | 1±0.1 | 1.36±0.17 | 1.49±0.26 | 2.82±0.16* |
| C3 | K02782 | 1±0.04 | 1.77±0.22* | 1.89±0.28* | 5.44±0.61* | 1±0.06 | 1.58±0.12* | 1.64±0.27* | 3.42±0.19* |
| Ccl8 | NM_021443 | 1±0.19 | 2.92±0.22* | 2.59±0.23* | 8.13±0.76* | 1±0.39 | 3.93±0.44* | 3.84±0.68* | 10.11±0.98* |
| Ccr5 | D83648 | 1±0.14 | 3.05±0.7* | 2.92±1.08 | 9.35±1.72* | 1±0.08 | 1.9±0.18* | 2.59±0.24* | 5.09±0.43* |
| Ecscr | BB736636 | 1±0.11 | 1.06±0.07 | 1.88±0.21* | 2.76±0.31* | 1±0.17 | 1.34±0.17 | 3.68±0.17* | 6.54±1.01* |
| Emb | BG064842 | 1±0.1 | 1.43±0.13* | 1.62±0.31 | 4.36±0.41* | 1±0.1 | 1.19±0.04 | 1.78±0.26* | 2.73±0.37* |
| Gbp2 | BE197524 | 1±0.19 | 2.8±0.42* | 7.1±0.48* | 9.33±0.4* | 1±0.13 | 2.61±0.17* | 6.22±1.13* | 10.53±0.75* |
| Ifitm6 | BB193024 | 1±0.15 | 1.58±0.26 | 0.92±0.82 | 2.81±0.39* | 1±0.17 | 1.28±0.24 | 1.14±0.27 | 2.42±0.37* |
| Ligp1 | BM239828 | 1±0.37 | 5.36±1.42* | 18.95±2.73* | 17.81±0.72* | 1±0.31 | 3.77±0.51* | 8.81±1.12* | 15.68±0.73* |
| Mmp13 | NM_008607 | 1±0.14 | 1±0.09 | 1.78±0.28* | 2.17±0.35* | 1±0.05 | 1.01±0.06 | 1.32±0.24 | 1.92±0.26* |
| Mmp3 | NM_010809 | 1±0.08 | 1.21±0.11 | 1.61±0.33 | 4.98±1.51* | 1±0.12 | 1.26±0.2 | 1.35±0.17 | 7.26±1.64* |
| Mpeg1 | L20315 | 1±0.11 | 1.15±0.07 | 1.73±0.2* | 3.78±0.32* | 1±0.12 | 1.05±0.04 | 1.22±0.12 | 3.34±0.41* |
| Ms4a4b | BB199001 | 1±0.28 | 4.34±1.46 | 8.85±1.73* | 24.15±5.52* | 1±0.25 | 3.18±0.31* | 5.59±1.2* | 15.04±1.17* |
| Ptk6 | NM_009184 | 1±0.21 | 0.73±0.08 | 1.95±0.18* | 2.01±0.18* | 1±0.16 | 0.81±0.04 | 1.66±0.47 | 1.6±0.13* |
| Ptprc | NM_011210 | 1±0.07 | 1.39±0.13* | 2.34±0.26* | 3.79±0.48* | 1±0.12 | 1.25±0.14 | 1.71±0.11* | 3.51±0.62* |
| Pyhin1 | BM241008 | 1±0.21 | 1.39±0.26 | 1.6±0.31 | 4.72±0.91* | 1±0.12 | 1.21±0.04 | 1.63±0.19 | 3.05±0.29* |
| S100a8 | NM_013650 | 1±0.17 | 2.9±0.3* | 6.1±1.16* | 39.06±10.26* | 1±0.18 | 4.67±0.8* | 10.93±4.45* | 45.68±24.26* |
| Wars | BB785450 | 1±0.02 | 1.34±0.11* | 3.01±0.21* | 3.58±0.13* | 1±0.04 | 1.07±0.06 | 2.00±0.40 | 2.66±0.08* |
Gene expression network analysis
We next performed gene network analysis using the Path Explorer function of Ingenuity Pathway Analysis software on the 1,775 differentially expressed genes. Twenty-five biological networks were generated for the 1,775 genes. Each network is composed of up to 35 highly related genes, and each network is assigned a significance score based on the fact that at least one gene is contained within that network (i.e. a score of 1). Thus, a score greater than 2 means that the resulting network is not generated by chance. The full list of networks, their significance scores, functions of the networks, as well as the contained genes are shown in supplementary table 1.
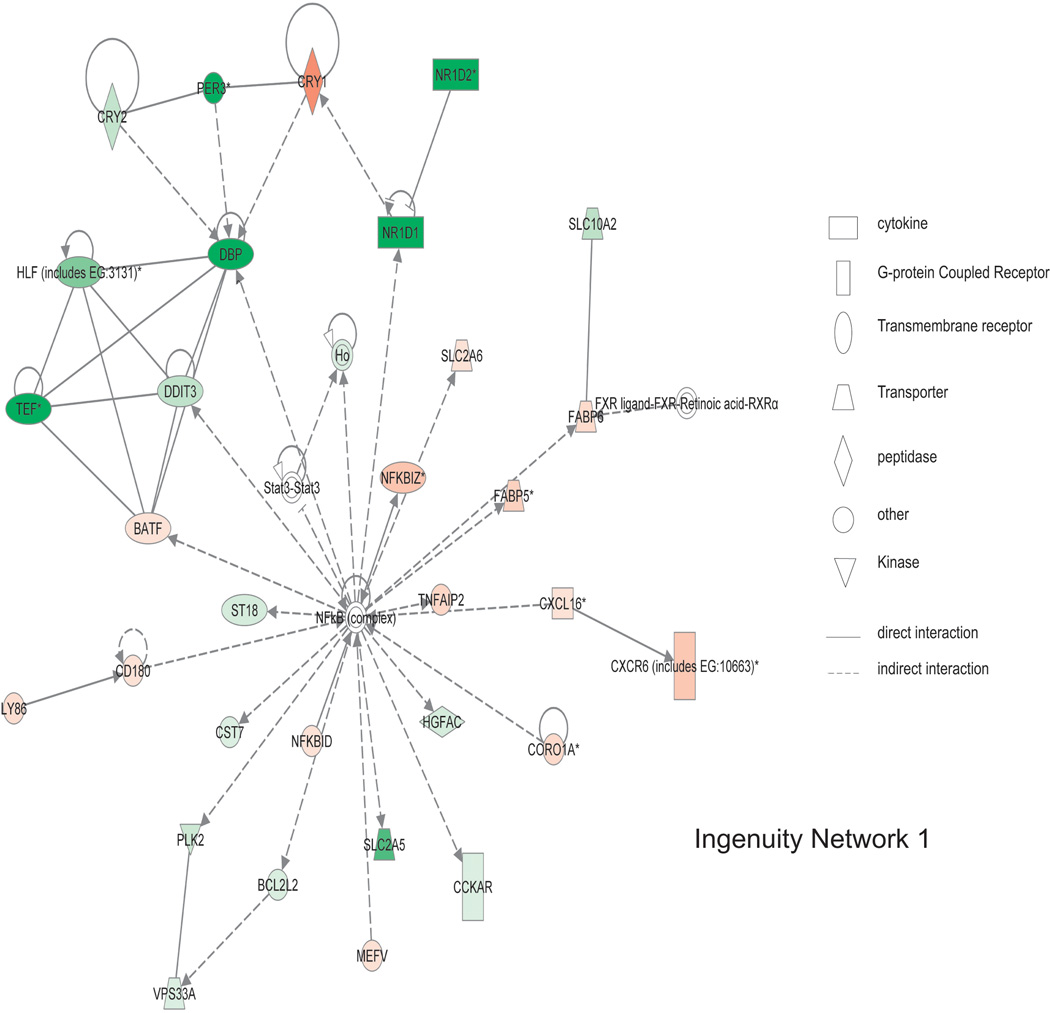
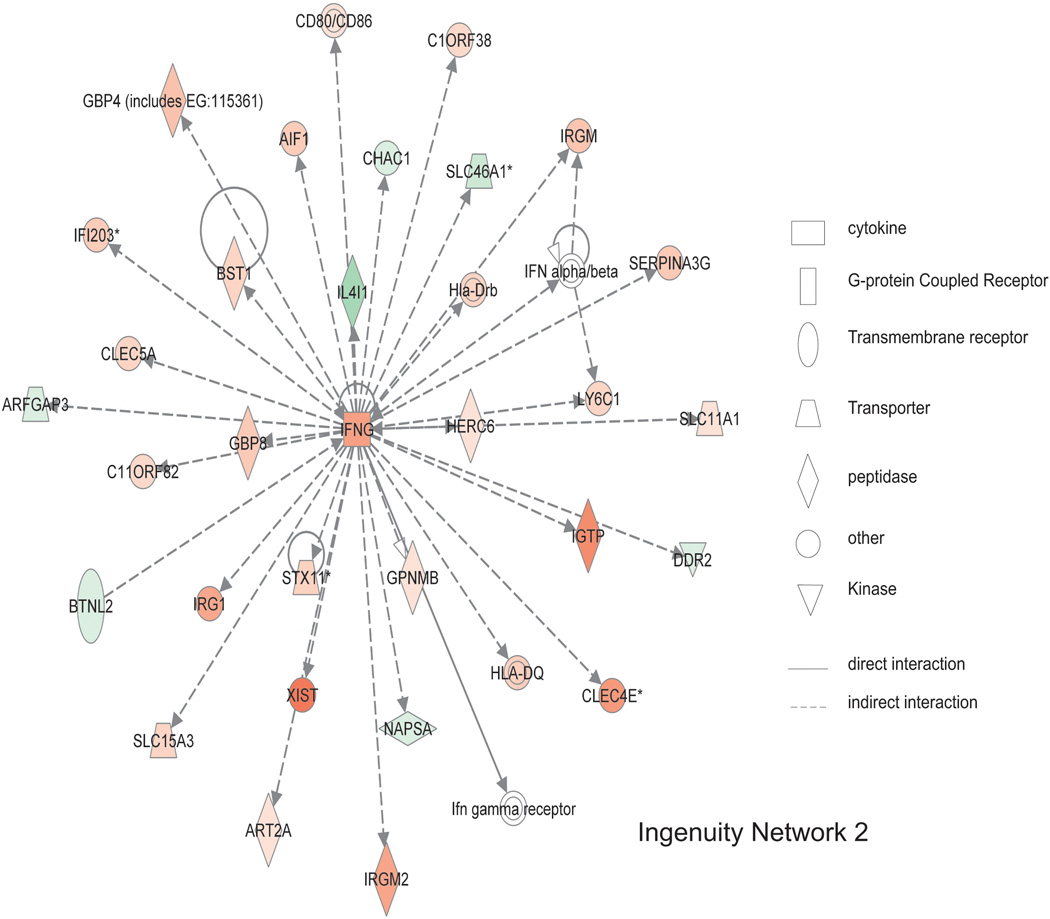
As shown in figure 6, the top scoring networks contain genes involved in antigen presentation, inflammatory responses, cell-to-cell signaling and interaction, and cell morphology. NFkB complex is in the center of network 1, which was directly regulated by NFKBID (weeks 2, 4 and 6 expression as 1.68, 1.93, 2.20 fold of week 0) and NFKBIZ (weeks 2, 4 and 6 expression as 1.75, 2.73, 2.63 fold of week 0). NFKBID is the nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta, and it binds and regulates NFkB, plays a role in cell proliferation, and responds to endotoxin shock (11). NFKBIZ is nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta, and is regulated by NFkB and plays a role in cell apoptosis (12). NFkB complex is also indirectly regulated by MEFV, CORA1A, CXCL16, and CD180. NFkB complex directly regulates NFKBIZ and indirectly interacts with 16 other genes. As shown in figure 7, in network 2, IFNG is centrally located. Its expression progressively increases over 6 weeks and indirectly interacts with the other 29 genes in the network suggesting IFNG up regulation is an important pathogenic mediator.
Figure 6. Ingenuity network 1 of gene expression changes from T cell mediated colitis.
Of 1,775 differentially expressed genes, a total of 25 statistically significant networks were identified. Network 1 has a score of 40 and involved 31 genes contained within that network. Major biological functions of network 1 are behavior, nervous system development and function, and gene expression. NFkB complex is centrally located within this network, showing a high degree of relationships with other molecules. Red means the expression value is greater than week 0, green indicates decreased expression compared to week 0. Symbol legend on right indicates the nature of the protein product and the type of relationship.
Figure 7. Ingenuity network 2 of gene expression changes from T cell mediated colitis.
Ingenuity network 2 has a score of 38 with 30 genes involved. Major biological functions of network 2 are antigen presentation, cell morphology, cell-to-cell signaling, and interaction. IFNG is centrally located within this network, indicating a high degree of relationships between this molecule and others. Red means the expression value is greater than week 0, green indicates decreased expression compared to week 0. Symbol legend on right indicates the nature of the protein product and the type of relationship.
Comparison of the T cell transfer colitis model and pediatric CD microarray data
While it is believed that the T cell transfer colitis model simulates CD better than other colitis models, relatively little is known of the differences and similarities between the T cell transfer model and clinical CD. Therefore, we compared our T cell transfer colitis microarray data to microarray data for pediatric CD colon tissue also generated from whole genome expression arrays.
Table 3 reports the KEGG pathways identified for T cell mediated colitis. Several different KEGG pathways were found that would have been expected to be associated with chronic inflammation such as cytokine-cytokine interactions, chemokine signalling, and cellular adhesion molecules. However, novel pathways such as metabolic, vascular smooth muscle contraction, and Jak-STAT signalling pathways were also identified. Genesifter analysis of the GSE9686 pediatric CD data showed that 242 genes were differentially expressed, with 173 genes up regulated and 69 genes down regulated. Likewise, analysis of GSE 10616 pediatric CD data showed that 298 genes were differentially expressed with 89 genes down regulated. Normalization using robust multichip averaging (RMA) of the two sets of pediatric CD microarray data showed that there were 117 genes up regulated and 50 genes down regulated in both data sets. In comparing the RMA normalized pediatric CD microarray data with the T cell transfer colitis microarray data, we found 13 commonly up regulated genes and 1 down regulated gene as shown in table 4. Supplementary tables 2 and 3 report the KEGG pathways identified in GSE9686 and GSE10616 pediatric CD microarray data, respectively. These results highlight the utility of array comparisons with which to identify potentially novel therapeutic molecular targets.
Table 3.
KEGG pathway classification of gene expression in T cell mediated colitis
| KEGG Pathway | List | Gene Set | z-score |
|---|---|---|---|
| Metabolic pathways | 63 | 1061 | −4.31 |
| Cytokine-cytokine receptor interaction | 54 | 232 | 7.49 |
| Chemokine signaling pathway | 42 | 173 | 6.89 |
| Cell adhesion molecules (CAMs) | 34 | 131 | 6.64 |
| Natural killer cell mediated cytotoxicity | 33 | 124 | 6.71 |
| Endocytosis | 30 | 201 | 2.79 |
| Systemic lupus erythematosus | 28 | 81 | 7.88 |
| Antigen processing and presentation | 24 | 71 | 7.15 |
| Chagas disease | 23 | 102 | 4.64 |
| Leishmaniasis | 22 | 66 | 6.75 |
| Viral myocarditis | 22 | 72 | 6.24 |
| Graft-versus-host disease | 21 | 38 | 9.78 |
| Jak-STAT signaling pathway | 20 | 138 | 2.12 |
| T cell receptor signaling pathway | 20 | 107 | 3.37 |
| Type I diabetes mellitus | 20 | 44 | 8.28 |
| Allograft rejection | 19 | 38 | 8.66 |
| Leukocyte transendothelial migration | 19 | 114 | 2.73 |
| Vascular smooth muscle contraction | 19 | 122 | 2.4 |
| Autoimmune thyroid disease | 18 | 49 | 6.63 |
| Neuroactive ligand-receptor interaction | 17 | 285 | −2.01 |
| Fc gamma R-mediated phagocytosis | 16 | 87 | 2.93 |
| Hematopoietic cell lineage | 16 | 79 | 3.37 |
| B cell receptor signaling pathway | 14 | 76 | 2.75 |
| Intestinal immune network for IgA production | 14 | 43 | 5.26 |
| Malaria | 13 | 47 | 4.34 |
| NOD-like receptor signaling pathway | 12 | 55 | 3.2 |
| Asthma | 9 | 25 | 4.6 |
| Primary immunodeficiency | 9 | 34 | 3.45 |
| Prion diseases | 9 | 34 | 3.45 |
| Alzheimer's disease | 7 | 154 | −2.07 |
| Circadian rhythm - mammal | 5 | 13 | 3.62 |
| Huntington's disease | 5 | 160 | −2.74 |
| Ubiquitin mediated proteolysis | 5 | 134 | −2.26 |
Table 4.
Similarly expressed genes between T cell mediated colitis and pediatric CD
| Gene name | Mouse gene accession NO. |
Human gene accession NO. |
T cell transfer colitis model | pediatric CD(GSE9686) | pediatric CD(GSE10616) | |||
|---|---|---|---|---|---|---|---|---|
| Colitis | P | CD | P | CD | P | |||
| ATP-binding cassette, sub-family B (MDR/TAP), member 1 | M30697 | AF016535 | 0.39±0.01 | 0.01 | 0.43±0.01 | 0.01 | 0.49±0.01 | 0.02 |
| Chemokine (C-X-C motif) ligand 1 | NM_008176 | NM_001511 | 14.62±0.09 | 0.01 | 5.6±0.01 | 0.01 | 4.66±0.01 | 0.01 |
| Chemokine (C-X-C motif) ligand 10 | NM_021274 | NM_001565 | 18.18±0.01 | 0.01 | 3.45±0.01 | 0.01 | 2.74±0.01 | 0.03 |
| Chemokine (C-X-C motif) ligand 2 | NM_009140 | M57731 | 17.97±0.09 | 0.01 | 3.35±0.01 | 0.01 | 3.39±0.01 | 0.01 |
| Chemokine (C-X-C motif) ligand 5 | NM_009141 | AK026546 | 32±0.05 | 0.01 | 6.04±0.08 | 0.02 | 6.49±0.06 | 0.01 |
| Chemokine (C-X-C motif) ligand 9 | NM_008599 | NM_002416 | 44.05±0.01 | 0.01 | 4.7±0.01 | 0.01 | 3.23±0.01 | 0.02 |
| Dual oxidase maturation factor 2 | NM_025777 | AI821606 | 3.88±0.01 | 0.01 | 2.24±0.01 | 0.01 | 3.79±0.02 | 0.01 |
| Guanylate binding protein 1, interferon-inducible, 67kDa | NM_010259 | NM_002053 | 3.61±0.01 | 0.01 | 2.66±0.01 | 0.01 | 2.32±0.01 | 0.01 |
| Interleukin 1, beta | BC011437 | NM_000576 | 8.55±0.01 | 0.01 | 3.32±0.01 | 0.01 | 3.37±0.01 | 0.01 |
| Matrix metallopeptidase 3 | NM_010809 | NM_002422 | 4.48±0.02 | 0.01 | 11.65±0.03 | 0.01 | 10.85±0.02 | 0.01 |
| Phosphoserine aminotransferase 1 | BC004827 | BC004863 | 2.57±0.01 | 0.01 | 3.49±0.02 | 0.01 | 2.49±0.01 | 0.01 |
| S100 calcium binding protein A8 | NM_013650 | NM_002964 | 39.06±10.26 | 0.01 | 6.09±0.02 | 0.02 | 5.66±0.01 | 0.01 |
| transglutaminase 2 | AF114266 | AL031651 | 2.13±0.01 | 0.01 | 2.47±0.01 | 0.01 | 2.52±0.01 | 0.01 |
| Tryptophanyl-tRNA synthetase (WARS) | BB785450 | M61715 | 2.61±0.01 | 0.01 | 2.22±0.01 | 0.01 | 1.89±0.01 | 0.02 |
Discussion
In this study, we found that the expression of 1,775 genes progressively changed during T cell mediated colitis providing valuable insight into possible mechanistic causes of T cell dependent colitis. It is difficult, if not impossible, to predict what these changes may mean from a pathophysiological standpoint but it is likely that they fall into separate categories that either: 1- may provide protection from colitis; 2- facilitate or hasten colitis progression; or 3- have no clear regulatory impact on colitis development and are a response to the disease process. Nonetheless, these data provide useful information with which to formulate new questions for future experiments using genetically altered mouse models or novel therapeutic agents to elucidate important pathophysiological mechanisms.
In comparing our T cell microarray data with pediatric CD microarray data, we found that TG2 (transglutaminase 2) expression is up regulated and could be a useful biomarker for pediatric CD as previously reported (13). Endothelial cells are a rich source of transgluminase-2 and Jones et al. (14) found that application of exogenous TG-2 blocks in vitro angiogenesis in a dose dependent manner without causing cell death. Thus, up regulation of TG-2 expression might be beneficial to pediatric CD by attenuating pathological angiogenesis.
Research in our laboratory (8) and others (15) have shown that pathological angiogenesis contributes to and perpetuates the development of colitis. Compared to our recently reported microarray data of DSS induced colitis, we found that the endothelial cell-specific chemotaxis receptor (Ecscr) was progressively upregulated during T cell mediated colitis. Ecscr, also known as endothelial cell-specific molecule 2 (ECSM-2) and Apoptosis Regulator through modulating IAp expression (ARIA), encodes a single-pass transmembrane protein of 205 amino acids. Ecscr expression has been reported to be restricted to endothelial cells and a marker for endothelial progenitor cells (16). Moreover, Ecscr-silenced cells show reduced VEGF-induced phosphorylation of KDR (17) suggesting it serves an important role in regulating pathological angiogenesis seen in the DSS and the T cell transfer colitis model.
We also found that the WARS gene is upregulated in both the DSS model, the T cell transfer model, and the 2 sets of pediatric CD microarray data (in pediatric CD microarray data, with t-test p<0.05). There are two forms of WARS protein products in human cells due to alternative mRNA splicing. The function of full length WARS (amino acids 1–471) is to carry out ligation of Trp amino acid to tRNATrp for protein synthesis, while its truncated form (mini TrpRS, amino acids 48–471, the natural alternative form of human TrpRS) inhibits angiogenesis (18). TrpRS also has a form of T2-TrpRS (composed of residues of 94–471) by proteolysis. Tzima et al. found that T2-TrpRS binds to the endothelial cell junction molecule VE-Cadherin, inhibiting angiogenesis related gene expression, such as AKT and endothelial cell NO synthase (19). Future research is needed to better understand the pathophysiological importance of WARS expression during experimental colitis. It is possible that this may represent a mechanism in which to inhibit pathological angiogenesis in pediatric CD.
Interestingly, we found that Ramp2, receptor activity-modifying protein 2, is progressively downregulated during the course of colitis, having only 13% mRNA compared with control by week 6. Ramp2 has also recently been demonstrated to be specifically expressed in endothelial cells and play a role in vascular growth and homeostasis (20). Ramp2−/− mice die in utero at midgestation due to blood vessel fragility resulting in severe edema and hemorrhage, while endothelial cells that overexpress RAMP2 showed enhanced capillary formation, firmer tight junctions, and reduced vascular permeability (21). These initial observations suggest that Ramp2 may act as a vascular stabilizing or maturity factor suggesting that augmentation of Ramp2 expression or function might attenuate vascular mediated pathogenic mechanisms during colitis that will require additional study.
The ATP-binding cassette, subfamily B member 1 (ABCB1), also known as multidrug resistance gene (MDR1) expression was significantly downregulated during the development of colitis compared to control. The ABCB1 gene encodes a membrane-bound efflux transporter P-glycoprotein 170 (PGP–170). Linkage analysis showed that the ABCB1 sequence polymorphism C3435T showed no significant association with Crohn's disease, suggesting that ABCB1 gene might serve as a disease marker versus a therapeutic target (22). However, Wu et al. found that in endoscopic biopsy samples ABCB1 was down regulated in both UC and CD (23). Together, these results indicate that ABCB1 may serve an important pathophysiological role during colitis that requires further study.
Phosphoserine aminotransferase was upregulated both in the T cell transfer model and in both sets of pediatric CD. Phosphoserine aminotransferase (PSAT) is the second enzyme in the L-serine synthetic pathway catalyzing the conversion of 3-phosphohydroxypyruvate into 3-phosphoserine that is dephosphorylated subsequently by phosphoserine phosphatase to form L-serine (24). Vie et al. have reported that overexpression of phosphoserine aminotransferase PSAT 1 stimulates colon cancer cell growth and is overexpressed in colon tumors (25). Since CD and experimental colitis are known to facilitate colon cancer development, PSAT1 might be an interesting target to investigate mechanisms of colitis progression of cellular transformation and carcinogenesis.
Our data also revealed a significant increase in Dual oxidase maturation factor 2 (DUOXa2) expression in T cell mediated colitis and pediatric CD. DUOXa2 is an endoplasmic reticulum-resident protein, and is absolutely necessary for post-translational processing and translocation of DUOX2 (an NADPH: O2 oxidoreductase flavoprotein) from the endoplasmic reticulum to the plasma membrane to initiate DUOX2 enzyme activity (26). Dual oxidases (DUOX1 and DUOX2) constitute the catalytic core of hydrogen peroxide generation, which either directly or indirectly by dismutation of superoxide. These findings implicate a specific redox modulatory related protein that may be important for increased oxidative stress and injury seen during colitis. As the complexity of redox regulation has grown, it will be interesting to determine whether DUOXa2 plays an important role in this pathophysiological response.
We also found that a member of the Small PRoline Rich (SPRR) family, Sprr2h, was progressively up regulated in the T cell transfer colitis model. Studies have shown that Sprr2h expression has been confined to epithelial and is a primary component of the cornified cell envelope (27, 28). Sprr2h expression is increased in Lor−/− mice to compensate for the deletion of loricrin, a major protein of the epidermal cornified cell envelope, thus preventing impairment of epidermal barrier function (29). Since intestinal epithelial cells critically regulate intestinal barrier function and control antigen traffic across the mucosa, it may be up regulation of Sprr2h is a compensatory mechanism to negate loss of epithelial barrier properties that are known to facilitate colitis development (30). Thus, it might be that induction of Sprr2h expression represents a protective response during T cell mediated colitis.
Lastly, a common dysregulated KEGG pathway between T cell mediated colitis and pediatric CD, such as cytokine-cytokine receptor interaction, might also be suitable for selection as targets for CD treatment. In the T cell transfer colitis model, among the 232 genes contained within the cytokine-cytokine receptor KEGG pathway 54 of them were significantly altered. In pediatric CD, among the 263 genes contained within cytokine-cytokine receptor KEGG pathway, 12 genes in the GSE 9686 data set and 15 genes in the GSE10616 data set were differentially expressed, and among these 3 data sets several chemokines were commonly up regulated including CXCL1, CXCL2, CXCL5, CXCL9 and CXCL10. CXCL1, CXCL2, and CXCL5 are well known neutrophil chemoattractants (31). In contrast, CXCL10 interaction with CXCR3 results in stimulation of monocyte, natural killer, and T-cell migration (32). Lastly, CXCL9 is known to stimulate disruption of the endothelial cell-cell barrier resulting in accelerated leukocyte trans-endothelial migration (TEM) and solute permeability (33). Importantly, the KEGG cytokine-cytokine receptor pathway is also dysregulated in the DSS induced colitis model with CXCL2, CXCL5, CXCL9, and CXCL10 being progressively up regulated in this model as well (7). These data suggest that chemokine expression and function is closely related to colitis pathophysiology and might be useful molecular targets to treat colitis.
In summary, our study provides comprehensive characterization of T cell mediated colitis colon tissue whole genome transcription profiles during the course of disease development. Several new and interesting molecules were identified that may be useful as potential colitis markers or therapeutic targets for clinical CD. Future studies are necessary to address the pathophysiological importance of these molecules for T cell mediated colitis through discrete genetic or pharmacological inhibition and manipulation experiments.
Supplementary Material
Acknowledgments
This work was supported by NIH grant DK43875-18 project 1 and 4 and Cores A, B, and C.
Footnotes
The authors have no conflict of interest to disclose.
References
- 1.Kugathasan S, Baldassano RN, Bradfield JP, et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viennot S, Deleporte A, Moussata D, et al. Colon cancer in inflammatory bowel disease: recent trends, questions and answers. Gastroenterol Clin Biol. 2009;33(Suppl 3):S190–S201. doi: 10.1016/S0399-8320(09)73154-9. [DOI] [PubMed] [Google Scholar]
- 3.Benchimol EI, Fortinsky KJ, Gozdyra P, et al. Epidemiology of pediatric inflammatory bowel disease: a systematic review of international trends. Inflamm Bowel Dis. 17:423–439. doi: 10.1002/ibd.21349. [DOI] [PubMed] [Google Scholar]
- 4.Ostanin DV, Bao J, Koboziev I, et al. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am J Physiol Gastrointest Liver Physiol. 2009;296:G135–G146. doi: 10.1152/ajpgi.90462.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okamoto S, Watanabe M, Yamazaki M, et al. A synthetic mimetic of CD4 is able to suppress disease in a rodent model of immune colitis. Eur J Immunol. 1999;29:355–366. doi: 10.1002/(SICI)1521-4141(199901)29:01<355::AID-IMMU355>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Augustin O, Merlos M, Zarzuelo A, et al. Disturbances in metabolic, transport and structural genes in experimental colonic inflammation in the rat: a longitudinal genomic analysis. BMC Genomics. 2008;9:490. doi: 10.1186/1471-2164-9-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang K, Bruce M, Pattillo CB, et al. Temporal genomewide expression profiling of DSS colitis reveals novel inflammatory and angiogenesis genes similar to ulcerative colitis. Physiol Genomics. 43:43–56. doi: 10.1152/physiolgenomics.00138.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chidlow JH, Jr, Langston W, Greer JJ, et al. Differential angiogenic regulation of experimental colitis. Am J Pathol. 2006;169:2014–2030. doi: 10.2353/ajpath.2006.051021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carey R, Jurickova I, Ballard E, et al. Activation of an IL-6:STAT3-dependent transcriptome in pediatric-onset inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:446–457. doi: 10.1002/ibd.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irizarry KJ. Bioinformatics detection and analysis of single nucleotide polymorphisms in the coding regions of the human genome. UCLA. 2003;2003:xviii. 153 leaves. [Google Scholar]
- 11.Kuwata H, Matsumoto M, Atarashi K, et al. IkappaBNS inhibits induction of a subset of Toll-like receptor-dependent genes and limits inflammation. Immunity. 2006;24:41–51. doi: 10.1016/j.immuni.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Raices RM, Kannan Y, Bellamkonda-Athmaram V, et al. A novel role for IkappaBzeta in the regulation of IFNgamma production. PLoS One. 2009;4:e6776. doi: 10.1371/journal.pone.0006776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galamb O, Gyorffy B, Sipos F, et al. Inflammation, adenoma and cancer: objective classification of colon biopsy specimens with gene expression signature. Dis Markers. 2008;25:1–16. doi: 10.1155/2008/586721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones RA, Kotsakis P, Johnson TS, et al. Matrix changes induced by transglutaminase 2 lead to inhibition of angiogenesis and tumor growth. Cell Death Differ. 2006;13:1442–1453. doi: 10.1038/sj.cdd.4401816. [DOI] [PubMed] [Google Scholar]
- 15.Danese S, Sans M, de la Motte C, et al. Angiogenesis as a novel component of inflammatory bowel disease pathogenesis. Gastroenterology. 2006;130:2060–2073. doi: 10.1053/j.gastro.2006.03.054. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda K, Nakano R, Uraoka M, et al. Identification of ARIA regulating endothelial apoptosis and angiogenesis by modulating proteasomal degradation of cIAP-1 and cIAP-2. Proc Natl Acad Sci U S A. 2009;106:8227–8232. doi: 10.1073/pnas.0806780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verma A, Bhattacharya R, Remadevi I, et al. Endothelial cell-specific chemotaxis receptor (ecscr) promotes angioblast migration during vasculogenesis and enhances VEGF receptor sensitivity. Blood. 115:4614–4622. doi: 10.1182/blood-2009-10-248856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otani A, Slike BM, Dorrell MI, et al. A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc Natl Acad Sci U S A. 2002;99:178–183. doi: 10.1073/pnas.012601899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tzima E, Reader JS, Irani-Tehrani M, et al. Biologically active fragment of a human tRNA synthetase inhibits fluid shear stress-activated responses of endothelial cells. Proc Natl Acad Sci U S A. 2003;100:14903–14907. doi: 10.1073/pnas.2436330100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallgard E, Larsson E, He L, et al. Identification of a core set of 58 gene transcripts with broad and specific expression in the microvasculature. Arterioscler Thromb Vasc Biol. 2008;28:1469–1476. doi: 10.1161/ATVBAHA.108.165738. [DOI] [PubMed] [Google Scholar]
- 21.Ichikawa-Shindo Y, Sakurai T, Kamiyoshi A, et al. The GPCR modulator protein RAMP2 is essential for angiogenesis and vascular integrity. J Clin Invest. 2008;118:29–39. doi: 10.1172/JCI33022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onnie CM, Fisher SA, Pattni R, et al. Associations of allelic variants of the multidrug resistance gene (ABCB1 or MDR1) and inflammatory bowel disease and their effects on disease behavior: a case-control and meta-analysis study. Inflamm Bowel Dis. 2006;12:263–271. doi: 10.1097/01.MIB.0000209791.98866.ba. [DOI] [PubMed] [Google Scholar]
- 23.Wu F, Dassopoulos T, Cope L, et al. Genome-wide gene expression differences in Crohn's disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13:807–821. doi: 10.1002/ibd.20110. [DOI] [PubMed] [Google Scholar]
- 24.Baek JY, Jun DY, Taub D, et al. Characterization of human phosphoserine aminotransferase involved in the phosphorylated pathway of L-serine biosynthesis. Biochem J. 2003;373:191–200. doi: 10.1042/BJ20030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vie N, Copois V, Bascoul-Mollevi C, et al. Overexpression of phosphoserine aminotransferase PSAT1 stimulates cell growth and increases chemoresistance of colon cancer cells. Mol Cancer. 2008;7:14. doi: 10.1186/1476-4598-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grasberger H, Refetoff S. Identification of the maturation factor for dual oxidase. Evolution of an eukaryotic operon equivalent. J Biol Chem. 2006;281:18269–18272. doi: 10.1074/jbc.C600095200. [DOI] [PubMed] [Google Scholar]
- 27.Song HJ, Poy G, Darwiche N, et al. Mouse Sprr2 genes: a clustered family of genes showing differential expression in epithelial tissues. Genomics. 1999;55:28–42. doi: 10.1006/geno.1998.5607. [DOI] [PubMed] [Google Scholar]
- 28.Patel S, Kartasova T, Segre JA. Mouse Sprr locus: a tandem array of coordinately regulated genes. Mamm Genome. 2003;14:140–148. doi: 10.1007/s00335-002-2205-4. [DOI] [PubMed] [Google Scholar]
- 29.Koch PJ, de Viragh PA, Scharer E, et al. Lessons from loricrin-deficient mice: compensatory mechanisms maintaining skin barrier function in the absence of a major cornified envelope protein. J Cell Biol. 2000;151:389–400. doi: 10.1083/jcb.151.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clayburgh DR, Shen L, Turner JR. A porous defense: the leaky epithelial barrier in intestinal disease. Lab Invest. 2004;84:282–291. doi: 10.1038/labinvest.3700050. [DOI] [PubMed] [Google Scholar]
- 31.Cai S, Batra S, Lira SA, et al. CXCL1 regulates pulmonary host defense to Klebsiella Infection via CXCL2, CXCL5, NF-kappaB, and MAPKs. J Immunol. 2010;185:6214–6225. doi: 10.4049/jimmunol.0903843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendez Samperio P, Trejo A, Miranda E. Role of type I interferon in the bacillus Calmette-Guerin-induced expression of CXCL10 from human monocytes. Mediators Inflamm. 2004;13:343–348. doi: 10.1155/S096293510400050X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amatschek S, Lucas R, Eger A, et al. CXCL9 induces chemotaxis, chemorepulsion and endothelial barrier disruption through CXCR3-mediated activation of melanoma cells. Br J Cancer. 104:469–479. doi: 10.1038/sj.bjc.6606056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.