Abstract
We have developed a genetic system, called degrakine, that specifically and stably inactivates chemokine receptors (CKR) by redirecting the ability of the HIV-1 protein, Vpu, to degrade CD4 in the endoplasmic reticulum (ER) via the host proteasome machinery. To harness Vpu’s proteolytic targeting capability to degrade new receptors, we fused a chemokine with the C terminal region of Vpu. The fusion protein, or degrakine, accumulates in the ER, trapping and functionally inactivating its target CKR. We have demonstrated that degrakines based on SDF-1 (CXCL12), MDC (CCL22) and RANTES (CCL5) specifically inactivate their respective receptor functions. Using a retroviral vector expressing the SDF-1 degrakine, we have established that CXCR4 is required for the homing of hematopoietic stem/progenitor cells (HSPC) to the bone marrow immediately after transplantation. Thus the degrakine provides an effective genetic tool to dissect receptor functions in a number of biological systems in vitro and in vivo.
CKRs are G protein–coupled receptors (GPCRs) that have critical roles in controlling the migration of HSPCs and mature leukocytes and that participate in important pathophysiological processes including inflammation, allograft rejection, autoimmunity, viral infection and lymphoid development1–5. Approximately 20 CKRs and over 40 chemokines are encoded in the mouse or human genome1,2. Chemokines are subdivided into CXC (alpha), CC (beta), C (gamma) and CX3C (delta) classes, depending on the position of conserved cystine residues in the polypeptide. Each CKR is specific to one or multiple chemokines, and each chemokine can bind to one or multiple CKRs. Despite recent interest in chemokines and CKRs, their involvement in HSPC and leukocyte migration remains poorly defined because of the large number of family members, cross-reactivity and a lack of efficient genetic tools to dissect their functions.
Gene targeting in mice has been done for a limited number of CKRs, but it is time-consuming and expensive to generate knockout mice. In addition, it is difficult to breed mice with mutations in multiple CKRs because the genes encoding them are closely linked at two chromosomal loci2. A common approach to studying CKR function is through the use of blocking antibodies, an approach that has limitations including toxicity and the lack of target specificity and in vivo efficacy6. Another method, the intrakine system, inhibits the level of CKR surface expression by sequestration of the target CKR in the ER7. However, the intrakine seems to have a paracrine activity, probably as a result of secretion of the intrakine protein out of the cell8, and this method may also lead to toxic accumulation of intrakine-CKR protein complexes in the ER. More recently, RNA interference technology has been used to knock down gene function in vitro and in vivo9–13.
Many viruses have intricate, naturally evolved mechanisms to interact with receptors on infected cells and thereby elude the host’s immune system and facilitate replication14,15. HIV-1 uses its envelope and Vpu proteins to specifically target CD4 for degradation in the ER14–16. The C terminal region of Vpu mediates this interaction by binding with the β-TrCP–Skp1 protein complex, which leads to CD4 ubiquitination and degradation16,17. Phosphorylation of the serine residues Ser52 and Ser56 in Vpu is critical for β-TrCP binding and CD4 degradation, although Vpu itself is resistant to ubiquitination and degradation16,18. To harness Vpu’s proteolytic targeting capability to degrade new receptors, we fused a chemokine with the C terminal region of Vpu. The chemokine–VpuC fusion protein (degrakine) is designed to specifically downregulate the surface expression and inhibit the function of its target CKR. Combining the degrakine with retroviral gene transfer, we have developed a genetic system to rapidly and specifically inactivate CKRs in either human or mouse cells in vitro and in vivo.
The prototype degrakine is designed with the chemokine SDF-1α (CXCL12), which specifically binds CXCR4 (refs. 19–22). SDF-1 is expressed from a single gene, but has two isoforms, SDF-1α and SDF-1β, that are generated by alternate splicing23. Although originally identified as a pre-B-cell growth factor expressed in mouse bone marrow21,23–25, SDF-1α elicits a potent migratory response from monocytes, T cells and HSPCs21,25,26. Mutations in either SDF-1 or CXCR4 in mice lead to embryonic lethality with neurological and hematological abnormalities27,28. Additionally, CXCR4 is required for stable engraftment of human HSPC cells into the bone marrow of NOD-SCID mice and of mouse fetal liver cells into lethally irradiated recipient mice28,29. However, it remains unclear whether CXCR4 mediates HSPC homing to the bone marrow, retention and proliferation within the bone marrow, or long-term maintenance of progenitor cells. Using the SDF-1 degrakine, we show that CXCR4 plays a role in HSPC homing to the bone marrow immediately after HSPC transplantation.
RESULTS
Degrakine design
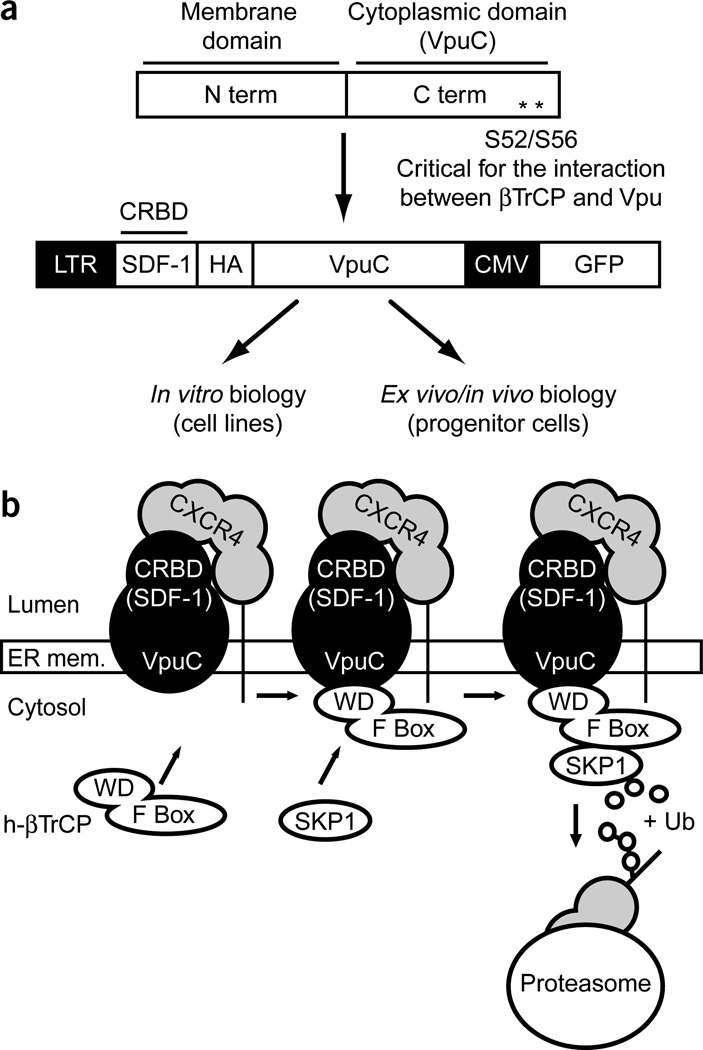
The degrakine protein comprises two domains. The C-terminal domain is aa28–81 (VpuC) of Vpu, which recruits the proteasome16. The N-terminal domain consists of a chemokine (or a chemokine receptor binding domain, CRBD) (Fig. 1a). The prototype degrakine is constructed with SDF-1 and a hemagglutinin tag as the CRBD. The murine stem cell virus long terminal repeat (MSCV LTR) drives expression of the SDF-1 degrakine fusion protein, and a cytomegalovirus (CMV)-driven enhanced green fluorescent protein (EGFP) marker is included in the retroviral vector (Fig. 1a). In the proposed SDF-1 degrakine pathway, the degrakine localizes to the ER, traps the target CKR (CXCR4) in the ER and recruits the proteasome to degrade the target CKR (Fig. 1b).
Figure 1.
Design and function of the degrakine system. (a) The SDF-1 degrakine consists of SDF-1 fused to the C terminus of Vpu (aa28–81). The SDF-1 degrakine was cloned into a MSCV retroviral vector, which also encodes EGFP under the control of a CMV promoter. (b) The SDF-1 degrakine localizes to the ER, binds CXCR4 and prevents CXCR4 translocation to the cell surface. Additionally, VpuC interacts with β-TrCP and targets CXCR4 for degradation via the proteasome. HA, hemagglutinin.
Degrakine ER localization, stable expression and noncytotoxicity
The SDF-1 degrakine protein was expressed and localized to the ER. Protein expression was confirmed by western blotting with anti-Vpu and anti-hemagglutinin (data not shown). ER localization was determined by coexpression of the ER-accumulated YFP-KDEL protein. Staining of human osteosarcoma U2OS cells with anti-hemagglutinin showed that the SDF-1 degrakine colocalized with YFP-KDEL in the ER (Fig. 2a).
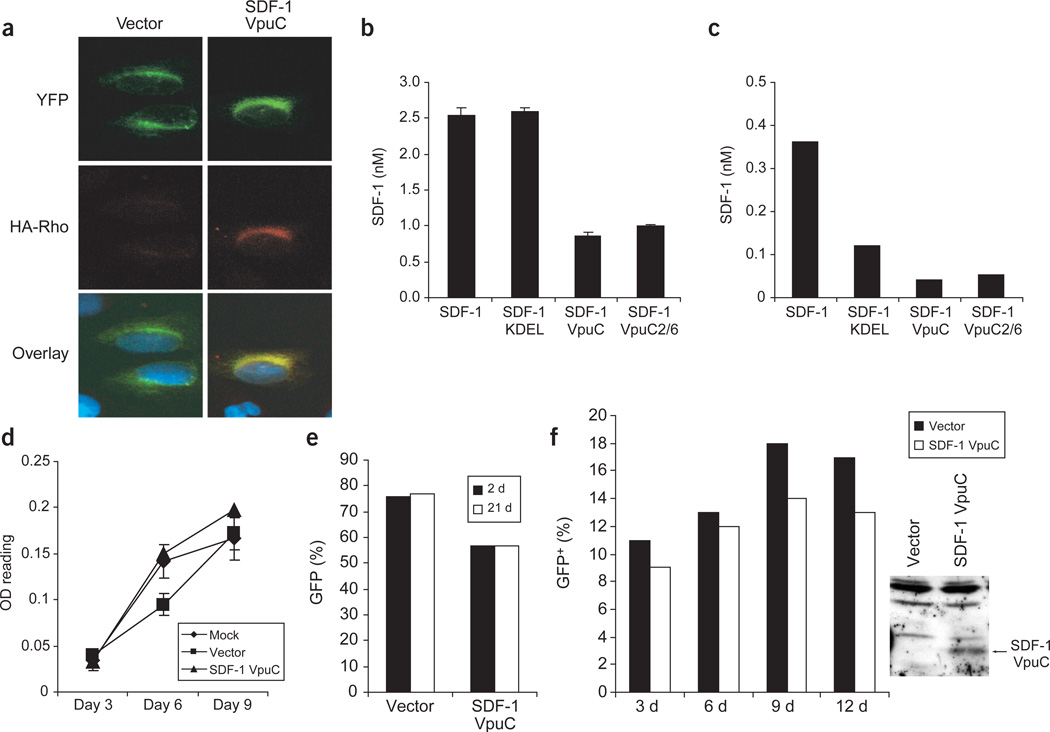
Figure 2.
The SDF-1 degrakine localizes to the ER and is stably expressed. (a) The EGFP in the SDF-1 degrakine vector was replaced with YFP-KDEL. U2OS cells were transfected, stained with anti-HA monoclonal antibody, followed by anti-mIgG rhodamine (degrakine) and DAPI and visualized by confocal microscopy. (b,c) 293T cells were transfected (b) and SupT1 cells were transduced (c) and monitored by ELISA for SDF-1 secretion into the medium. Similar results were observed in two independent experiments. (d) The proliferation of transduced SupT1 cells was measured at the indicated time points using a MTT assay (OD reading). Results are representative of two independent experiments with each sample in triplicate. (e) Transduced SupT1 cells assayed by FACS for EGFP percentage at 2 d and 3 weeks in culture. (f) GFP (%) Transduced cKit+ bone marrow cells were monitored for EGFP percentage at 3, 6, 9 and 12 d in vitro. At day 12, 1 × 106 cells were harvested and assayed for degrakine expression by western blotting (anti-Vpu).
SDF-1 contains a signal peptide sequence that directs its transport to the ER and into the secretory pathway21. Thus the SDF-1 degrakine could be secreted from the cell. To monitor this possibility, we analyzed human embryonic kidney 293T cells and SupT1 T-lymphocyte cells expressing the indicated vectors for SDF-1 secretion by enzyme-linked immunosorbent assay (ELISA). A control vector expressing SDF-1 was used to determine maximum secretion without ER retention. Additionally, a construct expressing the SDF-1 intrakine, SDF-1KDEL7, was used to demonstrate KDEL-mediated retention of SDF-1 within the ER. 293T cells transiently expressing SDF-1 and SDF-1KDEL both secreted SDF-1 efficiently into the medium (~2.5 nM, Fig. 2b). In contrast, SDF-1VpuC and SDF-1VpuC2/6 secreted 66% less SDF-1 (Fig. 2b). In stably transduced SupT1 cells, the presence of SDF-1VpuC construct resulted in an 89% and 67% decrease in secreted SDF-1 as compared with SDF-1 and SDF-1KDEL, respectively (Fig. 2c). The SDF-1VpuC2/6 protein with mutations inhibiting Vpu’s interaction with βTrCP was secreted in amounts similar to those of SDF-1VpuC (Fig. 2b,c). Thus, the SDF-1 degrakine is retained within the cell and may contain an unidentified ER retention motif because no known ER retention signal has been identified in the VpuC domain.
Expression of the SDF-1 degrakine did not affect cell viability or proliferation in a number of human cell lines, primary mouse thymocytes and mouse HSPCs. Comparable rates of proliferation were observed over 9 d in culture in parental SupT1 cells and cells transduced with vector or SDF-1VpuC (Fig. 2d). Cells transduced with vector or with SDF-1VpuC were also monitored by fluorescence-activated cell sorting (FACS) for EGFP expression at 2 d and 3 weeks after transduction. Constant levels of EGFP expression were observed at both time points (Fig. 2e), showing that the degrakine protein is not cytotoxic. In addition, stable expression of the degrakine was also observed in primary mouse bone marrow cells cultured for 12 d in vitro (Fig. 2f). Therefore, the degrakine fusion protein does not affect cell viability or proliferation in various types of cells.
Stable and specific suppression of CXCR4 expression
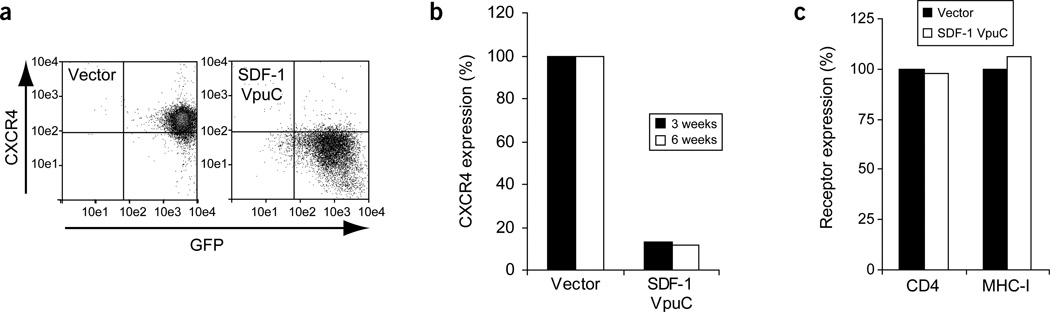
The SDF-1 degrakine induced rapid and stable downregulation of CXCR4 surface expression in SupT1 cells and Jurkat lymphoblastic leukemia cells. Three days after transduction, SupT1 cells showed a ~70% reduction in CXCR4 surface expression compared with the vector control, and after 1 week in culture, CXCR4 was downregulated by ~90% (Fig. 3a). At 3 and 6 weeks in culture, the SDF-1 degrakine maintained stable CXCR4 downregulation (Fig. 3b). Surface staining of CD4 and MHC-I confirmed that the VpuC-based degrakine did not alter the expression of CD4 or MHC-I as full-length Vpu14,17 (Fig. 3c).
Figure 3.
CXCR4 expression is stably and specifically downregulated by the SDF-1 degrakine. (a) A representative FACS plot of independent SupT1 infections showing CXCR4 surface expression 1 week after transduction. (b) The cells were reanalyzed for CXCR4 expression at 3 and 6 weeks after transduction. (c) CD4 and MHC-I expression was monitored at the 3-week time point. Identical results were observed in two independent experiments with SupT1 cells and Jurkat cells.
β-TrCP is involved in receptor downregulation
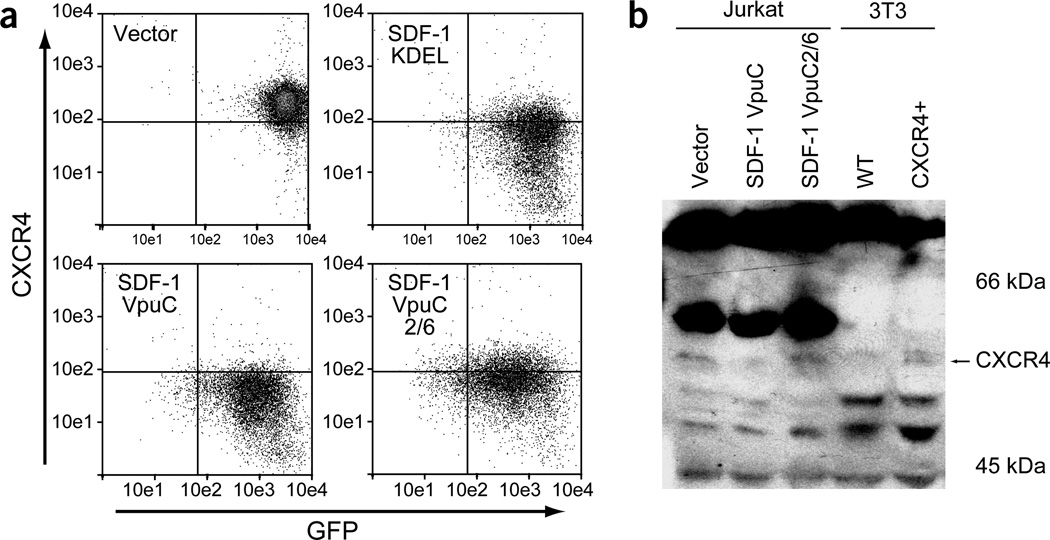
The degrakine system requires the interaction of VpuC with β-TrCP for efficient CKR downregulation. This was demonstrated using a VpuC mutant, VpuC2/6, which contains alanine substitutions at Ser52 and Ser56. These mutations block the interaction between Vpu and β-TrCP, which leads to the loss of Vpu-mediated CD4 degradation16,18. SDF-1VpuC2/6 therefore binds and sequesters CXCR4 in the ER, but cannot recruit the proteasome. We also used the SDF-1 intrakine, SDF-1KDEL7, as a control for CXCR4 downregulation by ER sequestration without degradation. Cells expressing vector, SDF-1KDEL, SDF-1VpuC or SDF-1VpuC2/6 were analyzed for CXCR4 expression by FACS. SDF-1VpuC inhibited CXCR4 expression by ~90% (Figs. 3a and 4a). In contrast, SDF-1VpuC2/6 and SDF-1KDEL (constructs that only sequester CXCR4 in the ER) showed a ~60–70% decrease in CXCR4 surface expression (Fig. 4a). These results demonstrate that the interaction between Vpu and β-TrCP is important in efficient CXCR4 downregulation by the SDF-1 degrakine.
Figure 4.
The interaction between VpuC and β-TrCP is important for efficient CXCR4 downregulation. (a) A representative FACS plot of independent SupT1 infections showing CXCR4 surface expression 1 week after transduction. Similar results were observed in Jurkat cells. (b) Total cytoplasmic lysates were analyzed by western blotting with a polyclonal anti-CXCR4. NIH3T3 cells stably expressing human CXCR4 were used as a positive control. Similar results were observed in two independent experiments.
To demonstrate VpuC-mediated degradation of CXCR4, we measured total CXCR4 protein in transduced Jurkat cells by western blotting. NIH3T3 cells stably expressing human CXCR4 were used as a positive control. Total levels of CXCR4 were similar in cells expressing vector or SDF-1VpuC2/6, but lower in cells expressing SDF-1VpuC (Fig. 4b). The expression levels of SDF-1VpuC and SDF-1VpuC2/6 were similar, as determined by western blotting with anti-VpuC (data not shown). Therefore, the interaction between VpuC and β-TrCP leads to the degradation of CXCR4 by the SDF-1 degrakine.
Functional inactivation of target CKRs by degrakines
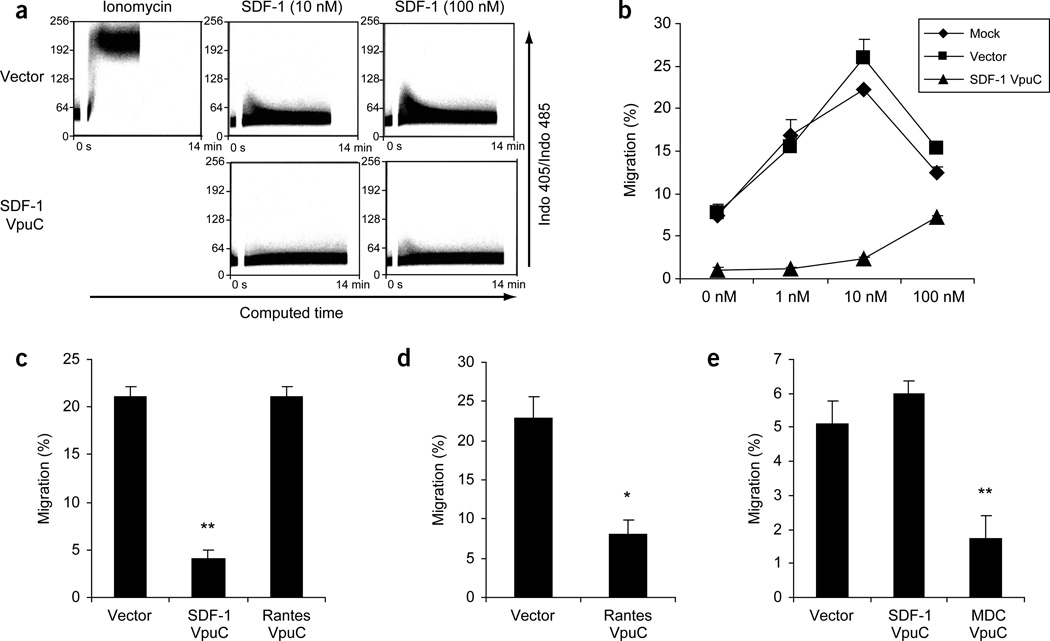
SDF-1 binding to CXCR4 resulted in the release of intracellular calcium into the cytosol (calcium influx)30. Intracellular calcium influx in response to SDF-1 in cells expressing the indicated proteins was measured by FACS. SDF-1VpuC inhibited the ability of SDF-1 to induce calcium influx even at 100 nM, indicating that the SDF-1 degrakine efficiently inactivated CXCR4 function (Fig. 5a).
Figure 5.
Degrakines specifically inhibit their target CKR function. (a) Transduced SupT1 cells were loaded with Indo-1AM, stimulated with SDF-1 and monitored by FACS for the change in Indo-1AM fluorescence. (b) Parental SupT1 cells (Mock), cells expressing vector or SDF-1VpuC were assayed for migration response to SDF-1 in a trans-well assay. Each sample was performed in triplicate and the assay is representative of two independent experiments. (c,d) THP-1 cells were transduced, cultured for 1 week and assayed for migration in response to 100 nM SDF-1 (c), or 100 nM MIP-1β (d) in a trans-well migration assay. (e) Mouse thymocytes were transduced, cultured for 1 week and measured for their migration response to 100 nM MDC in a trans-well assay. *, P < 0.05. **, P < 0.005.
CXCR4 mediated cell migration in response to SDF-1 (ref. 21,22). Mock or vector control cells migrated efficiently in response to 1 nM and 10 nM of SDF-1, with a typical oversaturation response at 100 nM. In contrast, cells expressing SDF-1VpuC showed no response to 1 nM and 10 nM of SDF-1 and only a low migration response to 100 nM of SDF-1 (Fig. 5b), a result consistent with low CXCR4 expression. The reduced migration of SDF-1VpuC cells in the absence of exogenous SDF-1 was probably a result of SDF-1-like activity in the culture medium. These experiments show that both CXCR4-mediated calcium influx and chemotaxis are inhibited by the SDF-1 degrakine.
Two additional degrakine constructs were tested to demonstrate the specificity and general utility of the system. RantesVpuC and MDCVpuC were designed to inhibit CCR5 and CCR4, respectively. Human acute monocytic leukemia THP-1 cells expressing both CXCR4 and CCR5 (ref. 31) were used to demonstrate the specific targeting of CXCR4 and CCR5 by SDF-1VpuC and RantesVpuC, respectively. The migration response to SDF-1 was inhibited by 81% only in THP-1 cells expressing SDF-1VpuC (Fig. 5c), whereas response to MIP-1β was reduced 65% in THP-1 cells expressing RantesVpuC (Fig. 5d). Similarly, MDCVpuC-transduced mouse thymocytes, which express CCR4 (ref. 32), showed a 66% reduction in migration response to 100 nM MDC as compared with vector or SDF-1VpuC (Fig. 5e). Thus, the degrakines specifically inactivated their target CKR functions. Taken together, these results suggest that the degrakine system is applicable to inactivating chemokine receptors in general.
CXCR4 is required for the homing of HSPC to the bone marrow
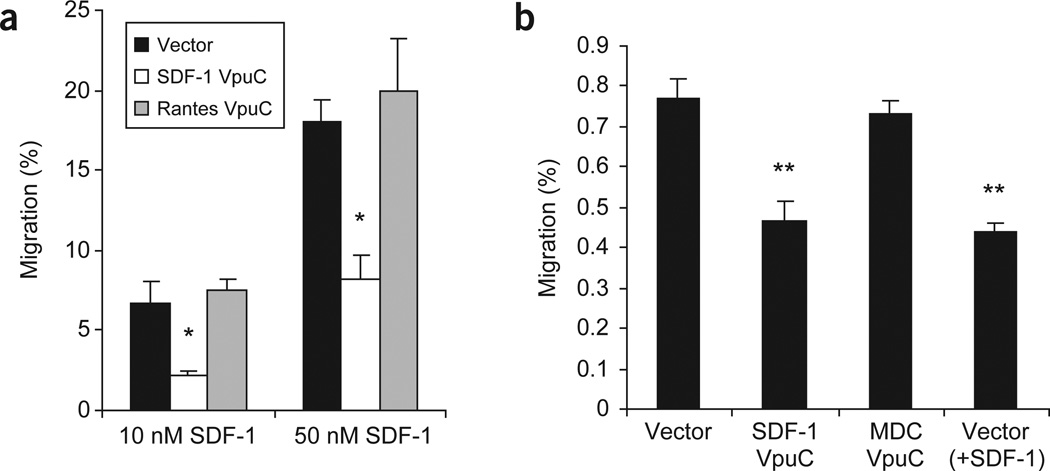
SDF-1 is a potent migratory chemokine for HSPC cells in vitro26,33,34. We used the SDF-1 degrakine to determine whether CXCR4 is required for the homing of HSPC to the bone marrow in vivo. Bone marrow cells from B6/Ly5.2 mice were enriched for cKit+ progenitor cells35 and transduced with the MSCV-based retroviral vectors. Stable EGFP expression in vector or SDF-1 degrakine–transduced progenitor cells was observed in vitro, and degrakine expression was confirmed by anti-Vpu western blotting 12 d after transduction (Fig. 2f). The SDF-1 degrakine is therefore not cytotoxic to mouse HSPCs. Progenitor cells expressing SDF-1 or RANTES degrakines were assayed for their migratory response to 10 nM and 50 nM of SDF-1. Cells expressing SDF-1VpuC showed a 60–70% reduction in migration response to SDF-1 whereas cells expressing the RANTES degrakine showed normal responses to SDF-1 (Fig. 6a). To investigate CXCR4’s role in HSPC homing during transplantation, we injected intravenously B6/Ly5.2 bone marrow cKit+ cells expressing vector, SDF-1VpuC or MDCVpuC into lethally irradiated B6/Ly5.1 recipient mice. Three hours after transplantation, bone marrow cells were harvested and analyzed by FACS for Ly5.2 (donor) and EGFP (degrakine-positive) expression. SDF-1VpuC inhibited short-term homing of progenitor cells by ~40% (Fig. 6b). As a control, vector-transduced progenitor cells were pulsed with SDF-1 (10 nM) before injection to downregulate and desensitize CXCR4. These cells showed the same homing defect as SDF-1VpuC-transduced HSPC (Fig. 6b). HSPCs expressing the MDC degrakine, which inhibited thymocyte migration to MDC in vitro (Fig. 5e), showed a bone marrow homing percentage similar to the control cells (Fig. 6b). This is consistent with the fact that CCR4 is not expressed on HSPC, and MDC is not expressed in the bone marrow34. These results demonstrate that CXCR4 plays an important role in HSPC homing to the bone marrow immediately after transplantation.
Figure 6.
The SDF-1 degrakine inhibits CXCR4-mediated HSPC migration in vitro and homing to the bone marrow in vivo. (a) Transduced HSPCs (cKit+) were assayed for migration to 10 nM and 50 nM of SDF-1 in a trans-well assay. Each sample was performed in triplicate and the results are representative of two independent experiments. (b) B6/Ly5.2+ HSPCs (cKit+) were transduced, injected into lethally irradiated B6/Ly5.1 recipients and harvested from the recipient’s bone marrow 3 h after transplantation. Vector + SDF-1 cells were vector-transduced cells pulsed with 10 nM of SDF-1 immediately before injection. Three mice were used for each vector and the assay is representative of two independent experiments. *, P < 0.005. **, P < 0.005.
DISCUSSION
A number of chemokines and their receptors are involved in normal hemato-lymphopoiesis and in the immune response to pathogens. We have developed a genetic approach for the specific functional inactivation of CKRs in various cell types in vitro and in vivo. In stably transduced cell lines and mouse thymocytes or HSPCs, the degrakine does not affect cell proliferation or viability. The functional inactivation of CXCR4 by the SDF-1 degrakine was verified by a reduction in CXCR4 expression, inhibition of SDF-1 induced calcium influx and blockage of chemotaxis in human T cell lines, mouse thymocytes and mouse HSPCs. The specificity and general utility of the degrakines were illustrated by the specific inactivation of their respective CKRs by SDF-1, MDC and RANTES degrakines. Furthermore, we used the SDF-1 degrakine to demonstrate that CXCR4 is involved in the homing of mouse HSPCs immediately after bone marrow transplantation, a finding that may partly explain the requirement of CXCR4 for the engraftment of human or mouse HSPCs in the mouse bone marrow compartment27–29.
The degrakine system offers a number of advantages for studying the function of CKRs in HSPC functions. First, it may be applicable in human HSPCs where conventional genetic approaches are not feasible. Second, multiple CKRs can be targeted by a single degrakine or by a combination of coexpressed degrakines. Finally, it is also feasible to combine the degrakine system with existing knockout mice to inactivate multiple CKRs.
Genetic manipulation of HSPCs from mature animals is directly relevant to clinical settings. The use of degrakines in these cells may elucidate the functions of specific CKRs during hemato-lymphopoiesis and immune responses in adults. In addition to blocking antibodies, which may have limited target specificity and in vivo efficacy, the intrakine system has also been used to inhibit CKR expression7. However, the intrakine secretes high levels of the intrakine protein out of the cell8 (Fig. 2b,c), is less efficient than the degrakine in downregulating CKR expression (Fig. 4a) and may cause an accumulation of intrakine-CKR proteins in the ER. In contrast, the degrakine is designed to trap the CKR in the ER and target it for degradation.
We envision several possible modifications of the degrakine system. First, one might be able to derive the CRBD from single-chain antibodies or peptides that specifically bind a target CKR. Such derivatives with enhanced CKR binding activity, but no agonistic or antagonistic activity, should substantially improve degrakine efficiency and potential applications. Second, the approach could be adapted to receptors other than chemokine receptors. Third, degrakines could be combined with stable hairpin RNAi in the retroviral system, so that the CKR is targeted for inactivation at both the mRNA and protein levels. This might overcome the variable success rate of RNAi alone. Finally, in combination with recent advances in in vivo imaging technology36,37, the degrakine system might be used to analyze the migration of HSPCs, T cells and dendritic cells in vitro and in vivo.
METHODS
Animals
All animals were housed at the University of North Carolina at Chapel Hill in a sterile animal facility. Total bone marrow was harvested from the tibias and femurs of 1- to 2-month-old B6/Ly5.2 animals. Three-month-old recipient B6/Ly5.1 animals were administered sterile pH 2.0 water supplemented with neomycin sulfate. Lethal irradiation of animals (1,000 rads) was administered with a cesium source irradiator. The Institutional Animal Care and Use Committee approved all experiments.
Recombinant DNA
SDF-1 was cloned by PCR from the SDF-1 KDEL7 template, so that it is in-frame with amino acids 28–81 of Vpu (forward primer: 5′-CGCGAATTCGCGCCATGAACGCCAAGGTCG-3′; reverse primer: 5′-CAT GCGGCCGCTAGCATAATCTGGAACATC-3′). Amino acids 28–81 of Vpu were cloned by PCR from the NL4-3 genome (forward primer: 5′-CTAGCGGCCGCGAATATAGGAAAATATTAAGAC-3′; reverse primer: 5′-CAGCTCGAGCACTACAGATCATCAATATCC-3′). VpuC2/6 was cloned with the same primers as VpuC, but using Vpu2/6 (ref. 18) as a template. All constructs were cloned into a high-titer retroviral vector derived from the MSCV LTR38.
Cell culture
SupT1 cells were cultured in RPMI 1640 with 10% (vol/vol) FBS and penicillin/streptomycin. U2OS cells and 293T cells were cultured in DMEM-H with 10% (vol/vol) FBS and penicillin/streptomycin. Primary bone marrow was flushed from femurs and tibias, enriched for cKit+ cells35, cultured in IMDM containing 20% FBS (Gibco), 50 ng/ml stem cell factor (SCF), 50 ng/ml interleukin-6 (IL-6) and 10 ng/ml IL-11 (Peprotech) for 48 h, and then transduced with the indicated retroviruses. HSPC cells were maintained in IMDM with 10 ng/ml of SCF, IL-6 and IL-11. Thymocytes were harvested and cultured in IMDM, 20% ES FBS, 50 ng/ml SCF and 15 ng/ml IL-7 (Peprotech).
Retrovirus production and infection
293T cells were transfected with plasmids encoding MSCV retroviral DNA, VSVg and gag/pol as described39. Retroviral infections by spinoculation were performed in 2 ml Eppendorf tubes with 1 × 106 cells with 500 µl RPMI 1640 with 10% (vol/vol) FBS, 500 µl of viral supernatant and 8 µg/ml polybrene. Cells were incubated at 22–24 °C for 20 min and centrifuged at 2000g for 3 h. Cells were either cultured as a pool or sorted for EGFP expression.
Immunofluorescence
The EGFP cassette in the degrakine vector was replaced with YFP-KDEL (Clontech). U2OS cells were transfected (Effectene, Qiagen) with the indicated DNAs. At 48 h after transfection, cells were fixed in 3% paraformaldehyde and permeabilized with 0.1% Triton X-100. Cells were stained with anti-hemagglutinin (12CA5) monoclonal and goat antimIgG– phycoerythrin (Pharmingen) and visualized by confocal microscopy (100×).
MTT assay
1 × 103 cells per well of EGFP-sorted SupT1 cells were plated into a 96-well plate (triplicate). At the indicated time points, the cells were transferred into a new 96-well plate containing MTT (methylthiazolydiphenyl tetrazolium bromide; Sigma) solution and incubated at 37 °C for 4 h. Acid-isopropanol was added and the wells were assayed with a microplate reader using a test wavelength of 570 nm, reference wavelength of 630 nm and calibration setting of 1.99 (ref. 40). All optical density (OD) readings were done in triplicate.
FACS
Each sample (1 × 105 cells) was washed in PBS with 2% (vol/vol) FBS and stained with the indicated antibody in PBS with 2% (vol/vol) FBS, and 1–2 × 104 cells were collected through a Becton Dickinson FACScan. CXCR4 expression was detected with anti-12G5 monoclonal antibody (NIH AIDS Reagent) and anti-mIgG–phycoerythrin (Caltag). Phycoerythrin–anti-CD4 and phycoerythrin–anti-MHC-I antibodies were purchased from Caltag. GFP-positive cells were sorted on a Cytomation MoFlo FACS. Calcium influx assays were performed using Indo-1 AM (Molecular Probes) as described41.
ELISA
Secreted SDF-1 was measured by ELISA (R&D Systems). 293T cells were transfected using Effectene (Qiagen) and cultured for 48 h. Stably transduced SupT1 cells were washed, plated at equal concentrations and cultured for 48 h.
Western blotting
Transduced mouse HSPCs (1 × 106 cells) or Jurkat cells (1 × 106 cells) were lysed and resolved by SDS-PAGE42. Gels were transferred to PVDF membranes (Amersham Pharmacia), blocked with 5% nonfat dry milk, probed with either anti-Vpu (1:1,000) or anti-CXCR442 (1:100) and anti-rabbit-IgG–horseradish peroxidase (1:10,000) (Amersham Pharmacia). Blots were visualized using an ECL Kit (Amersham Pharmacia).
In vitro migration assay
Sorted SupT1 cells (2 × 104/well in triplicate) were assayed for migration response to SDF-1 over 2 h using a 0.6 µm Neuroprobe ChemoTx 96-well assay plate and cell numbers were quantified with Packard’s ATPLite kit. 5 × 105 THP-1 cells, thymocytes or HSPCs were assayed for migration over a 3-h period (24-well Corning trans-well plate, 0.5 µM membrane) to 100 nM SDF-1 (NIH AIDS Reagent) and 100 nM Mip-1β (Peprotech). The percentage of migrating degrakine expressing cells was determined by FACS. (Migration % = migrating GFP+ cells/input GFP+ cells.)
HSPC homing assay in vivo
cKit+ B6/Ly5.2 cells were cultured for 3–6 d after retroviral transduction. In vivo homing to the bone marrow was determined by injection of 2 × 106 degrakines expressing cKit+ B6/Ly5.2 cells into lethally irradiated B6/Ly5.1 mice (1,000 rads, 24 h before injection). At 3 h after injection the bone marrow was flushed from femurs and tibias and monitored for Ly5.2 and GFP with a BD FACScan43. (Migration % = harvested Ly5.2+GFP+ cells/input Ly5.2+GFP+ cells.)
ACKNOWLEDGMENTS
We thank S. Chen, K. Strebel, H. Golding and M. Zaitseva for kindly providing the SDF-1 intrakine construct, the Vpu2/6 mutant, anti-CXCR4-4N and CXCR4 western protocol, respectively; J. Smith, Y. Kim, M. Townsend and A. Elms for technical assistance; E. Meissner and W. Helms for critical reading of the manuscript; R. Bagnell for assistance with confocal microscopy; and L. Arnold of the UNC Flow Cytometry Facility for assistance with FACS. Anti-Vpu and anti-CXCR4 (12G5) were obtained through the NIH AIDS Reagent program. The project was partially supported by grants from the US National Institutes of Health (nos. HL72240 and AI53804 to L.S.).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annu. Rev. Immunol. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Yoshie O, Imai T, Nomiyama H. Chemokines in immunity. Adv. Immunol. 2001;78:57–110. doi: 10.1016/s0065-2776(01)78002-9. [DOI] [PubMed] [Google Scholar]
- 3.Sallusto F, Mackay CR, Lanzavecchia A. The role of chemokine receptors in primary, effector, and memory immune responses. Annu. Rev. Immunol. 2000;18:593–620. doi: 10.1146/annurev.immunol.18.1.593. [DOI] [PubMed] [Google Scholar]
- 4.Nelson PJ, Krensky AM. Chemokines, chemokine receptors, and allograft rejection. Immunity. 2001;14:377–386. doi: 10.1016/s1074-7613(01)00118-2. [DOI] [PubMed] [Google Scholar]
- 5.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 6.Marsal J, et al. Involvement of CCL25 (TECK) in the generation of the murine small-intestinal CD8α α+CD3+ intraepithelial lymphocyte compartment. Eur. J. Immunol. 2002;32:3488–3497. doi: 10.1002/1521-4141(200212)32:12<3488::AID-IMMU3488>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 7.Chen JD, Bai X, Yang AG, Cong Y, Chen SY. Inactivation of HIV-1 chemokine co-receptor CXCR-4 by a novel intrakine strategy. Nat. Med. 1997;3:1110–1116. doi: 10.1038/nm1097-1110. [DOI] [PubMed] [Google Scholar]
- 8.Engel BC, et al. Intrakines—evidence for a trans-cellular mechanism of action. Mol. Ther. 2000;1:165–170. doi: 10.1006/mthe.2000.0026. [DOI] [PubMed] [Google Scholar]
- 9.McManus MT, et al. Small interfering RNA-mediated gene silencing in T lymphocytes. J. Immunol. 2002;169:5754–5760. doi: 10.4049/jimmunol.169.10.5754. [DOI] [PubMed] [Google Scholar]
- 10.McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 11.Cullen BR. RNA interference: antiviral defense and genetic tool. Nat. Immunol. 2002;3:597–599. doi: 10.1038/ni0702-597. [DOI] [PubMed] [Google Scholar]
- 12.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 13.Tiscornia G, Singer O, Ikawa M, Verma IM. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl. Acad. Sci. USA. 2003;27:27. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tortorella D, Gewurz BE, Furman MH, Schust DJ, Ploegh HL. Viral subversion of the immune system. Annu. Rev. Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch C, Ploegh HL. Intracellular targeting of the proteasome. Trends Cell Biol. 2000;10:268–272. doi: 10.1016/s0962-8924(00)01768-2. [DOI] [PubMed] [Google Scholar]
- 16.Margottin F, et al. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 17.Willey RL, Maldarelli F, Martin MA, Strebel K. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol. 1992;66:7193–7200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schubert U, Strebel K. Differential activities of the human immunodeficiency virus type 1-encoded Vpu protein are regulated by phosphorylation and occur in different cellular compartments. J. Virol. 1994;68:2260–2271. doi: 10.1128/jvi.68.4.2260-2271.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loetscher M, et al. Cloning of a human seven-transmembrane domain receptor, LESTR, that is highly expressed in leukocytes. J. Biol. Chem. 1994;269:232–237. [PubMed] [Google Scholar]
- 20.Bleul CC, et al. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 21.Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) J. Exp. Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oberlin E, et al. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 23.Shirozu M, et al. Structure and chromosomal localization of the human stromal cell-derived factor 1 (SDF1) gene. Genomics. 1995;28:495–500. doi: 10.1006/geno.1995.1180. [DOI] [PubMed] [Google Scholar]
- 24.Tashiro K, et al. Signal sequence trap: a cloning strategy for secreted proteins and type I membrane proteins. Science. 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- 25.Nagasawa T, et al. Molecular cloning and characterization of a murine pre-B-cell growth–stimulating factor/stromal cell-derived factor 1 receptor, a murine homolog of the human immunodeficiency virus 1 entry coreceptor fusin. Proc. Natl. Acad. Sci. USA. 1996;93:14726–14729. doi: 10.1073/pnas.93.25.14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aiuti A, Webb IJ, Bleul C, Springer T, Gutierrez-Ramos JC. The chemokine SDF-1 is a chemoattractant for human CD34+ hematopoietic progenitor cells and provides a new mechanism to explain the mobilization of CD34+ progenitors to peripheral blood. J. Exp. Med. 1997;185:111–120. doi: 10.1084/jem.185.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 28.Kawabata K, et al. A cell-autonomous requirement for CXCR4 in long-term lymphoid and myeloid reconstitution. Proc. Natl. Acad. Sci. USA. 1999;96:5663–5667. doi: 10.1073/pnas.96.10.5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peled A, et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science. 1999;283:845–848. doi: 10.1126/science.283.5403.845. [DOI] [PubMed] [Google Scholar]
- 30.D’Apuzzo M, et al. The chemokine SDF-1, stromal cell-derived factor 1, attracts early stage B cell precursors via the chemokine receptor CXCR4. Eur. J. Immunol. 1997;27:1788–1793. doi: 10.1002/eji.1830270729. [DOI] [PubMed] [Google Scholar]
- 31.Schols D, et al. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J. Exp. Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chantry D, et al. Macrophage-derived chemokine is localized to thymic medullary epithelial cells and is a chemoattractant for CD3+, CD4+, CD8low thymocytes. Blood. 1999;94:1890–1898. [PubMed] [Google Scholar]
- 33.Kim CH, Broxmeyer HE. In vitro behavior of hematopoietic progenitor cells under the influence of chemoattractants: stromal cell-derived factor-1, steel factor, and the bone marrow environment. Blood. 1998;91:100–110. [PubMed] [Google Scholar]
- 34.Wright DE, Bowman EP, Wagers AJ, Butcher EC, Weissman IL. Hematopoietic stem cells are uniquely selective in their migratory response to chemokines. J. Exp. Med. 2002;195:1145–1154. doi: 10.1084/jem.20011284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 36.Hardy J, et al. Bioluminescence imaging of lymphocyte trafficking in vivo. Exp. Hematol. 2001;29:1353–1360. doi: 10.1016/s0301-472x(01)00756-1. [DOI] [PubMed] [Google Scholar]
- 37.Contag PR, Olomu IN, Stevenson DK, Contag CH. Bioluminescent indicators in living mammals. Nat. Med. 1998;4:245–247. doi: 10.1038/nm0298-245. [DOI] [PubMed] [Google Scholar]
- 38.Cheng L, et al. Sustained gene expression in retrovirally transduced, engrafting human hematopoietic stem cells and their lympho-myeloid progeny. Blood. 1998;92:83–92. [PubMed] [Google Scholar]
- 39.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 41.Vilen BJ, Burke KM, Sleater M, Cambier JC. Transmodulation of BCR signaling by transduction-incompetent antigen receptors: implications for impaired signaling in anergic B cells. J. Immunol. 2002;168:4344–4351. doi: 10.4049/jimmunol.168.9.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lapham CK, Zaitseva MB, Lee S, Romanstseva T, Golding H. Fusion of monocyctes and macrophages with HIV-1 correlates with biochemical properties of CXCR4 and CCR5. Nature. 1999;5:303–308. doi: 10.1038/6523. [DOI] [PubMed] [Google Scholar]
- 43.Szilvassy SJ, Meyerrose TE, Ragland PL, Grimes B. Differential homing and engraftment properties of hematopoietic progenitor cells from murine bone marrow, mobilized peripheral blood, and fetal liver. Blood. 2001;98:2108–2115. doi: 10.1182/blood.v98.7.2108. [DOI] [PubMed] [Google Scholar]