Abstract
Irritable bowel syndrome (IBS) is a multifactorial functional disorder with no clearly defined etiology or pathophysiology. Modern culture-independent techniques have improved the understanding of the gut microbiota’s composition and demonstrated that an altered gut microbiota profile might be found in at least some subgroups of IBS patients. Research on IBS from a microbial perspective is gaining momentum and advancing. This review will therefore highlight potential links between the gut microbiota and IBS by discussing the current knowledge of the gut microbiota; it will also illustrate bacterial-host interactions and how alterations to these interactions could exacerbate, induce or even help alleviate IBS.
Keywords: Irritable bowel syndrome, Microbiota, Immunity, Dysbiosis, Probiotics
INTRODUCTION
Patients with functional bowel disorders (FBDs) have no clear structural or biochemical alterations on routine examinations, making diagnosis and treatment challenging. A number of FBDs affect the lower gastrointestinal (GI) tract with irritable bowel syndrome (IBS) being the most prevalent, affecting approximately 10% to 20% of the population in the Western world.1–3 IBS is characterized by abdominal discomfort or pain associated with disturbed bowel habits,4 but also other GI symptoms such as distension and bloating, with patients often reporting more stress and anxiety than the general population.5 Recent data supports the notion that there is a link between bacterial composition and gut wellbeing, therefore this review article will focus on gut microbiota in relation to IBS.
THE IMPORTANCE OF MICROBIOTA FOR GUT HOMEOSTASIS
Regarded by some as a neglected organ,6 the GI microbiota comprises around 400 species and greatly outnumbers the cell count of all other established organs combined.7,8 Commensal bacteria are seen to be necessary in healthy digestion, with roles such as producing enzymes and metabolites which help the body absorb otherwise unavailable essential nutrients and vitamins.9,10 Presence of bacteria is also important for normal development and function of the intestinal immune system which must be both tolerant to food antigens and commensal bacteria, but also able to mount a response to pathogens.11,12 Commensal bacteria also contribute to the maintenance of gut homeostasis by the secretion of bacteriocins,13 proteins that are able to inhibit bacterial toxins,14 and the pH lowering short-chain fatty acids15–17 which withhold an aggressive defence against colonization by noncommensal intruders. Finally, by outcompeting for resources and filling distinct colonization niches,18 commensal microbiota are able to block pathogenic organisms from gaining an all-important foothold in the intestinal microbiota ecosystem.
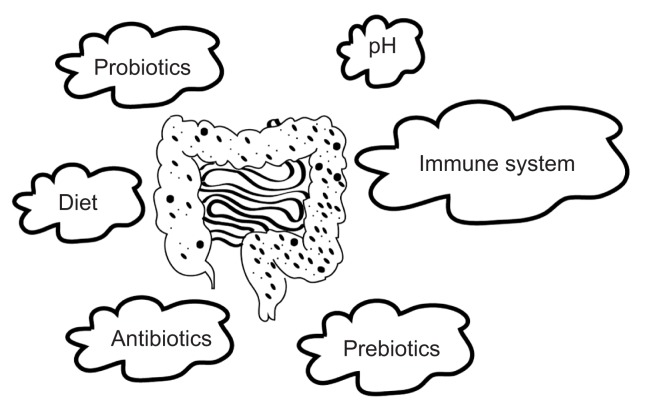
Generally, the intestinal microbiota composition of healthy individuals is relatively stable; however, changes in the microbiota community may lead to a permanent imbalance known as dysbiosis.19 Several factors, such as antibiotics, diet (including specific probiotic and prebiotic consumption), the host immune system and acidic milieu have been seen to affect the microbiota composition of the gut (Fig. 1). Disturbances to the gut microbiota ecosystem resulting in dysbiosis can lead to maladies of the GI tract20,21 with current research suggesting dysbiosis to have potential significance in IBS, but also other conditions such as obesity,22,23 diabetes,24,25 metabolic syndrome,26 cardiovascular disease,27 and IBD.28
Fig. 1.
Factors that might influence the composition of the gut microbiota. Consumption of antibiotics, prebiotics, and probiotics as well as dietary habits have been shown to affect the species of microbiota residing in the gut. These factors will generally dictate which species will become more prevalent. If an expansion of beneficial species occurs through probiotic or prebiotic use, the local pH level is likely to be altered by the beneficial commensals as a means to hamper pathogenic proliferation. Detrimentally, a bout of gastroenteritis in itself alerts the immune system which employs means to remove the offending species. However, the administration of antibiotics in an attempt to solve the problem has potential side effects by depleting levels of commensal microbiota, thus resulting in an opening for nonbeneficial microbiota to establish themselves.
SMALL INTESTINAL BACTERIAL OVERGROWTH IN IBS
In a healthy individual the small intestine contains a much lower density of bacteria than the large intestine. IBS has been suggested to be associated with small intestinal bacterial overgrowth (SIBO), defined as a bacterial density (colonic bacteria) of ≥105 colony-forming units (cfu) per mL of intestinal fluid,29 measured by the “gold standard” jejunal culture method.30,31 SIBO is, however, often diagnosed through culture-independent techniques such as glucose hydrogen and lactulose hydrogen breath tests.32–36 There has been much deliberation over these studies and the findings due to the validity of the techniques used.21,37,38 A meta-analysis study by Ford et al.35 concluded that although SIBO was seen in IBS patients at a higher rate when compared to controls, the prevalence to which this occurred varied substantially between studies and centres. Due to such discrepancy, the true influence SIBO has with regards to IBS symptom generation is still unclear.35,39
POSTINFECTIOUS IBS
Postinfectious IBS (PI-IBS) is likely the strongest evidence in the case of microbiota being important for the development of IBS, and may present after a bout of gastroenteritis caused by viral, parasitic or bacterial infections. Enteric pathogens such as Staphylococcus aureus, Shigella, Clostridium perfringens, Bacillus cereus, and Campylobacter species are potential culprits of PI-IBS,40–43 and could increase the risk of developing PI-IBS by at least six-fold.44–46 Other variables such as severity and duration of initial infection, as well as age and gender are additional risk factors of developing PI-IBS, with risk of development being higher amongst young females.
An episode of gastroenteritis will cause an inflammatory response of the gut, and may potentially lead to an intestinal dysbiosis. For example, C. jejuni and Shigella infections47,48 cause detrimental depletion of intestinal macrophages, which could potentially hamper the clearance of pathogens. Additionally, enterohemorrhagic E. coli have deleterious impacts on the epithelial barrier,49,50 which raise the risk for bacterial breach to the epithelial mucosa and subsequent inflammation.51 Thus, a previous gut infection may temporally cause changes to the immune system as well as the balance of the gut microbiota, resulting in an opening for nonbeneficial microbiota to establish themselves.
MICROBIOTA AND IMMUNITY IN IBS
The interplay of intestinal microbes and host immunity being widely acknowledged in promoting a normally functioning immune system7,12 is strengthened with studies suggesting that an altered gut microbiota composition may lead to an altered immune activity,52 potentially leading to low-grade inflammation in IBS.53–55 The putatively increased immune activity in IBS patients56 may be due to exogenous or endogenous triggers; however, the immune activity pattern of IBS is far from being fully understood and likely involves both the innate and adaptive immunity.
1. The innate immune system
Having a fundamental role in the innate immune system, mast cells are on the front line barrier between the host and the external environment. Numerous studies have reported an increase in number, level of activation and area occupied by mast cells in the intestinal mucosa of IBS patients when compared with healthy controls57–67 although other studies are not in agreement.68–71 These discrepancies potentially arise through methodological inconsistencies such as from which intestinal region the biopsy was taken from and the techniques used for detection and counting of the cells. Considering how intertwined the gut microbiota and immune system are72 and the reports of increase in mast cell numbers in IBS, it could be reasoned that a shift in gut microbiota composition could mediate an immune response leading to a subsequent mast cell increase, potentially contributing to visceral hypersensitivity.70 However, there is still no evidence supporting an association between an altered microbiota composition and levels of mast cells in IBS, so this putative link remains to be determined.
Another aspect of innate immunity is the phagocytic macrophages. Currently, the number of macrophages is still under discussion with reports of increased47,73 and decreased68 levels of this cell population seen in IBS. Potentially, for a subgroup of IBS patients, it could be proposed that the immune system may be compromised and therefore less capable to respond to pathogenic microorganisms. This theory is supported by the report of decreased levels of secreted chemoattractants such as CXCL-9 and MCP-1,74 known to recruit dendritic cells and other immune cells. However, data on the expression of intestinal chemoattractants is also under discussion as an elevation of MCP-1 has also been reported in IBS.75 Moreover, increased levels of proinflammatory serum cytokines such as interleukin (IL)-6, IL-8, and tumour necrosis factor α, tend to be found in IBS patients,61,75–79 once again evoking the idea of discord in the gut microbiota subsequently provoking an immune response to tackle any pathogenically caused disturbances.
Widely expressed by many cells, including but not limited to epithelial cells and macrophages are Toll-like receptors (TLRs). TLRs are utilized in the recognition of danger signals such as structures found on bacteria and viruses. Studies showing altered TLR expression in IBS patients compared to healthy controls80 with specific increases in TLR2, TLR4, and TLR581–83 and decreases in TLR7 and TLR8,81 present the notion that IBS is linked to an altered activation of the immune system in response to microorganisms of the gut. Also, defensins such as human β-defensin 2, antimicrobial peptides secreted by colon epithelial cells in response to proinflammatory cytokines or pathogenic microorganisms, have been shown to be increased in IBS patients.84
2. The adaptive immune system
Following the hypothesis of a low-grade immune activation occurring in IBS, the T cells of the adaptive immune system have been suggested to have an increased presence in IBS patients47,59,85–87 and subgroups such as PI-IBS;88 however, a contradictory study by Braak et al.68 suggests T cells to be decreased in IBS patients. Similar to an increase in activity of the innate immune system regarding IBS, an increased presence or activation of T cells may contribute to symptom generation. Therefore, the increased frequencies of blood T cells expressing activation markers such as CD69 and HLA-DR as well as the gut homing integrin a4β7, suggests that IBS patients display an activated T cell phenotype87 compared to healthy controls. Further, an increase in serum antibodies against flagellin has been seen in PI-IBS compared to non-PI-IBS and healthy controls.89,90 Also, our group has reported comparable levels of B cells in blood of IBS patients and healthy controls, although the frequency of B cells expressing IgG, indicating an activated status of the B cells, was increased in the patients.91 Furthermore, after bacterial cocktail stimulation of B cells, an impaired expression of costimulatory ligand CD80, thought to lead to restriction of T cell activation,92 was seen. This suggests that an altered B cell expression of costimulatory molecules could be a potential cause for the increased T cell numbers reported in IBS patients.91
Still, the evidence linking an increased immune activity and altered gut microbiota composition in IBS is limited93 and further research is therefore required.
GUT MICROBIOTA RELATED THERAPY AND TREATMENT OF IBS
1. Antibiotics
There is no clear consensus on the benefit of antibiotics to treat symptoms in IBS; however, several studies show that antibiotics alter the composition of the gut microbiota in a potentially deleterious way.94–97 The reported disruptive effect of antibiotics can diminish protective commensal bacteria populations, making it more likely for expansion of pathogenic species to occur,98,99 which may lead to dysbiosis and potentially even cause symptoms of IBS.100–103 However, nonabsorbable antibiotics such as neomycin104 and rifaximin105–107 have been seen to have beneficial effects, providing partial alleviation of IBS in general and bloating in particular. Findings showing reduction in IBS symptoms through the use of antibiotics further support the influence microbiota has on gut wellbeing and how the restoration of intestinal microbial normobiosis may help some patients with IBS.
2. Probiotics
The act of directly altering gut microbiota composition through the use of probiotics such as Bifidobacterium spp. and Lactobacillus spp.108 has been shown to have a positive effect on the symptoms of IBS,109–111 though this is not always the case.112–114 Most studies report no adverse events with the use of probiotics; however, these claims are often not well documented and inconsistent, with some studies even recording worsening of patient discomfort.115–117 Although probiotics may be beneficial in a subset of IBS patients, further research is required in order to elucidate the full efficacy as well as potential side effects, relevant strain or species cocktail118 and optimal dose in order to gain the full therapeutic effects in IBS patients.21,119
3. Prebiotics
If probiotics are like adding seeds of beneficial bacteria to the gut, prebiotics are the equivalent of fertilizers which affect only the favorable species already colonising the bowels. Commonly used and tested prebiotics are nondigestible oligosaccharides such as fructo-oligosaccharides and galacto-oligosaccharides. Currently, there have not been many randomized controlled trials regarding IBS and prebiotics. Supporting the link between microbiota composition and gut wellbeing, studies on prebiotic use and subsequent mitigation of IBS symptoms tend to show a similar beneficial effect as the use of probiotic strains such as Bifidobacterium spp. and Lactobacillus spp.120–124 Findings from the aforementioned studies show that the benefit from such treatment is strongly dependant on dose, whereby high levels of prebiotics were revealed to counteractively intensify problems such as bloating and flatulence.121 These results corroborate with up and coming research recommending a reduction of FOD-MAPs (fermentable oligosaccharides, disaccharides, monosaccharides, and polyols) intake for people suffering with IBS.125,126
4. Diet
The composition of the gut microbiota has been shown to be responsive and adaptable to the diet of the host organism.127–131 Since anything not absorbed by the host becomes a source of nutrients for the microbial community residing in the colon, differences have been seen in gut microbiota in favor of those species able to best adapt to and metabolise the primary nutrients like fat, protein, or carbohydrates present in the diet.132 This adaptability thus denotes the types and levels of metabolites produced, e.g., butyrate128 or methane133 and consequent promotion of gut health or IBS symptoms, respectively. Although there is no recommended IBS diet, a reduction in FODMAPs might subdue symptoms associated with IBS.134–137
In conclusion, the findings with regards to therapeutic methods for IBS, e.g., antibiotics, probiotics and prebiotics and restoration of the gut microbiota, adds to the rationale behind the suggested correlation of altered gut microbiota and IBS33,51,81,107,111,123,128,137 whereby treatment of the dysbiosis may help a subgroup of patients.
GUT MICROBIOTA AND ITS ASSOCIATION TO IBS
With increasing numbers of studies focusing on investigating the gut microbiota profile, data suggests that there is an altered diversity of gut microbiota in patients with IBS.138–140 Several studies suggest that fecal microbiota is altered in IBS, and present differences in microbiota composition between healthy controls and IBS patients as well as within the subgroups of IBS patients.141–146 Although the aforementioned endeavours141–146 are interesting and a definite step towards a better understanding of IBS from a microbiological point of view, it must be taken into consideration that the results are usually based on relatively small sample populations. Considering that IBS is a multifactorial disorder with many putative causes and broad symptom presentation, a conjecture could be made that results derived from these studies might not represent the IBS patient population as a whole, but rather a subgroup of patients. Also, microbiota can vary quite extensively even between healthy individuals, making a general inference on the microbiota composition of IBS, let alone of the subgroups, a difficult one.
The most easily obtainable material when sampling GI microbiota is fecal matter. For this reason it is used prevalently in gut microbiota research, as well as the less easily obtainable mucosal biopsies. Taken from various locations of the small and large intestine, biopsies can provide a more site specific view of the mucosa adherent microbes inhabiting the gut.
CURRENT KNOWLEDGE ABOUT MICROBIOTA COMPOSITION OF PATIENTS WITH IBS
Today, over 50 bacterial phyla have been defined147 with only 29 of these possible to culture.148 Ten of the known phyla have been discovered in the human gut149 with the majority of species attributed to one of the two largest phyla colonizing the human gut, Firmicutes and Bacteroidetes. Notably, since the advent and prominent use of culture-independent methods such as 16S sequencing in the last decade, numerous studies have focused on determining the gut microbiota in IBS patients. Table 1 gives a brief overview of the current findings on gut microbiota composition in patients with IBS in comparison to healthy controls with a more detailed review presented below.
Table 1.
Overview of Papers Studying the Microbiota of the Gut with Regards to Irritable Bowel Syndrome
| Phyla | Genus | IBS against healthy |
|---|---|---|
| Firmicutes | Lactobacillus | → Tana (2010)154; Carroll (2010)153; Carroll (2011)139 ↓ Balsari (1982)152; Kassinen (2007)141 ↔ Si (2004)158; Malinen (2005)159; Kerckhoffs (2009)160; Rajilić-Stojanović (2011)161 |
| Streptococcus spp. | → Kassinen (2007)141; Rajilić-Stojanović (2011)161 | |
| Dorea spp. | → Saulnier (2011)165; Rajilić-Stojanović (2011)161; Kassinen (2007)141 | |
| Ruminococcus spp. | → Kassinen (2007)141; Rajilić-Stojanović (2011)161; Jalanka-Tuovinen (2014)142 | |
| Eubacterium | ↔ Rajilić-Stojanović (2011)161 | |
| Faecalibacterium | ↔ Lopez-Siles (2014)175 | |
| Actinobacteria | Bifidobacterium | → Kassinen (2007)141 ↔ Balsari (1982)152; Si (2004)158; Kerckhoffs (2009)160; Rajilić-Stojanović (2011)161; Malinen (2005)159; Duboc (2012)173 |
| Bacteroidetes | Bacteroides spp. | → Jalanka-Tuovinen (2014)142 ↓ Compared to IBD Swidsinski (2005)190 |
| Proteobacteria | Escherichia | → Rajilić-Stojanović (2011)161; Duboc (2012)173; Si (2004)158 |
| Desulfovibrio spp. | ↓ Malinen (2005)159 | |
| Verrucomicrobia | Akkermansia | → Saulnier (2011)165 ↔ Rajilić-Stojanović (2011)161 |
| Euryarchaeota | Methanobrevibacter | ↓ Rajilić-Stojanović (2011)161; Jalanka-Tuovinen (2014)142; Rana (2009)187 |
1. Firmicutes
The most predominant phylum found in the gut microbiota of healthy individuals,150,151 the gram positive Firmicutes, consist of a number of genera which include the commonly known probiotic, Lactobacillus spp.108 As early as 1982152 studies have presented lowered levels of Lactobacillus spp. within IBS patients compared to healthy controls. However, studies are inconsistent in their results, presenting an increase,52,139,153–156 decrease,141 or no change157–161 in the prevalence of lactobacilli spp. within IBS patients. Also, comparisons between IBS subgroups and healthy controls show either decreased or normal abundance of Lactobacillus159,160 in the patients, thus it is still unclear if there are any IBS-subgroup-specific related effects of Lactobacillus. Regardless if Lactobacillus is occurring naturally or from probiotic administration its ability in alleviating symptoms of IBS such as visceral pain108,162 make probiotics, i.e., Lactobacillus, to be of potential benefit to some IBS patients.
Conversely, there might be a positive correlation between some potentially pathogenic species within the phylum of Firmicutes, such as Streptococcus spp. and IL-6 increase in IBS patients.142 This finding makes the significantly elevated levels of Streptococcus spp. seen in IBS patients141,161 particularly interesting and could potentially serve as evidence of the link between dysbiosis, occurring primarily from specific bacterial genera colonisation, and an altered immune activity in some IBS patients.
Within the Clostridia class, conflicting results have been reported with both an observed increase163 and decrease142,155,164 in IBS patients. Specifically, Ruminococcus spp. as well as Dorea spp., have both been shown to be increased in patients suffering with IBS,141,161,165 although no subgroup preferences have been defined. Interestingly, Rajilić-Stojanović et al.161 suggest the nonbutyrate producing phylotypes of Clostridium Group XIVa, related to R. gnavus and R. torques, known mucin degraders,166 to be indicators of IBS. These phylotypes are consistently found at elevated levels in IBS fecal samples,165 and also found in higher abundance in diarrhea predominant IBS167 and PI-IBS,142 conversely, however, in lower abundance in alternating IBS.161
Unlike R. gnavus and R. torques other members of Group XIVa, e.g., Roseburia spp.,168 produce beneficial short-chain fatty acids, such as butyrate. Butyrate is a preferred energy source for colonic epithelial cells and is suggested to reduce inflammation.169 Moreover, butyrate helps to maintain normal intestinal barrier function, through regulation of colon epithelial mucin gene MUC2170 and tight junction proteins,171 respectively, and might therefore have therapeutic effects in IBS patients.172 Although inconsistent, reduced levels in IBS patients141,161,173–175 of the butyrate producers Eubacterium, Faecalibacterium and Roseburia spp., known to inhibit the growth of potentially pathogenic species including Campylobacter spp., Salmonella spp., Shigella spp., and E. coli,176 could potentially be an ancillary cause for IBS symptom generation in some patients.
At this time, there is no clear consensus on the significance of alterations of Firmicutes in all IBS patients, although evidence suggests that Firmicutes, specifically the family Lachnospiraceae, are increased significantly enough in IBS-D as to make it discernible from other IBS subgroups.143 Nevertheless, although no consensus has been agreed upon, a weak tendency for a reduction in the beneficial bacteria of the gut, countered with an increase in pathogenic species is seen in IBS patients. This dysbiosis may potentially have influence on gut function whereby a degradation of the mucus layer by Ruminococcus spp. may allow infiltration of Streptococcus spp. or Staphylococcus aureus,43 thus provoking low-grade immune responses in a subgroup of IBS patients.177
2. Bacteroidetes
The second most abundant phylum in the human gut, gram negative Bacteroidetes are found to have a varying higher52,142,163 or lower143,145,161,178 presence and diversity in the gut microbiota of patients with IBS. Furthermore, increases in Bacteroides spp. have been reported in IBS142 although a net decrease in the Bacteroidetes phylum has also been observed.141 Our group has demonstrated that a majority of IBS patients had an altered microbiota composition with an increased abundance of Firmicutes and subsequent decrease in Bacteroidetes, whereas the remaining patients had a normal-type gut microbiota composition.145
The Bacteroidetes phylum harbours species with either beneficial or nonbeneficial traits, as shown in the comprehensive review by Wexler.179 The abundance of beneficial or nonbeneficial Bacteroidetes species may therefore be important to IBS, under the assumption that more nonbeneficial species might correlate with an increase in symptoms or severity, such as visceral pain.
3. Actinobacteria
Another of the main phyla of the human gut microbiota, the gram positive Actinobacteria includes the probiotic containing genera such as Bifidobacterium and Collinsella. Interestingly a decrease in Actinobacteria has been shown in the gut of patients with IBS161 with one study presenting a specific decrease among IBS-D patients.143 Notably, the reports on levels of the ubiquitous and recognised probiotic of the GI tract, Bifidobacterium spp., known to aid the gut mucosal barrier,180 are mixed. IBS patients are demonstrated to have increased141,152,158,160,161 or reduced156,159,173 levels of bifidobacteria. Although a specific reduction of B. catenulatum has been seen in IBS in some studies,159,160 Rajilić-Stojanović et al.161 suggested that it is in fact B. pseudocatenulatum which is significantly reduced in IBS. Some studies propose an increase in bifidobacteria in IBS; however, the overall inconsistency suggests that there may potentially be a subgroup of IBS patients where the beneficial traits of these probiotic species might be diminished which could account for symptom generation, this requires further research.
4. Proteobacteria
Increases of Proteobacteria165 in IBS, notably IBS-D,143 of the specific family Enterobacteriaceae155,158,161,173 which encompasses many gram negative pathogenic species, including other coliform bacteria157 such as E. coli159 with potential inflammation causing mechanisms, have been associated with IBS. Nevertheless, Malinen et al.159 recorded lower amounts of sulphate-reducing bacteria, Desulfovibrio spp. in IBS-D patients. When considering that species in this genus produce toxic sulphide, one would expect an increased abundance of Desulfovibrio spp., rather than reduced levels, as a plausible explanation for symptom generation in some IBS patients.
Since being found in both healthy controls159 and IBS patients43 there is no consensus on altered abundance of known, potentially pathogenic, genera such as Campylobacter spp. and Helicobacter spp. within IBS. Looking at the findings of pathogenic Proteobacteria alone, one could infer that an increase in pathogens could be a single element in the onset of symptoms in a subgroup of IBS patients. Thus, the increase of pathogenic species together with the previously mentioned reduction in probiotic species from genera such as Bifidobacterium and Lactobacillus and their metabolites, known to keep pathogens at bay, further support that a dysbiosis is occurring in a number of IBS patients.
5. Verrucomicrobia
Increased levels of the not so well documented Akkermansia spp.,161 may coincide with IBS. Since its known specialization for mucus degradation181 Akkermansia spp. could compromise the integrity of the mucus layer, and thus hamper the intestinal barrier in the gut. From this assumption, Akkermansia spp. may instigate a low grade inflammation in some IBS patients through degradation of the mucus layer, similar to R. gnavus and R. torques, enabling entry of pathogenic species to the epithelial mucosa.
6. Euryarchaeota
The methane generating archaea Methanogens convert hydrogen produced in the gut into methane. This gas was previously thought to be inert,182 but has now been shown to reduce gut transit.133,183 An increase of Methanogens in IBS, especially in those suffering of constipation predominant IBS, could possibly explain the slow gut transit in these patients and why methane is being found at increased volumes in IBS-C patients.184–186 Also, Methanobrevibacter smithii has been suggested to be the predominant Methanogen in IBS-C.186 These findings support the proposition that at least in some IBS-C patients a dysbiosis favoring Methanogens is potentially occurring which may be causing constipation. However, it might also be hypothesised that a reduction of Methanogens could potentially explain bloating symptoms in IBS, as hydrogen levels would not be reduced as efficiently. Studies which include Methanogens in their research present abundant levels of Methanogens in both healthy controls and IBS patients.142,161,185,187 Since an abundance of Methanogens is found also in healthy controls, further research is needed to explore the importance of Methanogens for generating IBS symptoms.
DIVERSITY AND LONG-TERM INSTABILITY OF GUT MICROBIOTA IN IBS
A temporal decreased stability of gut microbiota leading to a state of flux and dysbiosis may provide an explanation for the characteristic symptoms of certain IBS subgroups. The gut microbiota has the potential to affect and also be affected by the physiology of the gut, thus there is bidirectional communication. There are many factors which can potentially alter the normal gut microbiota composition. The use of antibiotics to treat infection is likely to have the side effect of depleting commensal bacterial, thus unintentionally allowing for later pathogenic infiltration of the gut. The consumption of probiotics and prebiotics, however, serve to increase beneficial bacteria of the gut. These beneficial bacteria employ various mechanisms, such as pH regulation, which alters their surroundings as to hinder the growth of noncommensals. The immune system works constantly in order to keep microbial homeostasis. Through this maintenance, alterations to the gut microbiota community occur by removing potentially pathogenic species. Additionally, diet can shape the composition of the gut microbiota through the shift in bacterial species which occurs when a gradual or radical change occurs in the food consumed by the host (Fig. 1). As levels of bacterial species fluctuate to adapt to these changes the shift may eventually favor certain species whereby a subsequent population expansion would likely occur. For example, an expansion of Methanogens in IBS could be linked to symptoms of IBS-C.185 A few studies have investigated gut microbiota stability through DNA157 and RNA analysis,164 showing that IBS patients have an instability of the gut microbiota composition over time as compared to healthy controls.157 Importantly, it must be acknowledged that the temporal fluctuation in at least some IBS patients, and healthy individuals for that matter, may be partially attributed to the administration of antibiotics.157
Several studies have demonstrated that IBS patients may have a diminished diversity of the gut microbiota composition,139,155,178,188 although, when focusing on specific groups such as Bacteroidetes and Lactobacillus, a broader diversity has been observed in IBS.52 In the gut microbiota ecosystem, much like in any other ecosystem, diversity and species richness is required for the system to flourish, where by all niches are filled and kept in check by neighboring beneficial or competing species. However, further studies with larger cohorts and potentially longer time periods are required in order to further investigate gut microbiota composition, instability and diversity in IBS.
GUT MICROBIOTA AND THE LINK TO IBS SYMPTOMS
Patients with IBS suffer from symptoms such as pain, constipation, diarrhea, abdominal distension, bloating and even psychiatric problems. The extent to which the gut microbiota influences these symptoms is not fully understood, especially not mechanistically. Interestingly, negative correlation between beneficial species of the gut and IBS symptoms have been demonstrated163 with Rajilić-Stojanović et al.161 reporting a reduction in Faecalibacterium spp., producing the anti-inflammatory metabolite butyrate, being associated with an increase in IBS symptoms. Conversely, yet equally as expectable, was a positive correlation between various Firmicute and Proteobacteria species and IBS symptoms.161 Also, an increase of R. torques has been associated with increased symptom severity such as emotional function, social function, systemic symptoms and bowel symptoms.177 Concordantly, IBS patients with R. torques in their fecal samples tended to present more frequently with self-reported symptoms.177 Moreover, our group has previously discovered that psychological symptoms such as clinically significant depression had a lower prevalence in patients with an altered microbiota profile, i.e., an increase of Firmicutes and subsequent decrease in Bacteroidetes, compared to those with normal microbiota composition.145 Additionally, Parkes et al.163 links altered, though primarily lower, bacterial mass and diversity to increased symptom severity, related to stool frequency, anxiety and pain. Although not fully understood, studies in rats have suggested that fecal bacteria, through specific bacterial metabolites such as sulphides, induce visceral hypersensitivity, which is often demonstrated in IBS patients.189 In conclusion, although gut microbiota might explain symptom generation in only a subgroup of IBS patients, this nevertheless helps towards a richer understanding of how symptoms of IBS might occur.
SUMMARY
Over the past decade the importance of gut microbiota in IBS has drawn increasing attention. Growing evidence suggest that at least subgroups of IBS patients have an altered gut microbiota composition or dysbiosis. Presented as an altered balance in beneficial or pathogenic bacterial species, dysbiosis is thought to have a bigger impact on gut wellbeing in IBS patients than previously thought, affecting such processes as intestinal barrier function and immune system regulation (Fig. 2). Therefore the use of therapeutic methods which interact with the microbiota continue to be an interesting option to both increase efficacy in hampering the growth of unwanted species whilst promoting beneficial bacteria in IBS. Improved understanding of the microbiota in respect to IBS may guide future therapeutic strategies with focus on the modulation of gut microbiota composition.
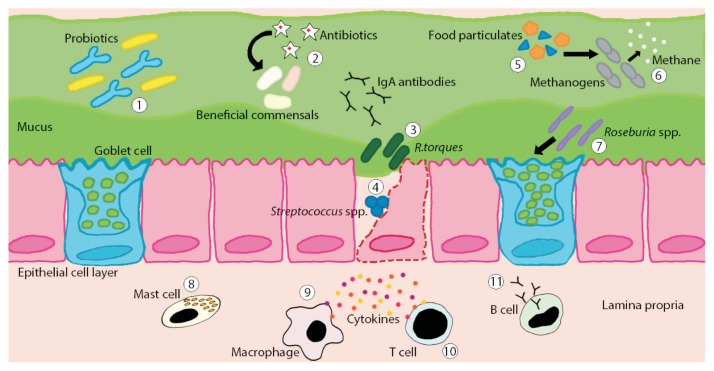
Fig. 2.
Illustration of host-microbiota interactions in the gut of irritable bowel syndrome (IBS) patients. Often recorded at lower levels in IBS patients, therapeutic administration of probiotic species such as Bifidobacterium spp. and Lactobacillus spp. have been shown to have positive effects on symptoms of IBS through their anti-inflammatory metabolites (1). Antibiotic use can have potential side effects such as depleting levels of beneficial commensal gut microbiota thus opening niches for nonspecific species to establish themselves (2). Species such as R. gnavus and R. torques are mucin degraders which may breach the mucus barrier allowing for potential pathogenic infiltration (3). Potential inflammation causing species including Streptococcus spp. or Staphylococcus aureus may enter into the epithelial layer and provoke an immune response (4). Diet plays a role in gut microbiota composition since nutrients not absorbed by the host become energy for both beneficial and nonbeneficial gut microbiota (5). Found to be increased in IBS patients, the nonbeneficial gut microbiota Methanogens produce methane which has been shown to slow down gut transit, potentially leading to constipation (6). Beneficial species such as Roseburia spp. produce butyrate, known to help to maintain normal intestinal barrier function through regulation of colon epithelial mucin gene MUC2, a primary component of mucus (7). A potential intestinal dysbiosis of IBS patients may lead to, or be the result of, an altered activity of the mucosal immune system. Although still under debate, increased density of activated mast cells in the mucosa might provoke symptoms (8). Altered macrophage density or function in IBS patients has been suggested leading to a hampered recognition of pathogenic microbiota (9). Possibly, an increased presence or activation of T cells may contribute to symptom generation (10). Also, higher levels of flagellin specific antibodies, as reported in IBS patients, suggests an increased B cell activity (11).
Footnotes
CONFLICTS OF INTEREST
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Talley NJ, Zinsmeister AR, Van Dyke C, Melton LJ., 3rd Epidemiology of colonic symptoms and the irritable bowel syndrome. Gastroenterology. 1991;101:927–934. doi: 10.1016/0016-5085(91)90717-y. [DOI] [PubMed] [Google Scholar]
- 2.Jones R, Lydeard S. Irritable bowel syndrome in the general population. BMJ. 1992;304:87–90. doi: 10.1136/bmj.304.6819.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drossman DA, Li Z, Andruzzi E, et al. U.S. householder survey of functional gastrointestinal disorders: prevalence, sociodemography, and health impact. Dig Dis Sci. 1993;38:1569–1580. doi: 10.1007/BF01303162. [DOI] [PubMed] [Google Scholar]
- 4.Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 5.Choung RS, Locke GR, 3rd, Zinsmeister AR, Schleck CD, Talley NJ. Psychosocial distress and somatic symptoms in community subjects with irritable bowel syndrome: a psychological component is the rule. Am J Gastroenterol. 2009;104:1772–1779. doi: 10.1038/ajg.2009.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baquero F, Nombela C. The microbiome as a human organ. Clin Microbiol Infect. 2012;18(Suppl 4):2–4. doi: 10.1111/j.1469-0691.2012.03916.x. [DOI] [PubMed] [Google Scholar]
- 7.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 8.Qin J, Li R, Raes J, Arumugam M, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burkholder PR, McVeigh I. Synthesis of vitamins by intestinal bacteria. Proc Natl Acad Sci U S A. 1942;28:285–289. doi: 10.1073/pnas.28.7.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi HN, Walker A. Bacterial colonization and the development of intestinal defences. Can J Gastroenterol. 2004;18:493–500. doi: 10.1155/2004/690421. [DOI] [PubMed] [Google Scholar]
- 13.Hammami R, Fernandez B, Lacroix C, Fliss I. Anti-infective properties of bacteriocins: an update. Cell Mol Life Sci. 2013;70:2947–2967. doi: 10.1007/s00018-012-1202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schamberger GP, Diez-Gonzalez F. Selection of recently isolated colicinogenic Escherichia coli strains inhibitory to Escherichia coli O157:H7. J Food Prot. 2002;65:1381–1387. doi: 10.4315/0362-028x-65.9.1381. [DOI] [PubMed] [Google Scholar]
- 15.Cherrington CA, Hinton M, Pearson GR, Chopra I. Short-chain organic acids at ph 5.0 kill Escherichia coli and Salmonella spp. without causing membrane perturbation. J Appl Bacteriol. 1991;70:161–165. doi: 10.1111/j.1365-2672.1991.tb04442.x. [DOI] [PubMed] [Google Scholar]
- 16.Shin R, Suzuki M, Morishita Y. Influence of intestinal anaerobes and organic acids on the growth of enterohaemorrhagic Escherichia coli O157:H7. J Med Microbiol. 2002;51:201–206. doi: 10.1099/0022-1317-51-3-201. [DOI] [PubMed] [Google Scholar]
- 17.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Momose Y, Hirayama K, Itoh K. Competition for proline between indigenous Escherichia coli and E. coli O157:H7 in gnotobiotic mice associated with infant intestinal microbiota and its contribution to the colonization resistance against E. coli O157:H7. Antonie Van Leeuwenhoek. 2008;94:165–171. doi: 10.1007/s10482-008-9222-6. [DOI] [PubMed] [Google Scholar]
- 19.Metchnikoff E, Mitchell PC. The prolongation of life: optimistic studies. New York: Putnam; 1908. [Google Scholar]
- 20.Ringel Y, Carroll IM. Alterations in the intestinal microbiota and functional bowel symptoms. Gastrointest Endosc Clin N Am. 2009;19:141–150. doi: 10.1016/j.giec.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Simrén M, Barbara G, Flint HJ, et al. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62:159–176. doi: 10.1136/gutjnl-2012-302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 24.Qin J, Li Y, Cai Z, Li S, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 25.Everard A, Cani PD. Diabetes, obesity and gut microbiota. Best Pract Res Clin Gastroenterol. 2013;27:73–83. doi: 10.1016/j.bpg.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 26.D’Aversa F, Tortora A, Ianiro G, Ponziani FR, Annicchiarico BE, Gasbarrini A. Gut microbiota and metabolic syndrome. Intern Emerg Med. 2013;8(Suppl 1):S11–S15. doi: 10.1007/s11739-013-0916-z. [DOI] [PubMed] [Google Scholar]
- 27.Karlsson FH, Fåk F, Nookaew I, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 29.Bardhan PK, Gyr K, Beglinger C, Vögtlin J, Frey R, Vischer W. Diagnosis of bacterial overgrowth after culturing proximal small-bowel aspirate obtained during routine upper gastrointestinal endoscopy. Scand J Gastroenterol. 1992;27:253–256. doi: 10.3109/00365529208999959. [DOI] [PubMed] [Google Scholar]
- 30.Quigley EM, Quera R. Small intestinal bacterial overgrowth: roles of antibiotics, prebiotics, and probiotics. Gastroenterology. 2006;130:S78–S90. doi: 10.1053/j.gastro.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 31.Posserud I, Stotzer PO, Björnsson ES, Abrahamsson H, Simrén M. Small intestinal bacterial overgrowth in patients with irritable bowel syndrome. Gut. 2007;56:802–808. doi: 10.1136/gut.2006.108712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bures J, Cyrany J, Kohoutova D, et al. Small intestinal bacterial overgrowth syndrome. World J Gastroenterol. 2010;16:2978–2990. doi: 10.3748/wjg.v16.i24.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pimentel M, Chow EJ, Lin HC. Eradication of small intestinal bacterial overgrowth reduces symptoms of irritable bowel syndrome. Am J Gastroenterol. 2000;95:3503–3506. doi: 10.1111/j.1572-0241.2000.03368.x. [DOI] [PubMed] [Google Scholar]
- 34.Pimentel M, Chow EJ, Lin HC. Normalization of lactulose breath testing correlates with symptom improvement in irritable bowel syndrome: a double-blind, randomized, placebo-controlled study. Am J Gastroenterol. 2003;98:412–419. doi: 10.1111/j.1572-0241.2003.07234.x. [DOI] [PubMed] [Google Scholar]
- 35.Ford AC, Spiegel BM, Talley NJ, Moayyedi P. Small intestinal bacterial overgrowth in irritable bowel syndrome: systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2009;7:1279–1286. doi: 10.1016/j.cgh.2009.06.031. [DOI] [PubMed] [Google Scholar]
- 36.Ghoshal UC, Park H, Gwee KA. Bugs and irritable bowel syndrome: the good, the bad and the ugly. J Gastroenterol Hepatol. 2010;25:244–251. doi: 10.1111/j.1440-1746.2009.06133.x. [DOI] [PubMed] [Google Scholar]
- 37.Simrén M, Stotzer PO. Use and abuse of hydrogen breath tests. Gut. 2006;55:297–303. doi: 10.1136/gut.2005.075127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanner S. The small intestinal bacterial overgrowth. Irritable bowel syndrome hypothesis: implications for treatment. Gut. 2008;57:1315–1321. doi: 10.1136/gut.2007.133629. [DOI] [PubMed] [Google Scholar]
- 39.Spiegel BM. Questioning the bacterial overgrowth hypothesis of irritable bowel syndrome: an epidemiologic and evolutionary perspective. Clin Gastroenterol Hepatol. 2011;9:461–469. doi: 10.1016/j.cgh.2011.02.030. [DOI] [PubMed] [Google Scholar]
- 40.DuPont AW. Postinfectious irritable bowel syndrome. Clin Infect Dis. 2008;46:594–599. doi: 10.1086/526774. [DOI] [PubMed] [Google Scholar]
- 41.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee BJ, Bak YT. Irritable bowel syndrome, gut microbiota and probiotics. J Neurogastroenterol Motil. 2011;17:252–266. doi: 10.5056/jnm.2011.17.3.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rinttilä T, Lyra A, Krogius-Kurikka L, Palva A. Real-time PCR analysis of enteric pathogens from fecal samples of irritable bowel syndrome subjects. Gut Pathog. 2011;3:6. doi: 10.1186/1757-4749-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halvorson HA, Schlett CD, Riddle MS. Postinfectious irritable bowel syndrome: a meta-analysis. Am J Gastroenterol. 2006;101:1894–1899. doi: 10.1111/j.1572-0241.2006.00654.x. [DOI] [PubMed] [Google Scholar]
- 45.Thabane M, Kottachchi DT, Marshall JK. Systematic review and meta-analysis: the incidence and prognosis of post-infectious irritable bowel syndrome. Aliment Pharmacol Ther. 2007;26:535–544. doi: 10.1111/j.1365-2036.2007.03399.x. [DOI] [PubMed] [Google Scholar]
- 46.Schwille-Kiuntke J, Enck P, Zendler C, et al. Postinfectious irritable bowel syndrome: follow-up of a patient cohort of confirmed cases of bacterial infection with Salmonella or Campylobacter. Neurogastroenterol Motil. 2011;23:e479–e488. doi: 10.1111/j.1365-2982.2011.01779.x. [DOI] [PubMed] [Google Scholar]
- 47.Spiller RC, Jenkins D, Thornley JP, et al. Increased rectal mucosal enteroendocrine cells, T lymphocytes, and increased gut permeability following acute Campylobacter enteritis and in post-dysenteric irritable bowel syndrome. Gut. 2000;47:804–811. doi: 10.1136/gut.47.6.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sasakawa C. A new paradigm of bacteria-gut interplay brought through the study of Shigella. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:229–243. doi: 10.2183/pjab.86.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roxas JL, Koutsouris A, Bellmeyer A, et al. Enterohemorrhagic E. coli alters murine intestinal epithelial tight junction protein expression and barrier function in a Shiga toxin independent manner. Lab Invest. 2010;90:1152–1168. doi: 10.1038/labinvest.2010.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flynn AN, Wang A, McKay DM, Buret AG. Apoptosis-inducing factor contributes to epithelial cell apoptosis induced by entero-pathogenic Escherichia coli. Can J Physiol Pharmacol. 2011;89:143–148. doi: 10.1139/Y11-002. [DOI] [PubMed] [Google Scholar]
- 51.Beatty JK, Bhargava A, Buret AG. Post-infectious irritable bowel syndrome: mechanistic insights into chronic disturbances following enteric infection. World J Gastroenterol. 2014;20:3976–3985. doi: 10.3748/wjg.v20.i14.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ponnusamy K, Choi JN, Kim J, Lee SY, Lee CH. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J Med Microbiol. 2011;60:817–827. doi: 10.1099/jmm.0.028126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohman L, Simrén M. Pathogenesis of IBS: role of inflammation, immunity and neuroimmune interactions. Nat Rev Gastroenterol Hepatol. 2010;7:163–173. doi: 10.1038/nrgastro.2010.4. [DOI] [PubMed] [Google Scholar]
- 54.Ringel Y, Maharshak N. Intestinal microbiota and immune function in the pathogenesis of irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2013;305:G529–G541. doi: 10.1152/ajpgi.00207.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karantanos T, Markoutsaki T, Gazouli M, Anagnou NP, Karamanolis DG. Current insights in to the pathophysiology of irritable bowel syndrome. Gut Pathog. 2010;2:3. doi: 10.1186/1757-4749-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liebregts T, Adam B, Bredack C, et al. Immune activation in patients with irritable bowel syndrome. Gastroenterology. 2007;132:913–920. doi: 10.1053/j.gastro.2007.01.046. [DOI] [PubMed] [Google Scholar]
- 57.Weston AP, Biddle WL, Bhatia PS, Miner PB., Jr Terminal ileal mucosal mast cells in irritable bowel syndrome. Dig Dis Sci. 1993;38:1590–1595. doi: 10.1007/BF01303164. [DOI] [PubMed] [Google Scholar]
- 58.O’Sullivan M, Clayton N, Breslin NP, et al. Increased mast cells in the irritable bowel syndrome. Neurogastroenterol Motil. 2000;12:449–457. doi: 10.1046/j.1365-2982.2000.00221.x. [DOI] [PubMed] [Google Scholar]
- 59.Dunlop SP, Jenkins D, Spiller RC. Distinctive clinical, psychological, and histological features of postinfective irritable bowel syndrome. Am J Gastroenterol. 2003;98:1578–1583. doi: 10.1111/j.1572-0241.2003.07542.x. [DOI] [PubMed] [Google Scholar]
- 60.Spiller RC. Overlap between irritable bowel syndrome and inflammatory bowel disease. Dig Dis. 2009;27(Suppl 1):48–54. doi: 10.1159/000268121. [DOI] [PubMed] [Google Scholar]
- 61.Barbara G, Cremon C, Carini G, et al. The immune system in irritable bowel syndrome. J Neurogastroenterol Motil. 2011;17:349–359. doi: 10.5056/jnm.2011.17.4.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Akbar A, Yiangou Y, Facer P, Walters JR, Anand P, Ghosh S. Increased capsaicin receptor TRPV1-expressing sensory fibres in irritable bowel syndrome and their correlation with abdominal pain. Gut. 2008;57:923–929. doi: 10.1136/gut.2007.138982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cremon C, Gargano L, Morselli-Labate AM, et al. Mucosal immune activation in irritable bowel syndrome: gender-dependence and association with digestive symptoms. Am J Gastroenterol. 2009;104:392–400. doi: 10.1038/ajg.2008.94. [DOI] [PubMed] [Google Scholar]
- 64.Guilarte M, Santos J, de Torres I, et al. Diarrhoea-predominant IBS patients show mast cell activation and hyperplasia in the jejunum. Gut. 2007;56:203–209. doi: 10.1136/gut.2006.100594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park JH, Rhee PL, Kim HS, et al. Mucosal mast cell counts correlate with visceral hypersensitivity in patients with diarrhea predominant irritable bowel syndrome. J Gastroenterol Hepatol. 2006;21:71–78. doi: 10.1111/j.1440-1746.2005.04143.x. [DOI] [PubMed] [Google Scholar]
- 66.Wang SH, Dong L, Luo JY, et al. Decreased expression of serotonin in the jejunum and increased numbers of mast cells in the terminal ileum in patients with irritable bowel syndrome. World J Gastroenterol. 2007;13:6041–6047. doi: 10.3748/wjg.13.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barbara G, Wang B, Stanghellini V, et al. Mast cell-dependent excitation of visceral-nociceptive sensory neurons in irritable bowel syndrome. Gastroenterology. 2007;132:26–37. doi: 10.1053/j.gastro.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 68.Braak B, Klooker TK, Wouters MM, et al. Mucosal immune cell numbers and visceral sensitivity in patients with irritable bowel syndrome: is there any relationship? Am J Gastroenterol. 2012;107:715–726. doi: 10.1038/ajg.2012.54. [DOI] [PubMed] [Google Scholar]
- 69.Cenac N, Andrews CN, Holzhausen M, et al. Role for protease activity in visceral pain in irritable bowel syndrome. J Clin Invest. 2007;117:636–647. doi: 10.1172/JCI29255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barbara G, Stanghellini V, De Giorgio R, et al. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology. 2004;126:693–702. doi: 10.1053/j.gastro.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 71.Törnblom H, Lindberg G, Nyberg B, Veress B. Full-thickness biopsy of the jejunum reveals inflammation and enteric neuropathy in irritable bowel syndrome. Gastroenterology. 2002;123:1972–1979. doi: 10.1053/gast.2002.37059. [DOI] [PubMed] [Google Scholar]
- 72.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim HS, Lim JH, Park H, Lee SI. Increased immunoendocrine cells in intestinal mucosa of postinfectious irritable bowel syndrome patients 3 years after acute Shigella infection: an observation in a small case control study. Yonsei Med J. 2010;51:45–51. doi: 10.3349/ymj.2010.51.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Macsharry J, O’Mahony L, Fanning A, et al. Mucosal cytokine imbalance in irritable bowel syndrome. Scand J Gastroenterol. 2008;43:1467–1476. doi: 10.1080/00365520802276127. [DOI] [PubMed] [Google Scholar]
- 75.Darkoh C, Comer L, Zewdie G, Harold S, Snyder N, Dupont HL. Chemotactic chemokines are important in the pathogenesis of irritable bowel syndrome. PLoS One. 2014;9:e93144. doi: 10.1371/journal.pone.0093144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dinan TG, Quigley EM, Ahmed SM, et al. Hypothalamic-pituitary-gut axis dysregulation in irritable bowel syndrome: plasma cytokines as a potential biomarker? Gastroenterology. 2006;130:304–311. doi: 10.1053/j.gastro.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 77.Rodríguez-Fandiño O, Hernández-Ruiz J, Schmulson M. From cytokines to toll-like receptors and beyond - current knowledge and future research needs in irritable bowel syndrome. J Neurogastroenterol Motil. 2010;16:363–373. doi: 10.5056/jnm.2010.16.4.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scully P, McKernan DP, Keohane J, et al. Plasma cytokine profiles in females with irritable bowel syndrome and extra-intestinal co-morbidity. Am J Gastroenterol. 2010;105:2235–2243. doi: 10.1038/ajg.2010.159. [DOI] [PubMed] [Google Scholar]
- 79.Chang L, Adeyemo M, Karagiannides I, et al. Serum and colonic mucosal immune markers in irritable bowel syndrome. Am J Gastroenterol. 2012;107:262–272. doi: 10.1038/ajg.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McKernan DP, Gaszner G, Quigley EM, Cryan JF, Dinan TG. Altered peripheral toll-like receptor responses in the irritable bowel syndrome. Aliment Pharmacol Ther. 2011;33:1045–1052. doi: 10.1111/j.1365-2036.2011.04624.x. [DOI] [PubMed] [Google Scholar]
- 81.Brint EK, MacSharry J, Fanning A, Shanahan F, Quigley EM. Differential expression of toll-like receptors in patients with irritable bowel syndrome. Am J Gastroenterol. 2011;106:329–336. doi: 10.1038/ajg.2010.438. [DOI] [PubMed] [Google Scholar]
- 82.Belmonte L, Beutheu Youmba S, Bertiaux-Vandaële N, et al. Role of toll like receptors in irritable bowel syndrome: differential mucosal immune activation according to the disease subtype. PLoS One. 2012;7:e42777. doi: 10.1371/journal.pone.0042777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ohman L, Stridsberg M, Isaksson S, Jerlstad P, Simrén M. Altered levels of fecal chromogranins and secretogranins in IBS: relevance for pathophysiology and symptoms? Am J Gastroenterol. 2012;107:440–447. doi: 10.1038/ajg.2011.458. [DOI] [PubMed] [Google Scholar]
- 84.Langhorst J, Junge A, Rueffer A, et al. Elevated human beta-defensin-2 levels indicate an activation of the innate immune system in patients with irritable bowel syndrome. Am J Gastroenterol. 2009;104:404–410. doi: 10.1038/ajg.2008.86. [DOI] [PubMed] [Google Scholar]
- 85.Chadwick VS, Chen W, Shu D, et al. Activation of the mucosal immune system in irritable bowel syndrome. Gastroenterology. 2002;122:1778–1783. doi: 10.1053/gast.2002.33579. [DOI] [PubMed] [Google Scholar]
- 86.Ohman L, Isaksson S, Lundgren A, Simrén M, Sjövall H. A controlled study of colonic immune activity and beta7+ blood T lymphocytes in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol. 2005;3:980–986. doi: 10.1016/S1542-3565(05)00410-6. [DOI] [PubMed] [Google Scholar]
- 87.Ohman L, Isaksson S, Lindmark AC, et al. T-cell activation in patients with irritable bowel syndrome. Am J Gastroenterol. 2009;104:1205–1212. doi: 10.1038/ajg.2009.116. [DOI] [PubMed] [Google Scholar]
- 88.Dunlop SP, Jenkins D, Neal KR, Spiller RC. Relative importance of enterochromaffin cell hyperplasia, anxiety, and depression in postinfectious IBS. Gastroenterology. 2003;125:1651–1659. doi: 10.1053/j.gastro.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 89.Schoepfer AM, Schaffer T, Seibold-Schmid B, Müller S, Seibold F. Antibodies to flagellin indicate reactivity to bacterial antigens in IBS patients. Neurogastroenterol Motil. 2008;20:1110–1118. doi: 10.1111/j.1365-2982.2008.01166.x. [DOI] [PubMed] [Google Scholar]
- 90.Cremon C, Pallotti F, Bacchilega M, Stanghellini V, Corinaldesi R, Barbara G. Antiflagellin antibodies suggest infective participation in irritable bowel syndrome pathogenesis. Expert Rev Gastroenterol Hepatol. 2008;2:735–740. doi: 10.1586/17474124.2.6.735. [DOI] [PubMed] [Google Scholar]
- 91.Ohman L, Lindmark AC, Isaksson S, et al. B-cell activation in patients with irritable bowel syndrome (IBS) Neurogastroenterol Motil. 2009;21:644–650. doi: 10.1111/j.1365-2982.2009.01272.x. [DOI] [PubMed] [Google Scholar]
- 92.Egen JG, Kuhns MS, Allison JP. CTLA-4: new insights into its biological function and use in tumor immunotherapy. Nat Immunol. 2002;3:611–618. doi: 10.1038/ni0702-611. [DOI] [PubMed] [Google Scholar]
- 93.Parkes GC, Rayment N, Woodman I, et al. Increases in ileal mast cells in patients with diarrhoea predominant irritable bowel syndrome may be due to a relative reduction in mucosa-associated Lactobacilli. Gut. 2011;60(Suppl 1):A39. doi: 10.1136/gut.2011.239301.77. [DOI] [Google Scholar]
- 94.Löfmark S, Jernberg C, Jansson JK, Edlund C. Clindamycin-induced enrichment and long-term persistence of resistant Bacteroides spp. and resistance genes. J Antimicrob Chemother. 2006;58:1160–1167. doi: 10.1093/jac/dkl420. [DOI] [PubMed] [Google Scholar]
- 95.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 96.De La Cochetière MF, Durand T, Lalande V, Petit JC, Potel G, Beaugerie L. Effect of antibiotic therapy on human fecal microbiota and the relation to the development of Clostridium difficile. Microb Ecol. 2008;56:395–402. doi: 10.1007/s00248-007-9356-5. [DOI] [PubMed] [Google Scholar]
- 97.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Parvez S, Malik KA, Ah Kang S, Kim HY. Probiotics and their fermented food products are beneficial for health. J Appl Microbiol. 2006;100:1171–1185. doi: 10.1111/j.1365-2672.2006.02963.x. [DOI] [PubMed] [Google Scholar]
- 99.Wiström J, Norrby SR, Myhre EB, et al. Frequency of antibiotic-associated diarrhoea in 2462 antibiotic-treated hospitalized patients: a prospective study. J Antimicrob Chemother. 2001;47:43–50. doi: 10.1093/jac/47.1.43. [DOI] [PubMed] [Google Scholar]
- 100.Mendall MA, Kumar D. Antibiotic use, childhood affluence and irritable bowel syndrome (IBS) Eur J Gastroenterol Hepatol. 1998;10:59–62. doi: 10.1097/00042737-199801000-00011. [DOI] [PubMed] [Google Scholar]
- 101.Maxwell PR, Rink E, Kumar D, Mendall MA. Antibiotics increase functional abdominal symptoms. Am J Gastroenterol. 2002;97:104–108. doi: 10.1111/j.1572-0241.2002.05428.x. [DOI] [PubMed] [Google Scholar]
- 102.De La Cochetière MF, Durand T, Lepage P, Bourreille A, Galmiche JP, Doré J. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol. 2005;43:5588–5592. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Villarreal AA, Aberger FJ, Benrud R, Gundrum JD. Use of broad-spectrum antibiotics and the development of irritable bowel syndrome. WMJ. 2012;111:17–20. [PubMed] [Google Scholar]
- 104.Pimentel M, Chatterjee S, Chow EJ, Park S, Kong Y. Neomycin improves constipation-predominant irritable bowel syndrome in a fashion that is dependent on the presence of methane gas: sub-analysis of a double-blind randomized controlled study. Dig Dis Sci. 2006;51:1297–1301. doi: 10.1007/s10620-006-9104-6. [DOI] [PubMed] [Google Scholar]
- 105.American College of Gastroenterology Task Force on Irritable Bowel Syndrome. Brandt LJ, Chey WD, et al. An evidence-based position statement on the management of irritable bowel syndrome. Am J Gastroenterol. 2009;104(Suppl 1):S1–S35. doi: 10.1038/ajg.2008.122. [DOI] [PubMed] [Google Scholar]
- 106.Pimentel M, Morales W, Chua K, et al. Effects of rifaximin treatment and retreatment in nonconstipated IBS subjects. Dig Dis Sci. 2011;56:2067–2072. doi: 10.1007/s10620-011-1728-5. [DOI] [PubMed] [Google Scholar]
- 107.Menees SB, Maneerattannaporn M, Kim HM, Chey WD. The efficacy and safety of rifaximin for the irritable bowel syndrome: a systematic review and meta-analysis. Am J Gastroenterol. 2012;107:28–35. doi: 10.1038/ajg.2011.355. [DOI] [PubMed] [Google Scholar]
- 108.Gomes AM, Malcata FX. Bifidobacterium spp. and Lactobacillus acidophilus: biological, biochemical, technological and therapeutical properties relevant for use as probiotics. Trends Food Sci Technol. 1999;10:139–157. doi: 10.1016/S0924-2244(99)00033-3. [DOI] [Google Scholar]
- 109.Sinn DH, Song JH, Kim HJ, et al. Therapeutic effect of Lactobacillus acidophilus-SDC 2012, 2013 in patients with irritable bowel syndrome. Dig Dis Sci. 2008;53:2714–2718. doi: 10.1007/s10620-007-0196-4. [DOI] [PubMed] [Google Scholar]
- 110.Kruis W, Chrubasik S, Boehm S, Stange C, Schulze J. A double-blind placebo-controlled trial to study therapeutic effects of probiotic Escherichia coli Nissle 1917 in subgroups of patients with irritable bowel syndrome. Int J Colorectal Dis. 2012;27:467–474. doi: 10.1007/s00384-011-1363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sisson G, Ayis S, Sherwood RA, Bjarnason I. Randomised clinical trial: a liquid multi-strain probiotic vs. placebo in the irritable bowel syndrome: a 12 week double-blind study. Aliment Pharmacol Ther. 2014;40:51–62. doi: 10.1111/apt.12787. [DOI] [PubMed] [Google Scholar]
- 112.Simrén M, Ohman L, Olsson J, et al. Clinical trial: the effects of a fermented milk containing three probiotic bacteria in patients with irritable bowel syndrome--a randomized, double-blind, controlled study. Aliment Pharmacol Ther. 2010;31:218–227. doi: 10.1111/j.1365-2036.2009.04183.x. [DOI] [PubMed] [Google Scholar]
- 113.Ligaarden SC, Axelsson L, Naterstad K, Lydersen S, Farup PG. A candidate probiotic with unfavourable effects in subjects with irritable bowel syndrome: a randomised controlled trial. BMC Gastroenterol. 2010;10:16. doi: 10.1186/1471-230X-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ludidi S, Jonkers DM, Koning CJ, et al. Randomized clinical trial on the effect of a multispecies probiotic on visceroperception in hypersensitive IBS patients. Neurogastroenterol Motil. 2014;26:705–714. doi: 10.1111/nmo.12320. [DOI] [PubMed] [Google Scholar]
- 115.Niv E, Naftali T, Hallak R, Vaisman N. The efficacy of Lactobacillus reuteri ATCC 55730 in the treatment of patients with irritable bowel syndrome: a double blind, placebo-controlled, randomized study. Clin Nutr. 2005;24:925–931. doi: 10.1016/j.clnu.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 116.Guyonnet D, Chassany O, Ducrotte P, et al. Effect of a fermented milk containing Bifidobacterium animalis DN-173 010 on the health-related quality of life and symptoms in irritable bowel syndrome in adults in primary care: a multicentre, randomized, double-blind, controlled trial. Aliment Pharmacol Ther. 2007;26:475–486. doi: 10.1111/j.1365-2036.2007.03362.x. [DOI] [PubMed] [Google Scholar]
- 117.O’Sullivan MA, O’Morain CA. Bacterial supplementation in the irritable bowel syndrome: a randomised double-blind placebo-controlled crossover study. Dig Liver Dis. 2000;32:294–301. doi: 10.1016/S1590-8658(00)80021-3. [DOI] [PubMed] [Google Scholar]
- 118.Chapman CM, Gibson GR, Rowland I. Health benefits of probiotics: are mixtures more effective than single strains? Eur J Nutr. 2011;50:1–17. doi: 10.1007/s00394-010-0166-z. [DOI] [PubMed] [Google Scholar]
- 119.Cash BD. Emerging role of probiotics and antimicrobials in the management of irritable bowel syndrome. Curr Med Res Opin. 2014;30:1405–1415. doi: 10.1185/03007995.2014.908278. [DOI] [PubMed] [Google Scholar]
- 120.Hunter JO, Tuffnell Q, Lee AJ. Controlled trial of oligofructose in the management of irritable bowel syndrome. J Nutr. 1999;129:1451S–1453S. doi: 10.1093/jn/129.7.1451S. [DOI] [PubMed] [Google Scholar]
- 121.Olesen M, Gudmand-Hoyer E. Efficacy, safety, and tolerability of fructooligosaccharides in the treatment of irritable bowel syndrome. Am J Clin Nutr. 2000;72:1570–1575. doi: 10.1093/ajcn/72.6.1570. [DOI] [PubMed] [Google Scholar]
- 122.Paineau D, Payen F, Panserieu S, et al. The effects of regular consumption of short-chain fructo-oligosaccharides on digestive comfort of subjects with minor functional bowel disorders. Br J Nutr. 2008;99:311–318. doi: 10.1017/S000711450779894X. [DOI] [PubMed] [Google Scholar]
- 123.Silk DB, Davis A, Vulevic J, Tzortzis G, Gibson GR. Clinical trial: the effects of a trans-galactooligosaccharide prebiotic on faecal microbiota and symptoms in irritable bowel syndrome. Aliment Pharmacol Ther. 2009;29:508–518. doi: 10.1111/j.1365-2036.2008.03911.x. [DOI] [PubMed] [Google Scholar]
- 124.Roberfroid M, Gibson GR, Hoyles L, et al. Prebiotic effects: metabolic and health benefits. Br J Nutr. 2010;104(Suppl 2):S1–S63. doi: 10.1017/S0007114510003363. [DOI] [PubMed] [Google Scholar]
- 125.Halmos EP, Power VA, Shepherd SJ, Gibson PR, Muir JG. A diet low in FODMAPs reduces symptoms of irritable bowel syndrome. Gastroenterology. 2014;146:67–75. doi: 10.1053/j.gastro.2013.09.046. [DOI] [PubMed] [Google Scholar]
- 126.Simrén M. Diet as a therapy for irritable bowel syndrome: progress at last. Gastroenterology. 2014;146:10–12. doi: 10.1053/j.gastro.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 127.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 128.Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007;73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Muegge BD, Kuczynski J, Knights D, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Furnari M, Savarino E, Bruzzone L, et al. Reassessment of the role of methane production between irritable bowel syndrome and functional constipation. J Gastrointestin Liver Dis. 2012;21:157–163. [PubMed] [Google Scholar]
- 134.Staudacher HM, Whelan K, Irving PM, Lomer MC. Comparison of symptom response following advice for a diet low in fermentable carbohydrates (FODMAPs) versus standard dietary advice in patients with irritable bowel syndrome. J Hum Nutr Diet. 2011;24:487–495. doi: 10.1111/j.1365-277X.2011.01162.x. [DOI] [PubMed] [Google Scholar]
- 135.Staudacher HM, Lomer MC, Anderson JL, et al. Fermentable carbohydrate restriction reduces luminal bifidobacteria and gastrointestinal symptoms in patients with irritable bowel syndrome. J Nutr. 2012;142:1510–1518. doi: 10.3945/jn.112.159285. [DOI] [PubMed] [Google Scholar]
- 136.Shepherd SJ, Lomer MC, Gibson PR. Short-chain carbohydrates and functional gastrointestinal disorders. Am J Gastroenterol. 2013;108:707–717. doi: 10.1038/ajg.2013.96. [DOI] [PubMed] [Google Scholar]
- 137.Staudacher HM, Irving PM, Lomer MC, Whelan K. Mechanisms and efficacy of dietary FODMAP restriction in IBS. Nat Rev Gastroenterol Hepatol. 2014;11:256–266. doi: 10.1038/nrgastro.2013.259. [DOI] [PubMed] [Google Scholar]
- 138.Codling C, O’Mahony L, Shanahan F, Quigley EM, Marchesi JR. A molecular analysis of fecal and mucosal bacterial communities in irritable bowel syndrome. Dig Dis Sci. 2010;55:392–397. doi: 10.1007/s10620-009-0934-x. [DOI] [PubMed] [Google Scholar]
- 139.Carroll IM, Ringel-Kulka T, Keku TO, et al. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2011;301:G799–G807. doi: 10.1152/ajpgi.00154.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jeffery IB, Quigley EM, Öhman L, Simrén M, O’Toole PW. The microbiota link to irritable bowel syndrome: an emerging story. Gut Microbes. 2012;3:572–576. doi: 10.4161/gmic.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kassinen A, Krogius-Kurikka L, Mäkivuokko H, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 142.Jalanka-Tuovinen J, Salojärvi J, Salonen A, et al. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut. 2014;63:1737–1745. doi: 10.1136/gutjnl-2013-305994. [DOI] [PubMed] [Google Scholar]
- 143.Krogius-Kurikka L, Lyra A, Malinen E, et al. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Durbán A, Abellán JJ, Jiménez-Hernández N, et al. Instability of the faecal microbiota in diarrhoea-predominant irritable bowel syndrome. FEMS Microbiol Ecol. 2013;86:581–589. doi: 10.1111/1574-6941.12184. [DOI] [PubMed] [Google Scholar]
- 145.Jeffery IB, O’Toole PW, Öhman L, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997–1006. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]
- 146.Kerckhoffs AP, Ben-Amor K, Samsom M, et al. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J Med Microbiol. 2011;60:236–245. doi: 10.1099/jmm.0.022848-0. [DOI] [PubMed] [Google Scholar]
- 147.Rappé MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 148.Euzéby JP. List of bacterial names with standing in nomenclature: a folder available on the Internet. Int J Syst Bacteriol. 1997;47:590–592. doi: 10.1099/00207713-47-2-590. [DOI] [PubMed] [Google Scholar]
- 149.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tap J, Mondot S, Levenez F, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 151.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metage-nomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Balsari A, Ceccarelli A, Dubini F, Fesce E, Poli G. The fecal microbial population in the irritable bowel syndrome. Microbiologica. 1982;5:185–194. [PubMed] [Google Scholar]
- 153.Carroll IM, Chang YH, Park J, Sartor RB, Ringel Y. Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog. 2010;2:19. doi: 10.1186/1757-4749-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tana C, Umesaki Y, Imaoka A, Handa T, Kanazawa M, Fukudo S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:512–519. doi: 10.1111/j.1365-2982.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- 155.Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:521–530. doi: 10.1111/j.1365-2982.2012.01891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Rigsbee L, Agans R, Shankar V, et al. Quantitative profiling of gut microbiota of children with diarrhea-predominant irritable bowel syndrome. Am J Gastroenterol. 2012;107:1740–1751. doi: 10.1038/ajg.2012.287. [DOI] [PubMed] [Google Scholar]
- 157.Mättö J, Maunuksela L, Kajander K, et al. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome: a longitudinal study in IBS and control subjects. FEMS Immunol Med Microbiol. 2005;43:213–222. doi: 10.1016/j.femsim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 158.Si JM, Yu YC, Fan YJ, Chen SJ. Intestinal microecology and quality of life in irritable bowel syndrome patients. World J Gastroenterol. 2004;10:1802–1805. doi: 10.3748/wjg.v10.i12.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Malinen E, Rinttilä T, Kajander K, et al. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–382. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 160.Kerckhoffs AP, Samsom M, van der Rest ME, et al. Lower Bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World J Gastroenterol. 2009;15:2887–2892. doi: 10.3748/wjg.15.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rajilić-Stojanović M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 162.Wang H, Gong J, Wang W, et al. Are there any different effects of Bifidobacterium, Lactobacillus and Streptococcus on intestinal sensation, barrier function and intestinal immunity in PI-IBS mouse model? PLoS One. 2014;9:e90153. doi: 10.1371/journal.pone.0090153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Parkes GC, Rayment NB, Hudspith BN, et al. Distinct microbial populations exist in the mucosa-associated microbiota of subgroups of irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:31–39. doi: 10.1111/j.1365-2982.2011.01803.x. [DOI] [PubMed] [Google Scholar]
- 164.Maukonen J, Satokari R, Mättö J, Söderlund H, Mattila-Sandholm T, Saarela M. Prevalence and temporal stability of selected clostridial groups in irritable bowel syndrome in relation to predominant faecal bacteria. J Med Microbiol. 2006;55:625–633. doi: 10.1099/jmm.0.46134-0. [DOI] [PubMed] [Google Scholar]
- 165.Saulnier DM, Riehle K, Mistretta TA, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Leitch EC, Walker AW, Duncan SH, Holtrop G, Flint HJ. Selective colonization of insoluble substrates by human faecal bacteria. Environ Microbiol. 2007;9:667–679. doi: 10.1111/j.1462-2920.2006.01186.x. [DOI] [PubMed] [Google Scholar]
- 167.Lyra A, Rinttilä T, Nikkilä J, et al. Diarrhoea-predominant irritable bowel syndrome distinguishable by 16S rRNA gene phylotype quantification. World J Gastroenterol. 2009;15:5936–5945. doi: 10.3748/wjg.15.5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Duncan SH, Hold GL, Barcenilla A, Stewart CS, Flint HJ. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int J Syst Evol Microbiol. 2002;52:1615–1620. doi: 10.1099/ijs.0.02143-0. [DOI] [PubMed] [Google Scholar]
- 169.Barcenilla A, Pryde SE, Martin JC, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–1661. doi: 10.1128/AEM.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hatayama H, Iwashita J, Kuwajima A, Abe T. The short chain fatty acid, butyrate, stimulates MUC2 mucin production in the human colon cancer cell line, LS174T. Biochem Biophys Res Commun. 2007;356:599–603. doi: 10.1016/j.bbrc.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 171.Peng L, He Z, Chen W, Holzman IR, Lin J. Effects of butyrate on intestinal barrier function in a Caco-2 cell monolayer model of intestinal barrier. Pediatr Res. 2007;61:37–41. doi: 10.1203/01.pdr.0000250014.92242.f3. [DOI] [PubMed] [Google Scholar]
- 172.Załęski A, Banaszkiewicz A, Walkowiak J. Butyric acid in irritable bowel syndrome. Prz Gastroenterol. 2013;8:350–353. doi: 10.5114/pg.2013.39917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Duboc H, Rainteau D, Rajca S, et al. Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:513–520. doi: 10.1111/j.1365-2982.2012.01893.x. [DOI] [PubMed] [Google Scholar]
- 174.Chassard C, Dapoigny M, Scott KP, et al. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment Pharmacol Ther. 2012;35:828–838. doi: 10.1111/j.1365-2036.2012.05007.x. [DOI] [PubMed] [Google Scholar]
- 175.Lopez-Siles M, Martinez-Medina M, Busquets D, et al. Mucosa-associated Faecalibacterium prausnitzii and Escherichia coli co-abundance can distinguish irritable bowel syndrome and inflammatory bowel disease phenotypes. Int J Med Microbiol. 2014;304:464–475. doi: 10.1016/j.ijmm.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 176.Chen CC, Walker WA. Probiotics and prebiotics: role in clinical disease states. Adv Pediatr. 2005;52:77–113. doi: 10.1016/j.yapd.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 177.Malinen E, Krogius-Kurikka L, Lyra A, et al. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J Gastroenterol. 2010;16:4532–4540. doi: 10.3748/wjg.v16.i36.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Noor SO, Ridgway K, Scovell L, et al. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 2010;10:134. doi: 10.1186/1471-230X-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Moratalla A, Gómez-Hurtado I, Santacruz A, et al. Protective effect of Bifidobacterium pseudocatenulatum CECT7765 against induced bacterial antigen translocation in experimental cirrhosis. Liver Int. 2014;34:850–858. doi: 10.1111/liv.12380. [DOI] [PubMed] [Google Scholar]
- 181.Derrien M, van Passel MW, van de Bovenkamp JH, Schipper RG, de Vos WM, Dekker J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes. 2010;1:254–268. doi: 10.4161/gmic.1.4.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.King TS, Elia M, Hunter JO. Abnormal colonic fermentation in irritable bowel syndrome. Lancet. 1998;352:1187–1189. doi: 10.1016/S0140-6736(98)02146-1. [DOI] [PubMed] [Google Scholar]
- 183.Pimentel M, Lin HC, Enayati P, et al. Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1089–G1095. doi: 10.1152/ajpgi.00574.2004. [DOI] [PubMed] [Google Scholar]
- 184.Pimentel M, Mayer AG, Park S, Chow EJ, Hasan A, Kong Y. Methane production during lactulose breath test is associated with gastrointestinal disease presentation. Dig Dis Sci. 2003;48:86–92. doi: 10.1023/A:1021738515885. [DOI] [PubMed] [Google Scholar]
- 185.Chatterjee S, Park S, Low K, Kong Y, Pimentel M. The degree of breath methane production in IBS correlates with the severity of constipation. Am J Gastroenterol. 2007;102:837–841. doi: 10.1111/j.1572-0241.2007.01072.x. [DOI] [PubMed] [Google Scholar]
- 186.Kim G, Deepinder F, Morales W, et al. Methanobrevibacter smithii is the predominant methanogen in patients with constipation-predominant IBS and methane on breath. Dig Dis Sci. 2012;57:3213–3218. doi: 10.1007/s10620-012-2197-1. [DOI] [PubMed] [Google Scholar]
- 187.Rana SV, Sharma S, Sinha SK, Kaur H, Sikander A, Singh K. Incidence of predominant methanogenic flora in irritable bowel syndrome patients and apparently healthy controls from North India. Dig Dis Sci. 2009;54:132–135. doi: 10.1007/s10620-008-0315-x. [DOI] [PubMed] [Google Scholar]
- 188.Durbán A, Abellán JJ, Jiménez-Hernández N, et al. Structural alterations of faecal and mucosa-associated bacterial communities in irritable bowel syndrome. Environ Microbiol Rep. 2012;4:242–247. doi: 10.1111/j.1758-2229.2012.00327.x. [DOI] [PubMed] [Google Scholar]
- 189.Crouzet L, Gaultier E, Del’Homme C, et al. The hypersensitivity to colonic distension of IBS patients can be transferred to rats through their fecal microbiota. Neurogastroenterol Motil. 2013;25:e272–e282. doi: 10.1111/nmo.12103. [DOI] [PubMed] [Google Scholar]
- 190.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]