Abstract
This review considers the role of bacterial antizyme in the regulation of polyamine biosynthesis and gives new perspectives on the involvement of antizyme in other significant cellular mechanisms. Antizyme is a protein molecule induced by the end product of the enzymic reaction that it inhibits, in a non-competitive manner. The bacterial ornithine decarboxylase is regulated by nucleotides, phosphorylation and antizyme. The inhibition of ornithine decarboxylase by antizyme can be relieved to different degrees by DNA or by a variety of synthetic nucleic acid polymers, attributed to a specific interaction between nucleic acid and antizyme. Recently, this interplay between bacterial antizyme and nucleic acid was determined by discerning an additional function to antizyme that proved to be the atoC gene product, encoding the response regulator of the bacterial two-component system AtoS-AtoC. The gene located just upstream of atoC encodes the sensor kinase, named AtoS, that modulates AtoC activity. AtoC regulates expression of atoDAEB operon which is involved in short-chain fatty acid metabolism. Antizyme is thus referred to as AtoC, functioning both as a post-translational and transcriptional regulator. Also, the AtoS-AtoC signal transduction system in E. coli has a positive regulatory role on poly-(R)-3-hydroxybutyrate biosynthesis. The properties and gene structural similarities of antizymes from different organisms were compared. It was revealed that conserved domains are present mostly in the C-domain of all antizymes. BLAST analysis of the E. coli antizyme protein (AtoC) showed similarities around 69–58% among proteobacteria, g-proteobacteria, enterobacteria and the thermophilic bacterium Thermus thermophilus. A working hypothesis is proposed for the metabolic role of antizyme (AtoC) describing the significant biological implications of this protein molecule. Whether antizymes exist to other enzymes in different tissues, meeting the criteria discussed in the text remains to be elucidated.
Review
In 1978 Seymour Cohen, the father of the field of polyamines posed the question "what do the polyamines do?" in his excellent article [1]. Since then, thousands of papers appeared in the literature concerning the metabolic role of polyamines. We will not attempt to discuss all the recent progress in the field, which has been well documented in book form [2-8] as well as in a number of excellent reviews [9-15]. Instead, we shall only consider the role of antizyme (Az) in the regulation of ornithine decarboxylase (L-ornithine carboxylyase, EC 4.1.1.17, ODC), its involvement in the bacterial two-component signal transduction system AtoS-AtoC [16], as well as its implication in other significant cellular functions.
The role of Az in various organisms
Az was discovered by Canellakis and co-workers in rat liver and several cell lines. It is a 26.5 kDa protein, induced by polyamines, the product of ODC action. Az forms complex with ODC, inhibiting that way the enzyme activity in a stoichiometric manner, non-competitively [17,18]. The inactive ODC-Az complex can be dissociated by high salt concentrations. The induction of Az by exogenously added polyamines is inhibited by cycloheximide or puromycin, but not by actinomycin D indicating that polyamines induce antizyme by stimulating the translation of its mRNA [13]. This explains an old paradoxical finding that ODC activity was stimulated in rat liver when puromycin was administered to animals [19].
Since then, three different Azs have been identified in eukayotic cells, possessing different roles at the cellular levels: a) Az1 inhibits ODC and then directs its degradation by the 26 S proteosome. ODC when bound to Az1 is efficiently degraded by the proteosome and Az1 is usually recycled to act again [13,20]. This type of degradation of ODC usually occurs in an ATP-dependent, but ubiquitin-independent manner. Polyamines can trigger a +1 translational frameshift on Az mRNA, allowing the complete Az1 protein to be expressed [13,21]. ODC can be released from Az1 by another protein called "anti-antizyme", which liberates ODC in the presence of growth stimuli having higher affinity for Az1 than for ODC [22]. b) Az2 shares similar properties with Az1, including the regulatory frameshifting. It does not stimulate, however, degradation of ODC under certain conditions, but alters polyamine homeostasis, by down-regulating polyamine uptake independently of the effects on ODC, thus playing a negative role in the regulation of polyamine transport [23,24]. c) Az3 is expressed to a limited extend in testis germ cells, at a particular stage of spermatogenesis [25,26]. The pattern of Az3 expression suggests that it acts by sharply limiting polyamine accumulation in cells that have finished DNA synthesis and meiotic reduction and are about to be remodeled into mature spermatozoa [25].
Searches on genomic databases have revealed that the Azs comprise a widespread family of conserved homologues [27]. In humans, five non-allelic Az homologues have been detected. Two copies are presented in zebra fish Danio rerio. Az has also been cloned from fruit flies (Drosophila melanogaster and Drosophila virillis) and is found in a number of invertebrate species (Bombyx mori, Caenorhabditis elegans, Onchocerca volvulus, Haemonchus contortus and Pristionchus pacificus). Moreover, Az activity has been detected in a wide variety of organisms, from plants to eubacteria [8,9,14,15,23-32].
The question is why a cell or an organism needs more than one type of Az with distinct functions? Their distinct functions may be due to their special time of expression, compartmentalization or different place and way of degradation of ODC.
Regulation of polyamine biosynthesis in bacteria
Regulation of ODC allosterically by nucleotides
Two ODCs, the biosynthetic and the biodegradative have been characterized in E. coli [33,34]. The biodegradative ODC is induced by low pH and by the presence of ornithine in the growth medium. The activity of the biosynthetic ODC is modulated by a number of positive and negative effectors [11]. The positive effectors include nucleotides, GTP being the more effective [34], while ppGpp acts as a negative effector [35]. The requirement for GTP in protein synthesis and the accumulation of ppGpp during starvation of E. coli that are under stringent control, suggest that changes in polyamine pools may be responsible for the stringent effect in E. coli [35]. The accumulation of ppGpp leads to the cessation of stable RNA synthesis and appears to have a bearing on the fidelity of protein synthesis [36]. Treating recombinant ODC with calf intestine alkaline phosphatase leads to inactivation of ODC that can be allosterically reversed only by guanosyl or uridyl phosphate analogues at a concentration of 10-4 or 10-3 M [37]. Nucleotides are effective in activating ODC, in the order (G,U)TP>(G,U)DP>(G,U)MP. [8-3H]GTP binds specifically to ODC since cold GTP but not ATP can dissociate the radioactive analogue. High concentration of GTP can dissociate the ODC-Az complex and either reactivate or liberate ODC [37].
Regulation of ODC by phosphorylation
Biosynthetic ODC of E. coli can be phosphorylated both in vivo as well as in vitro [37]. An ODC-phosphorylating kinase was purified from middle-log growing E. coli K-12 strain MG1655, containing plasmid pODC [38]. The kinase requires 10 mM Ca++ for optimal activity.
In vivo phosphorylation of ODC
E. coli K-12 strain MG 1655+pODC, was labeled in vivo with [32P] orthophosphate, ODC was immunoprecipitated by ODC antibody and proteins were then analyzed by SDS-PAGE followed by autoradiography. In the autoradiogram one radioactive band appeared at 82 kDa, corresponding to the Mr of the subunit of the biosynthetic ODC [37].
In vitro phosphorylation of ODC
The in vitro phosphorylation of ODC depends on the state of phosphorylation of the ODC molecule [37]. Therefore, the difficulties of the negative data on the ODC phosphorylation in different bacterial systems can be explained by the above observation. A partially purified kinase from E. coli was capable to phosphorylate the dephosphorylated preparation of ODC (ODCb). This was the first indication that a homologous kinase can in vitro phosphorylate ODC in bacteria.
Regulation of ODC by Az
Two macromolecular effectors of ODC were identified in the ODC- E. coli mutant, MA255 [28]. One was an inhibitor of ODC, with characteristics similar to the Az of eukaryotic cells. It was a noncompetitive inhibitor of ODC; its complex with ODC could be dissociated with salt to yield active ODC and active ODC inhibitor. This E. coli inhibitor also inhibited ODC activity of eukaryotic cells. The other effector was a thermostable, nondialyzable molecule that activated the ODC of E. coli 6- to 7-fold. Similar macromolecular inhibitors and activators have been identified in the parent ODC+ strain, MA197. Those results suggested that ODC activity might be controlled post-translationally by positive and negative macromolecular effectors whose intracellular levels also fluctuate in response to the extracellular putrescine, the end product of the ODC reaction [28].
The mode of Az induction
The induction of macromolecular inhibitors of ODC by putrescine, spermidine, and spermine, as well as by other diamines, has been now amply verified by a number of laboratories, which has been reviewed elsewhere [9,15]. Initial attempts to demonstrate the existence of Az, by mixing extracts, each of which had ODC activity failed [39]. The reasons of failure which have been presented in detail [40], rest upon the fact that the bound inhibitor cannot be assayed by activity measurements. Only excess or free unbound inhibitor can be assayed by such mixing experiments; therefore, in order to assay for free Az, the test sample should have no measurable ODC activity.
Following the published methodology from Canellakis' laboratory for the induction of Az [9,17,18,28], extensive purifications of rat liver and of E. coli Az have been accomplished [9]. Increasing concentrations of diaminooctane progressively inhibit ODC activity and eventually produce free assayable ODC antizyme. Since diaminooctane is an analogue of spermidine-compare NH2(CH2)3NH(CH2)4NH2 and NH2(CH2)8NH2- a possible explanation is that the very low concentrations of diaminooctane compete for certain spermidine sites and release the inhibition exerted by spermidine; the higher diaminooctane concentrations then inhibit ODC activity through the production of Az, as do all diamines and polyamines that have been tested [9,40].
The evidence in favor of Az participating, at least in part, in the regulation of ODC is as follows:
a) Free Az can be induced by 10-7 – 10-6 M putrescine in cells that have been depleted of ODC activity. Originally, it was necessary to use higher concentrations of putrescine in order to elicit free Az in cells that either contained ODC activity or were being induced concurrently for ODC activity. Higher concentrations of the inducer were required, because large amounts of Az were necessary to neutralize the existing intracellular ODC activity [40].
b) Az is a normal component of the nuclei and sub cellular particles of uninduced rat liver cells [41,42].
c) Putrescine also induces an Az in E. coli, which cross-reacts with rat liver ODC and functions like the eukaryotic Az1 [28,29].
Overproduction of Az by E. coli transformed with a plasmid carrying the Az gene is inhibitory to ODC and ADC (arginine decarboxylase). Indeed the ODC of an inactivated ODC-Az complex can be reactivated by the addition of ADC; the converse also occurs [43]. This technique enables the demonstration of much inactive ODC and ADC in E. coli extracts.
Reversal of Az inhibition by nucleic acids
Huang et al. [44] found that the inhibition of E. coli ODC by Az from the same source is relieved to different degrees by DNA and by a variety of synthetic nucleic acid polymers, including ribo- and deoxyribo-nucleotide polymers. Preferences for certain nucleic acid sequences were observed. The inhibition of rat liver ODC by the E. coli Az was also relieved by nucleic acids.
The ability of E. coli genomic DNA or other nucleic acids to relieve the inhibition of ODC by the acidic Az [28] was originally attributed to possible binding of Az to certain sequences of DNA [45]. For this reason the relief of the Az inhibition of ODC activity was used as an assay of the relative effectiveness of various nucleic acid sequences. Furthermore, the relative effectiveness of single-stranded deoxynucleotide and ribonucleotide homopolymers in relieving the inhibition of E. coli ODC by Az was compared. Since both the acidic Az and the nucleic acids are negatively charged at the assay pH, these interactions could not be ascribed to charge effects alone. Moreover, the nucleic acids were not equivalent in their abilities to relieve the inhibition of ODC by antizyme. The differences in the degree of relief of inhibition exerted by various segments of E. coli DNA, suggested that there was a specificity of interaction between nucleic acids and Az. The main property that differentiates Az from many of the double or single-stranded DNA or RNA binding proteins is that Az also binds and inhibits ODC and subsequently, upon its binding to DNA, this inhibition of ODC is reversed.
Recently, the interplay between the bacterial Az (AtoC) and nucleic acids has been determined by attributing an additional physiological function to Az, that of the response regulator of the bacterial two-component signal transduction system AtoS-AtoC regulating expression of atoDAEB operon encoded enzymes [16,46-53].
The two-component regulatory system AtoS-AtoC
The E. coli Az gene has been identified and found to share significant homology with bacterial transcriptional activators of the two-component regulatory system family [16]. These systems consist of a "sensor" kinase and a response regulator, which often is a transcriptional factor [54-56]. By sensing an appropriate signal, the sensor kinase autophosphorylates on a histidine residue. Following physical contact of the phosphorylated sensor kinase with the response regulator, the phosphoryl group is transferred, usually to an aspartate residue of the response regulator, which leads to its activation.
Recently, it has been reported that Az is the atoC gene product [47,48,53] possessing a second function as the regulator of the expression of genes encoding enzymes involved in short chain fatty acid metabolism (atoDAEB operon) [49-51]. Today, the protein is referred to as AtoC, functioning both as a transcriptional and post-translational regulator. The gene located just upstream of atoC encodes the sensor kinase that modulates AtoC activity [16] and has since been named atoS, to reflect the role of its product, AtoS, as a sensor kinase [48,52,53]. Recently, direct biochemical data have been obtained proving that the gene products of atoS and atoC constitute a two-component signal transduction system involved in the regulation of the atoDAEB operon [46]. Moreover, the DNA binding sequences of AtoC on atoDAEB operon promoter have been determined [46,57]. Thus, the novel physiological function that has been ascribed to bacterial Az (AtoC) as a transcriptional regulator is in accordance with Az's DNA specific binding on the E. coli genome and can clearly explain the fore mentioned data describing the relief of ODC inhibition upon Az binding to DNA.
The role of the AtoS-AtoC signal transduction system in E. coli on the positive regulation of the levels of poly-(R)-3-hydroxybutyrate (PHB) biosynthesis has been recently identified [58]. Increased amounts of PHB are synthesized in E. coli upon exposure of the cells to acetoacetate, the inducer of the AtoS-AtoC two-component signal transduction system, when both components of the system are overproduced (our unpublished data).
A recent transcriptome analysis of the E. coli two-component systems [59] revealed possible roles, positive and/or negative, of the AtoS-AtoC system in a number of processes including flagellar synthesis and chemotactic behaviour of E. coli. The same study also highlights the interplay between two-component systems, as mutations in the ompR-envZ system were also found to affect the expression of atoC. Moreover, a phenotype microarray analysis of E. coli mutants with deletions of all two-component systems showed that atoSC mutants presented susceptibility to specific osmolarity conditions, some membrane agents, aminoglycoside and a respiration inhibitor indicating other putative roles for the two-component system under study [60].
Gene structural analysis of antizymes
Eukaryotic Az genes require ribosomal frame shifting for their expression. Twelve nucleotides around the frame shift site are identical between S. pombe and the mammalian counterparts. The core element for this frame shifting is likely to have been present in the last common ancestor of yeast, nematodes and mammals [24], but not in E. coli [61]. The existence of homology between the mammalian antizymes, Az-1, -2 and -3, and the bacterial antizyme AtoC was more thoroughly investigated. It was revealed that conserved domains are present at the C-terminal parts of all antizymes (data obtained and will appear shortly) as it has been similarly reported for the S. pombe counterpart [62]. This region is responsible for binding and inactivating ODC, while it cannot accelerate its degradation by the 26S proteosome [13,20]. AtoC was found to belong to the NtrC-NifA family of sigma54-RNA polymerase transcriptional activators [52], while it presents 40% identities and 58% positives with the E. coli NtrC transcriptional regulator [63-67]. The E. coli AtoC protein which has been biochemically characterized [46] was found to have homologues in the proteobacteria, g-proteobacteria and enterobacteria as indicated by BLAST analysis [68-88], while an AtoC homologue was also present in T. thermophilus, whose complete genome analysis was recently published [69] [Table 1]. The latter is in agreement with previous biochemical data reporting the presence of antizyme activity in T. thermophilus [31]. BLAST analysis also indicated that AtoC structurally comprises three conserved domains, i.e. the N-terminal receiver domain containing the phosphorylation site, the central sigma54 interaction domain and the C-terminal DNA-binding domain.
Table 1.
Best hits of BLAST analysis of the E. coli K-12 atoC gene against the genomes of all microorganisms
| Locus or Accession No | Genome annotation |
Microorganism [Taxa] |
Hits a | Id./Sim.b(%/%) | Ref. |
| Q06065 ATOC ECOLI | Acetoacetate metabolism regulatory protein atoC (Ornithine/arginine decarboxylase inhibitor) (Ornithine decarboxylase antizyme) | Escherichia coli K12 [enterobacteria] | 2 | 95/95 | 16, 47, 48, 70 |
| NP 754649.1 | Acetoacetate metabolism Regulatory protein atoC | Escherichia coli CFT073 [enterobacteria] | 3 | 95/95 | 71 |
| ZP 00015409.1 | COG2204: Response regulator containing CheY-like receiver, AAA-type ATPase and DNA-binding domains | Rhodospirillum rubrum [a-proteobacteria] | 1 | 52/69 | 72 |
| NP 953090.1 | Sigma-54 dependent DNA binding response regulator | Geobacter sulfurreducens PCA [d-proteobacteria] | 14 | 45/61 | 73 |
| NP 972572.1 | sigma-54 dependent transcriptional regulator/response regulator | Treponema denticola ATCC 35405 [spirochetes] | 1 | 44/64 | 74 |
| ZP 00079669.1 | COG2204: Response regulator containing CheY-like receiver, AAA-type ATPase and DNA-binding domains | Geobacter metallireducens [d-proteobacteria] | 13 | 42/63 | 72 |
| YP 012430.1 | sigma-54 dependent transcriptional regulator/response regulator | Desulfovibrio vulgaris subsp. vulgaris str. Hildenborough [d-proteobacteria] | 5 | 43/61 | 75 |
| NP 218960.1 | response regulatory protein (atoC) | Treponema pallidum subsp. pallidum str. Nichols [spirochetes] | 1 | 43/63 | 76 |
| AAQ63912.1 | mutant NtrC-like activator | Myxococcus xanthus [d-proteobacteria] | 8 | 44/62 | 77 |
| YP 008363.1 | probable two-component response regulator | Parachlamydia sp. UWE25 [chlamydias] | 1 | 44/62 | 78 |
| ZP 00128642.1 | COG2204: Response regulator containing CheY-like receiver, AAA-type ATPase and DNA-binding domains | Desulfovibrio desulfuricans G20 [d-proteobacteria] | 4 | 42/61 | 72 |
| NP 968400.1 | regulator protein pilR | Bdellovibrio bacteriovorus HD100 [d-proteobacteria] | 3 | 40/63 | 79 |
| AAL27375.1 | HydG | Yersinia pestis [enterobacteria] | 1 | 43/62 | 80 |
| NP 709798.1 | response regulator of hydrogenase 3 activity (sensor HydH) | Shigella flexneri 2a str. 301 [enterobacteria] | 2 | 44/61 | 81 |
| NP 992358.1 | Response regulator containing CheY-like receiver, AAA-type ATPase and DNA-binding domains | Yersinia pestis biovar Medievalis str. 91001 [enterobacteria] | 1 | 43/63 | 82 |
| NP 290636.1 | response regulator of hydrogenase 3 activity (sensor HydH) | Escherichia coli O157:H7 EDL933 [enterobacteria] | 2 | 44/60 | 83 |
| NP 243814.1 | two-component response regulator in acetoacetate metabolism | Bacillus halodurans C-125 [eubacteria] | 1 | 41/62 | 84 |
| Q9APD9 ZRAR KLEOX | Transcriptional regulatory protein zraR | Klebsiella oxytoca [enterobacteria] | 1 | 43/61 | 85 |
| NP 312817.1 | GlnG; response regulator for gln | Escherichia coli O157:H7 [enterobacteria] | 1 | 40/58 | 86 |
| P25852 ZRAR SALTY | Transcriptional regulatory protein zraR | Salmonella typhimurium LT2 [enterobacteria] | 2 | 43/60 | 87 |
| NP 458044.1 | Two-component system, response regulator | Salmonella enterica subsp. enterica serovar Typhi str. CT18 [enterobacteria] | 2 | 40/58 | 88 |
| YP 005107.1* | two-component response regulator | Thermus thermophilus HB27 [eubacteria] | 22 | 33/58 | 69 |
aHits: Number of homologues found in each microorganism (only the one with the highest homology is presented). bId./Sim.: Identities/Similarities. * The E. coli K-12 atoC gene was aligned against the T. thermophilus genome independently since its homology was not detected among the first 100 best BLAST hits (threshold 0.005).
A working hypothesis for the metabolic role of Az (AtoC)
A working hypothesis for the bacterial Az (AtoC) role and the cascade of reactions that are triggered by its activation/induction is shown in Fig. 1. Acetoacetate or its metabolic counterpart acetoacetyl-CoA, is the signal upon which the kinase AtoS is autophosphorylated [49-52,46]. Subsequently, the phosphoryl group is transferred by protein-protein interaction to the response regulator AtoC. AtoC is thus activated and able to bind to the atoDAEB operon promoter at two "dyad symmetry" sequences [46], as it has been reported for the NtrC response regulator [63-67]]. Thereafter, AtoC is probably oligomerized, a process that triggers its ATPase activity (our unpublished data). Upon ATP hydrolysis, the "closed" form of the RNA-sigma54 holoenzyme is transformed into an "open" form [67]. The activated open form is able to induce atoDAEB operon transcription which leads to short chain fatty acid catabolism and activation of the PHB biosynthetic pathway, via the AtoDA molecule (acetoacetyl-CoA trasnferase) [58, our unpublished data]. A functional AtoS-AtoC system has also been reported to affect E. coli chemotactic behavior [59], while some other putative roles are still under investigation [60]. AtoC whose production is stimulated by polyamines in E. coli as well, plays a central role in ODC regulation by binding to ODC and inhibiting it non-competitively [28]. We still don't know whether the ODC inhibiting activity of AtoC/Az is activated upon AtoC/Az phosphorylation and thus affected by the two-component system under study. Moreover, native ODC can be reversibly inactivated by a dephosphorylation-phosphorylation reaction. Therefore, phosphorylation of the bacterial ODC is one of the possible regulating mechanisms for this enzyme, as was proved for the mammalian and protozoan ODC [15,37]. Finally, nucleotides can convert allosterically the inactive, phosphorylated ODCb to an active form of ODCb, suggesting that nucleotides may be physiological regulators of ODC in E. coli. Active ODC can be recovered from the ODC-Az complex by high concentrations of salts, DNA or GTP [28,37,44].
Figure 1.
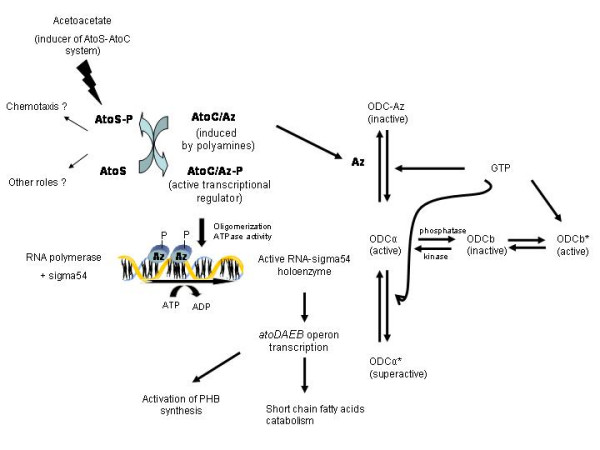
The involvement of antizyme (AtoC) in various regulatory mechanisms in E. coli.
Do antizymes exist for other enzymes?
The antizyme may constitute part of a normal control mechanism that defines the levels of ODC activity. However, it is a noncompetitive protein inhibitor and cannot be detected in free form when there is measurable ODC activity. Consequently, to prove the participation of the Az in the normal control of ODC, methodology has been developed permitting the isolation of the ODC-Az complex, separation of the complex into the component parts and assaying the respective activities [9,28,29].
We do know that the cell will produce free Az in the presence of high diamine or polyamine concentrations. Under these conditions, the tissue culture cells are in a relatively "unhappy" state; in the rat, the amount of putrescine required to elicit high levels of Az makes the animal extremely sick. Consequently, in the search for Azs to other enzymes, we believe the following criteria should be kept in mind:
1. The product should be added in high enough concentrations so that no enzyme activity can be detected.
2. The inhibition should be maintained over relatively long periods of time to maximize the amount of Az.
3. Inhibition, by the protein which will be induced by the end product of the enzymic reaction must be of a non-competitive manner.
We believe that under these extreme conditions the cell is responding to the excess product by what may be its last available defense mechanism producing additional non-competitive inhibitory proteins to neutralize the enzyme in order to lower the level of the product. So far, not any antizyme to other enzyme has appeared in the literature. The above question needs to be addressed at the biochemical as well as genetic level.
Conclusions and future directions
This review has evaluated some of the information available for the ornithine decarboxylase antizyme and has attempted to summarize the regulatory mechanisms in which it may be involved. Our understanding of the functions of Az derives from different biological sources. In eukaryotes three antizymes have been detected: i) Az1 functions as an inhibitory protein that targets its enzyme for degradation as well as a negative regulator of polyamine transport, ii) Az2 possesses very similar properties to Az1, except that it does not stimulate degradation of ODC and iii) Az3 is expressed only during spermatogenesis. The bacterial Az (AtoC) functions as an inhibitory protein to ODC, as well as transcriptional regulator for the two component AtoS-AtoC system. This two component system regulates the expression of atoDAEB operon encoded enzymes, participates in the mechanism of chemotaxis and regulates the biosynthesis of polyhydroxybutyrate, while some other possible roles remain to be elucidated. Today, under the term antizyme one can find 22 entries in the SwissProt database and 21 in the TrEMBL database, respectively. We have proposed possible functions of this unique protein, but a lot of work must still be done, in order to understand its pleotropic role from mammals to archaea and eubacteria. For example, we still don't know if Az has a function as a transcriptional regulator in higher organisms and whether its nuclear localization can be ascribed to such an activity. Whether phosphorylation of Az plays a role in ODC regulation is also under investigation. Also, a possible role of polyamines or other catabolic products in activating the two-component system AtoS-AtoC, which will bring into interplay two apparently unrelated biological pathways, i.e short chain fatty acid catabolism and polyamine biosynthesis, remains to be elucidated. Uncertainties within some areas concerning the actual role of Az must be further clarified before it can be considered worthwhile to extend the scope of research in this particular area.
Abbreviations
Az, antizyme; ODC, ornithine decarboxylase; ADC, arginine decarboxylase
Acknowledgments
Acknowledgements
The authors are grateful to B. Klonaridou for her excellent technical assistance and Professor J. Georgatsos for his valuable discussion.
Contributor Information
Efthimia E Lioliou, Email: elioliou@chem.auth.gr.
Dimitrios A Kyriakidis, Email: kyr@chem.auth.gr.
References
- Cohen SS. What do the polyamines do. Nature. 1978;274:209–210. doi: 10.1038/274209a0. [DOI] [PubMed] [Google Scholar]
- Bachrach U. Function of Naturally Occurring Polyamines. New York: Academic Press; 1973. [Google Scholar]
- Campbell RA, Morris DR, Bartos D, Daves GD, Bartos F. Advances in Polyamine Research. 1 and 2. New York: Raven Press; 1978. [Google Scholar]
- Caldarera CM, Zappia V, Bachrach U. Advances in Polyamine Research. Vol. 3. New York: Raven Press; 1981. [Google Scholar]
- Bachrach U, Kaye A, Chayen R. Advances in Polyamine Research. Vol. 4. New York: Raven Press; 1982. [Google Scholar]
- Cohen SS. Introduction to the Polyamines. New Jersey: Prentice Hall; 1971. [Google Scholar]
- Russell DH. Polyamines in Normal and Neoplastic Growth. New York: Raven Press; 1973. [Google Scholar]
- Cohen SS. A Guide to the Polyamines. New York: Oxford University Press; 1998. [Google Scholar]
- Canellakis ES, Viceps-Madore D, Kyriakidis DA, Heller JS. The regulation and function of ornithine decarboxylase and of the polyamines. In: Horecker BL, Stadtman ER, editor. In Current Topics in Cellular Regulation. Vol. 15. New York: Academic Press Inc; 1979. pp. 155–202. [PubMed] [Google Scholar]
- Tabor CW, Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem J. 1986;234:249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE. Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res. 1988;48:759–774. [PubMed] [Google Scholar]
- Hayashi SI, Murakami Y. Rapid and regulated degradation of ornithine decarboxylase. Biochem J. 1995;306:1–10. doi: 10.1042/bj3060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace HM, Fraser AV, Hughes A. A perspective of polyamine metabolism. Biochem J. 2003;376:1–14. doi: 10.1042/BJ20031327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi SI, Canellakis ES. Ornithine decarboxylase antizymes. In: Hayashi SI, editor. In International Encyclopaedia of Pharmacology and Therapeutics. Vol. 129. New York: Pergamon Press Inc; 1989. pp. 47–58. [Google Scholar]
- Canellakis ES, Paterakis AA, Huang SC, Panagiotidis CA, Kyriakidis DA. Identification, cloning, and nucleotide sequencing of the ornithine decarboxylase antizyme gene of Escherichia coli. Proc Natl Acad Sci. 1993;90:7129–7133. doi: 10.1073/pnas.90.15.7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong WF, Heller JS, Canellakis ES. The appearance of an ornithine decarboxylase inhibitory protein upon the addition of putrescine to cell cultures. Biochim Biophys Acta. 1976;428:456–465. doi: 10.1016/0304-4165(76)90054-4. [DOI] [PubMed] [Google Scholar]
- Heller JS, Fong WF, Canellakis ES. Induction of a protein inhibitor of ornithine decarboxylase by the end products of its reaction. Proc Natl Acad Sci. 1976;73:1858–1862. doi: 10.1073/pnas.73.6.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck WT, Bellantone RA, Canellakis ES. Puromycin stimulation of rat liver ornithine decarboxylase activity. Nature. 1973;241:275–277. doi: 10.1038/241275a0. [DOI] [PubMed] [Google Scholar]
- Coffino P. Degradation of ornithine decarboxylase. In: Peters J-M, Harris JR, Finley D, editor. In Ubiquitin and the biology of the cell. New York: Plenum; 1998. pp. 411–427. [Google Scholar]
- Miyazaki Y, Matsufuji S, Hayashi S. Cloning and characterization of a rat gene encoding ornithine decarboxylase antizyme. Gene. 1992;113:191–197. doi: 10.1016/0378-1119(92)90395-6. [DOI] [PubMed] [Google Scholar]
- Fujita K, Murakami S, Hayashi S. A macromolelular inhibitor of the antizyme to ornithine decarboxylase. Biochem J. 1982;204:647–653. doi: 10.1042/bj2040647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Gesteland RF, Atkins JF. A second mammalian antizyme: conservation programmed ribosomal frameshifting. Genomics. 1998;52:119–129. doi: 10.1006/geno.1998.5434. [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Matsufuji S, Murakami Y, Gesteland RF, Atkins JF. Conservation of polyamine regulation by translational frameshifting from yeast to mammals. EMBO J. 2000;19:1907–1917. doi: 10.1093/emboj/19.8.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Rohrwasser A, Terreros DA, Gesteland RF, Atkins JF. Discovery of spermatogenesis, stage specific, ornithine decarboxylase antizyme: antizyme3. Proc Nath Acad Sci USA. 2000;97:4808–4813. doi: 10.1073/pnas.070055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosaka Y, Tanaka H, Yano Y, Masai K, Nozaki M, Yomogida K, Otani S, Nojima H, Nishimune Y. Identification and characterization of testis specific ornithine decarboxylase antizyme (OAZ-t) gene: expression in haploid germ cells and polyamine-induced frameshifting. Genes Cells. 2000;5:265–76. doi: 10.1046/j.1365-2443.2000.00324.x. [DOI] [PubMed] [Google Scholar]
- Coffino P. Polyamines in spermiogenesis: Not now, darling. Proc Natl Acad Sci USA. 2000;97:4421–4423. doi: 10.1073/pnas.97.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakidis DA, Heller JS, Canellakis ES. Modulation of ornithine decarboxylase activity in Escherichia coli by positive and negative effectors. Proc Natl Acad Sci. 1978;75:4699–4703. doi: 10.1073/pnas.75.10.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakidis DA, Heller JS, Canellakis ES. Purification of ornithine decarboxylase antizymes (Escherichia coli) Meth Enzymol. 1983;94:193–199. doi: 10.1016/S0076-6879(83)94032-6. [DOI] [PubMed] [Google Scholar]
- Kyriakidis DA. Effect of plant growth hormone and polyamines on ornithine decarboxylase activity during the germination of barley seeds. Plant Physiol. 1983;57:499–508. [Google Scholar]
- Pantazaki AA, Anagnostopoulos CG, Lioliou EE, Kyriakidis DA. Characterization of ornithine decarboxylase and regulation by its antizyme in Thermus thermophilus. Mol Cell Biochem. 1999;195:55–64. doi: 10.1023/A:1006984618465. [DOI] [PubMed] [Google Scholar]
- Sklaviadis TK, Georgatsos JG, Kyriakidis DA. Purification and properties of ornithine decarboxylase from Tetrahymena pyriformis. Biochem Biophys Acta. 1985;831:288–296. doi: 10.1016/0167-4838(85)90109-8. [DOI] [PubMed] [Google Scholar]
- Moris DR, Pardee AB. Multiple pathways of putrescine biosynthesis in Escherichia coli. J Biol Chem. 1966;241:3129–3135. [PubMed] [Google Scholar]
- Holtta E, Janne J, Pispa J. Ornithine decarboxylase from Escherichia coli : stimulation of the enzyme activity by nucleotides. Biochem Biophys Res Commun. 1972;47:1165–1171. doi: 10.1016/0006-291x(72)90957-6. [DOI] [PubMed] [Google Scholar]
- Holtta E, Janne J, Pispa J. The regulation of polyamine synthesis during the stringent control in Escherichia coli. Biochem Biophys Res Commun. 1974;59:1104–1111. doi: 10.1016/s0006-291x(74)80092-6. [DOI] [PubMed] [Google Scholar]
- Kurland CG. Translational accuracy in vitro. Cell. 1982;28:201–202. doi: 10.1016/0092-8674(82)90336-1. [DOI] [PubMed] [Google Scholar]
- Anagnostopoulos CG, Kyriakidis DA. Regulation of the Escherichia coli biosynthetic ornithine decarboxylase activity by phosphorylation and nucleotides. Biochem Biophys Acta. 1996;1297:228–234. doi: 10.1016/S0167-4838(96)00107-0. [DOI] [PubMed] [Google Scholar]
- Boyle SM, Markham GD, Hafner EW, Wright JM, Tabor H, Tabor CW. Expression of the cloned genes encoding the putrescine biosynthetic enzymes and methionine adenosyltransferase of Escherichia coli (speA, speB, speC and metK) Gene. 1984;30:129–136. doi: 10.1016/0378-1119(84)90113-6. [DOI] [PubMed] [Google Scholar]
- Insel PA, Fenno J. Cyclic AMP-dependent protein kinase mediates a cyclic AMP-stimulated decrease in ornithine and S-adenosylmethionine decarboxylase activities. Proc Natl Acad Sci USA. 1978;75:862–865. doi: 10.1073/pnas.75.2.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller JS, Chen KY, Kyriakidis DA, Fong WF, Canellakis ES. The modulation of the induction of ornithine decarboxylase by spermine, spermidine and diamines. J Cell Physiol. 1978;96:225–234. doi: 10.1002/jcp.1040960211. [DOI] [PubMed] [Google Scholar]
- Heller JS, Canellakis ES. Minimal requirements for the induction of the antizyme to ornithine decarboxylase. In: Gaugas JM, editor. In Polyamines in Biomedical Research. New York: Wiley J; 1980. pp. 135–145. [Google Scholar]
- Gritli-Linde A, Nilsson J, Bohlooly M, Heby YO, Linde A. Nuclear translocation of antizyme and expression of ornithine decarboxylase and antizyme are developmentally regulated. Dev Dyn. 2001;220:259–275. doi: 10.1002/1097-0177(20010301)220:3<259::AID-DVDY1100>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Panagiotidis CA, Huang S, Canellakis ES. Post-translational and transcriptional regulation of polyamine biosynthesis in Escherichia coli. Int J Biochem. 1994;26:991–1001. doi: 10.1016/0020-711X(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Huang SC, Kyriakidis DA, Rinehart CA, Canellakis ES. Reversal of the antizyme inhibition of ornithine decarboxylase by nucleic acids. Biochem Pharmacol. 1984;33:1383–1386. doi: 10.1016/0006-2952(84)90200-4. [DOI] [PubMed] [Google Scholar]
- Canellakis ES, Kyriakidis DA, Rinehart CA, Huang SC, Panagiotidis CA, Fong WF. Regulation of polyamine biosynthesis by antizyme and some recent developments relating the induction of polyamine biosynthesis to cell growth. Bioscience Reports. 1985;5:189–204. doi: 10.1007/BF01119588. [DOI] [PubMed] [Google Scholar]
- Lioliou EE. PhD Thesis. Aristotle University of Thessaloniki, Chemistry Department; 2003. Polyamine biosynthesis regulation and metabolic control of a two-component signal transduction system. [Google Scholar]
- Chen C, Cooke PA, Rudd KE, Shanley MS. Direct submission EMBL/GenBank/DDBJ Databases. Accession number Q06065. 1994.
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley MCollado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Pauli G, Overath P. ato operon: a highly inducible system for acetoacetate and butyrate degradation in Escherichia coli. Eur J Biochem. 1972;29:553–562. doi: 10.1111/j.1432-1033.1972.tb02021.x. [DOI] [PubMed] [Google Scholar]
- Jenkins LS, Nunn WD. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli : the ato system. J Bacteriol. 1987;169:42–52. doi: 10.1128/jb.169.1.42-52.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins LS, Nunn WD. Regulation of the ato operon by the atoC gene in Escherichia coli. J Bacteriol. 1987;169:2096–2102. doi: 10.1128/jb.169.5.2096-2102.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DP, Cronan JE. Two-carbon compounds and fatty acids as carbon sources. In: Neidhardt FC, editor. In Escherichia coli and Salmonella typhimurium Cellular and Molecular Biology. Vol. 1. Washigton DC: Am Soc Microbiol; 1996. pp. 343–357. [Google Scholar]
- Berlyn MKB. Linkage map of Escherichia coli K-12, edition 10: The traditional map. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS, Kofoid EC. Communication modules in bacterial signalling proteins. Ann Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/S1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signalling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/S0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- Lioliou EE, Theodorou MC, Panagiotidis CA, Kyriakidis DA. Phosphorylation of antizyme by a membrane protein in E.coli [abstract] Eur J Biochem. 2001;268:PS3–102. [Google Scholar]
- Rhie HG, Dennis D. Role of fadR and atoC(Con) mutations in poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthesis in recombinant pha+ Escherichia coli. Appl Environ Microbiol. 1995;61:2487–2492. doi: 10.1128/aem.61.7.2487-2492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. Transcriptome analysis of all two component regulatory system mutants of Escherichia coli K-12. Mol Microbiol. 2002;46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- Zhou L, Lei XH, Bochner BR, Wanner BL. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J Bacteriol. 2003;185:4956–4972. doi: 10.1128/JB.185.16.4956-4972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Gesteland RF, Atkins JF. Does antizyme exist in Escherichia coli? Mol Microbiol. 1998;29:1521–1522. doi: 10.1046/j.1365-2958.1998.01032.x. [DOI] [PubMed] [Google Scholar]
- Zhu C, Karplus K, Grate L, Coffino P. A homolog of mammalian antizyme is present in fission yeast Schizosaccharomyces pombe but not detected in budding yeast Saccharomyces cerevisiae. Bioinformatics. 2000;16:478–481. doi: 10.1093/bioinformatics/16.5.478. [DOI] [PubMed] [Google Scholar]
- Reitzer LJ, Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986;45:785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- Ninfa AJ, Reitzer LJ, Magasanik B. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell. 1987;50:1039–1046. doi: 10.1016/0092-8674(87)90170-X. [DOI] [PubMed] [Google Scholar]
- Chen P, Reitzer LJ. Active contribution of two domains to cooperative DNA binding of the enhancer-binding protein nitrogen regulator I (NtrC) of Escherichia coli : Stimulation by phosphorylation and the binding of ATP. J Bacteriol. 1995;177:2490–2496. doi: 10.1128/jb.177.9.2490-2496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SC, North AK, Kustu S. Mechanism of transcriptional activation by NtrC. In: Hoch JA, Silhavy TJ, editor. In Two-component signal transduction. Washington DC: American Society for Microbiology Press; 1995. pp. 147–158. [Google Scholar]
- Yan D, Kustu S. "Switch I" mutant forms of the bacterial enhancer-binding protein NtrC that perturb the response to DNA. Proc Natl Acad Sci USA. 1999;96:13142–13146. doi: 10.1073/pnas.96.23.13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul , Stephen F, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne A, Bruggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H, Johann A, Lienard T, Gohl O, Martinez-Arias R, Jacobi C, Starkuviene V, Schlenczeck S, Dencker S, Huber R, Klenk HP, Kramer W, Merkl R, Gottschalk G, Fritz HJ. The genome sequence of the extreme thermophile Thermus thermophilus. Nat Biotechnol. 2004;22:547–553. doi: 10.1038/nbt956. [DOI] [PubMed] [Google Scholar]
- Itoh T, Aiba H, Baba T, Fujita K, Hayashi K, Inada T, Isono K, Kasai H, Kimura S, Kitakawa M, Kitagawa M, Makino K, Miki T, Mizobuchi K, Mori H, Mori T, Motomura K, Nakade S, Nakamura Y, Nashimoto H, Nishio Y, Oshima T, Saito N, Sampei G, Seki Y, Sivasundaram S, Tagami H, Takeda J, Takemoto K, Wada C, Yamamoto Y, Horiuchi T. A 460-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 40.1–50.0 min region on the linkage map. DNA Res. 1996;3:379–392. doi: 10.1093/dnares/3.6.379. [DOI] [PubMed] [Google Scholar]
- Welch RA, Burland V, Plunkett GD, III, Redford P, Roesch P, Rasko DA, Buckles EL, Liou S-R, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HLT, Donnenberg MS, Blattner FR. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Microbial Genomes Annotation Project Direct Submission 07-NOV-2002. National Center for Biotechnology Information, NIH, Bethesda, MD 20894, USA.
- Methe BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ, Dodson RJ, Madupu R, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Gwinn M, Kolonay JF, Sullivan SA, Haft DH, Selengut J, Davidsen TM, Zafar N, White O, Tran B, Romero C, Forberger HA, Weidman J, Khouri H, Feldblyum TV, Utterback TR, Van Aken SE, Lovley DR, Fraser CM. Genome of Geobacter sulfurreducens : metal reduction in subsurface environments. Science. 2003;302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, Davidsen TM, DeBoy RT, Fouts DE, Haft DH, Selengut J, Ren Q, Brinkac LM, Madupu R, Kolonay JF, Durkin AS, Daugherty SC, Shetty J, Shvartsbeyn A, Gebregeorgis E, Geer K, Tsegaye G, Malek JA, Ayodeji B, Shatsman S, McLeod MP, Majs D, Howell JK, Pal S, Amin A, Vashisth P, McNeill TZ, Weinstock GM, Norris SJ, Fraser CM, Paulsen IT. Direct Submission (29-JAN-2004) The Institute for Genomic Research, 9712 Medical Center Dr, Rockville, MD 20850 USA.
- Heidelberg JF, Seshadri R, Haveman SA, Hemme CL, Paulsen IT, Kolonay JF, Eisen JA, Ward N, Methe B, Brinkac LM, Daugherty SC, DeBoy RT, Dodson RJ, Durkin AS, Madupu R, Nelson WC, Sullivan SA, Fouts DE, Haft DH, Selengut J, Peterson JD, Davidsen TM, Zafar N, Zhou L, Radune D, Dimitrov G, Hance M, Tran K, Khouri HM, Gill J, Utterback TR, Feldblyum TV, Wall JD, Voordouw G, Fraser CM. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat Biotechnol. 2004;22:554–559. doi: 10.1038/nbt959. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Norris SJ, Weinstock GM, White O, Sutton GG, Dodson R, Gwinn M, Hickey EK, Clayton R, Ketchum KA, Sodergren E, Hardham JM, McLeod MP, Salzberg S, Peterson J, Khalak H, Richardson D, Howell JK, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton MD, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Watthey L, Weidman J, Smith HO, Venter JC. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- Caberoy NB, Welch RD, Jakobsen JS, Slater SC, Garza AG. Global mutational analysis of NtrC-Like activators in Myxococcus xanthus : Identifying activator mutants defective for motility and fruiting body development. J Bacteriol. 2003;185:6083–6094. doi: 10.1128/JB.185.20.6083-6094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, Brandt P, Nyakatura GJ, Droege M, Frishman D, Rattei T, Mewes H, Wagner M. Direct submission (15-JAN-2003) Department of Microbial Ecology, University of Vienna, Althanstr 14, A-1090 Wien, Austria, Complete annotation. http://mips.gsf.de/services/genomes/uwe25
- Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, Meyer F, Sockett RE, Schuster SC. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- Radnedge L, Agron PG, Worsham PL, Andersen GL. Genome plasticity in Yersinia pestis. Microbiology. 2002;148:1687–1698. doi: 10.1099/00221287-148-6-1687. [DOI] [PubMed] [Google Scholar]
- Jin Q, Yuan ZH, Xu JG, Wang Y, Shen Y, Lu WC, Wang JH, Liu H, Yang J, Yang F, Qu D, Zhang XB, Zhang JY, Yang GW, Wu HT, Dong J, Sun LL, Xue Y, Zhao AL, Gao YS, Zhu JP, Kan B, Chen SX, Yao ZJ, He BK, Chen RS, Ma DL, Qiang BQ, Wen YM, Hou YD, Yu J. Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 2002;30:4432–4441. doi: 10.1093/nar/gkf566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Microbial Genomes Annotation Project Direct Submission (25-JUN-2001) National Center for Biotechnology Information, NIH, Bethesda, MD 20894 USA.
- Perna NT, Plunkett G, III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta E, Potamousis K, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki Y, Masui N, Fuji F, Hirama C, Nakamura Y, Ogasawara N, Kuhara S, Horikoshi K. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhartsberger S, Huber A, Lottspeich F, Bock A. The hydH/G genes from Escherichia coli code for a zinc and lead responsive two-component regulatory system. J Mol Biol. 2001;307:93–105. doi: 10.1006/jmbi.2000.4451. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han C-G, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. Complete genome sequence of Salmonella enterica serovar typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MTG, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O'Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]


