Abstract
We approached the problems of event detection, argument identification, and negation and speculation detection in the BioNLP’09 information extraction challenge through concept recognition and analysis. Our methodology involved using the OpenDMAP semantic parser with manually written rules. The original OpenDMAP system was updated for this challenge with a broad ontology defined for the events of interest, new linguistic patterns for those events, and specialized coordination handling. We achieved state-of-the-art precision for two of the three tasks, scoring the highest of 24 teams at precision of 71.81 on Task 1 and the highest of 6 teams at precision of 70.97 on Task 2. We provide a detailed analysis of the training data and show that a number of trigger words were ambiguous as to event type, even when their arguments are constrained by semantic class. The data is also shown to have a number of missing annotations. Analysis of a sampling of the comparatively small number of false positives returned by our system shows that major causes of this type of error were failing to recognize second themes in two-theme events, failing to recognize events when they were the arguments to other events, failure to recognize nontheme arguments, and sentence segmentation errors. We show that specifically handling coordination had a small but important impact on the overall performance of the system. The OpenDMAP system and the rule set are available at http://bionlp.sourceforge.net.
Keywords: event recognition, conceptual analysis, natural language processing, text mining, BioNLP
1. INTRODUCTION
We approached the problem of biomedical event recognition as one of concept recognition and analysis. Concept analysis is the process of taking a textual input and building from it an abstract representation of the concepts that are reflected in it. Concept recognition can be equivalent to the named entity recognition task when it is limited to locating mentions of particular semantic types in text, or it can be more abstract when it is focused on recognizing predicative relationships, e.g., events and their participants. A short description of our methodology can be found inCohen et al. (2009). Here we give additional details on the OpenDMAP system and the ontology that it used and include analyses of the BioNLP’09 shared task data and of our own false positives.
1.1. The BioNLP’09 Shared Task
The event types selected for inclusion in the BioNLP’09 shared task were done so based on their frequency and annotation quality within the GENIA corpus, and represented biological events of central importance to biology and therefore research biologists. For the event detection and characterization task, nine molecular biology events were identified. They included gene-centric processes such as “expression” and “translation” (different yet related terms describing the process through which genes encoded in the DNA of a cell function as blueprints for the creation of functioning proteins), protein-centric events including “protein catabolism” (the biochemical process of degradation or the breaking down of a protein) and “protein localization” (which included where a protein resided, in addition to where a protein was moving from or to), as well as interaction or modification events such as binding (protein binding to another protein, or a protein “binding” to DNA) and “phosphorylation” (the process of adding a phosphate group to a protein). Event terms describing the control of such events were also included, and were modifiers of the previous six events; “regulation,” “positive regulation,” and “negative regulation.” The correct identification of these events required not only the identification of the core theme (i.e., phosphorylation) but also when the information was available, all the participants in the event (i.e., that Protein A was being phosphorylated and it was Protein B whose function it was to do the phosphorylating). It is important to note that the BioNLP’09 shared task made no effort to distinguish between gene and protein mentions, and in addition protein families and protein complexes were considered beyond the scope of the task. Such exclusions should not be seen as indication of lesser importance (understanding and identifying these entities are critical if we are to fully exploit knowledge captured in biological texts), but rather is indicative of the incredibly challenging nature of these concepts. The official event descriptions as used for this task are available at http://www-tsujii.is.s.u-tokyo.ac.jp/GENIA/SharedTask/detail.shtml#event.
Our system was entered into all three of the BioNLP’09 (Kim et al. 2009) shared tasks:
Event detection and characterization This task requires recognition of nine biological events: gene expression, transcription, protein catabolism, protein localization, binding, phosphorylation, regulation, positive regulation, and negative regulation. It requires identification of the core THEME and/or CAUSE participants in the event, i.e., the protein(s) being produced, broken down, bound, regulated, etc.
Event argument recognition This task builds on the previous task, adding in additional arguments of the events, such as the site (protein or DNA region) of a binding event, or the location of a protein in a localization event.
Recognition of negations and speculations This task requires identification of negations of events (e.g., event X did not occur), and speculation about events (e.g., We claim that event X should occur).
1.2. Related Work
OpenDMAP is distinguished from a significant body of work on information extraction of biological events in that it uses an ontology as its organizing structure, and uses the declarations of classes and slots in that ontology as semantic constraints that must be validated during extraction. While other work in information extraction focuses on surface patterns (Blaschke and Valencia 2001, 2002) or syntactic structures (Yakushiji et al. 2001; Saetre et al. 2009), OpenDMAP is more closely aligned with work in the context of the semantic web that takes advantage of ontology structures in semantic markup of text (e.g., COHSE (Bechhofer et al. 2008) and OntoMat (Handschuh et al. 2001)), and is directly related to work that explicitly couples linguistic patterns with ontological constraints, such as the MnM system (Vargas-Vera et al. 2002) and MedScan (Daraselia et al. 2004). In the broadest sense, the major dividing line between our work and that of others is in whether an ontology is involved, as it intimately is in our work, or whether it is not.
Comparing with other ontology-based systems, several are construed more as semantic annotation tools than information extraction systems. Concept recognition in COHSE, for instance, is limited to term and synonym lookup from source vocabularies, while we go beyond this to recognize relationships between concepts. Onto Mat does not support information extraction pattern definition, but rather is limited to (manual) semantic annotation using concepts from ontologies. The MnM system, however, is closer to OpenDMAP in that it is coupled with the Amilcare learning system (Ciravegna and Wilks 2003) to support pattern acquisition. Amilcare induces generalized rules from a training corpus for tagging segments of text with a given role label, using the local context. In contrast, OpenDMAP is oriented towards recognizing complete relation structures in one step by defining the linguistic context of an event predicate along with its arguments (class plus slot fillers). MedScan is the closest system in terms of approach to OpenDMAP. As compared to MedScan, OpenDMAP allows but does not require a full syntactic analysis of a sentence, instead allowing patterns to be defined that specify more surface-level features in combination with the semantic constraints, which makes the overall approach less dependent on the performance of the underlying syntactic analysis.
The shared task was defined as an event extraction task. It is not clear that this definition is actually linguistically accurate, because by definition events occur at some time t, and the “events” under discussion in the molecular biology literature are closer to statements about probabilistic tendencies in populations of molecules. For this reason, we do not review the general literature on event recognition here. However, a number of systems from the shared task provide instructive comparisons to our own work. Most, although not all, approaches have involved a syntactic parse, with dependency parses predominating. Many systems have used rule-based approaches, although most of the top-ranking systems on this task used a machine learning application; the top-ranking system used two machine learning passes followed by the operation of a rule-based semantic processing component (Björne et al. 2009). The top-ranking system is representative of many alternative approaches. In an initial step, it performed sentence splitting, tokenization, and syntactic parsing. The parse tree was then transformed into an alternative graph representation. A multi-class support vector machine was used to identify trigger words, and then applied again to locate arguments. A rule-based post processing step then performed a number of operations, including pruning invalid edges and ensuring the correct number of arguments (Björne et al. 2009).
A detailed comparison of the various participating systems can be found in Kim et al. (2009).
2. OUR APPROACH
We used the OpenDMAP system developed at the University of Colorado School of Medicine (Hunter et al. 2008) for our submission to the BioNLP’09 Shared Task on Event Extraction. OpenDMAP is an ontology-driven, integrated concept analysis system that supports information extraction from text through the use of patterns represented in a classic form of “semantic grammar,” freely mixing text literals, semantically typed basal syntactic constituents, and semantically defined classes of entities. Our approach is to take advantage of the high-quality ontologies available in the biomedical domain to formally define entities, events, and constraints on slots within events and to develop patterns for how concepts can be expressed in text that take advantage of both semantic and linguistic characteristics of the text. We manually built patterns for each event type by examining the training data and by using native-speaker intuitions about likely ways of expressing relationships, similar to the technique described inCohen et al. (2004). The patterns characterize the linguistic expression of that event and identify the arguments (participants) of the events according to (a) occurrence in a relevant linguistic context and (b) satisfaction of appropriate semantic constraints, as defined by our ontology. Our solution results in very-high-precision information extraction, although the current rule set has limited recall.
2.1. The Reference Ontology
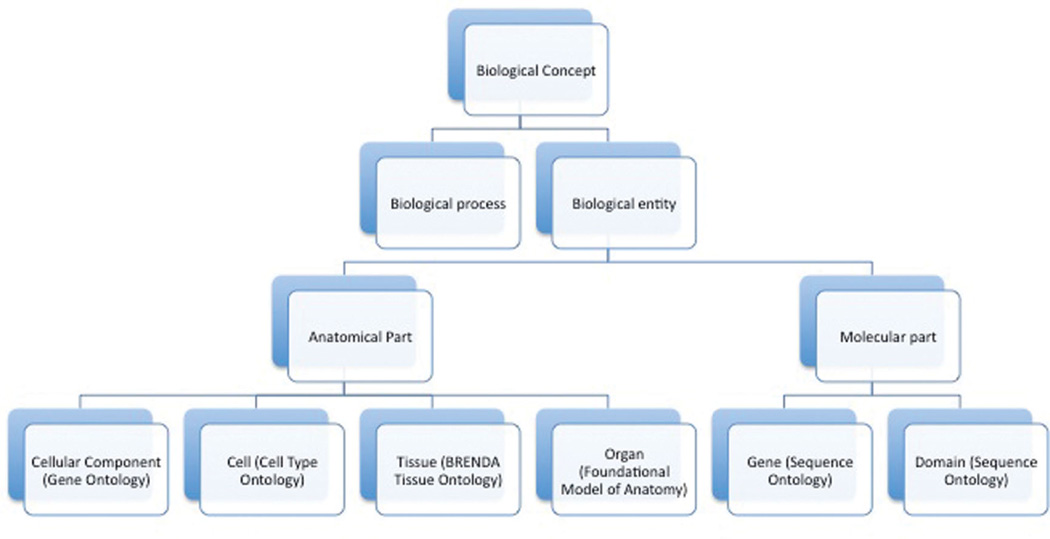
The central organizing structure of an OpenDMAP project is an ontology. We built the ontology for this project by combining elements of several community-consensus ontologies—the Gene Ontology (GO) (The Gene Ontology Consortium 2000; Consortium 2001), Cell Type Ontology (CTO) (Bard, Rhee, and Ashburner 2005), BRENDA Tissue Ontology (BTO; Schomburg et al. 2004), Foundational Model of Anatomy (FMA; Rosse and Mejino 2003), and Sequence Ontology (SO; Eilbeck et al. 2005)—and a small number of additional concepts to represent task-specific aspects of the system, such as event trigger words. Combining the ontologies was done with the Prompt plug-in for Protégé. A partial view of the ontology is shown in Figure 1.
Figure 1.
A partial view of the central organizing ontology for our system. Biological Concept, Anatomical Part, and Biological Entity were added to organize other elements of the ontology, but the other elements are community-consensus, independently constructed ontologies.
The ontology included concepts representing each event type. These were represented as frames, with slots for the things that needed to be returned by the system—the trigger word and the various slot fillers. All slot fillers were constrained to be concepts in some community-consensus ontology. The core event arguments were constrained in the ontology to be of type protein from the Sequence Ontology (except in the case of higher-order events, where biological events themselves could satisfy the THEME role), while the type of the other event arguments varied. For instance, the AtLoc argument of a gene expression event was constrained to be one of tissue (from BTO), cell type (from CTO), or cellular component (from GO-Cellular Component), while the binding argument of a binding event was constrained to be one of binding_site, DNA, domain, or chromosome (all from the SO and all tagged by LingPipe). Table 1 lists the various types.
Table 1.
Semantic Restrictions on Task 2 Event Arguments. Ontology abbreviations are defined in the text.
| Event Type | Site | AtLoc | ToLoc |
|---|---|---|---|
| Binding | protein domain (SO), binding site (SO), DNA (SO), chromosome (SO) |
||
| Gene expression | gene (SO), biological, entity |
tissue (BTO), cell type (CTO), cellular component (GO) |
|
| Localization | cellular component (GO) |
cellular component (GO) | |
| Phosphorylation | amino acid (FMA), polypeptide region (SO) |
||
| Protein catabolism | cellular component (GO) |
||
| Transcription | gene (SO), biological entity |
2.2. The OpenDMAP Semantic Parser
As indicated above, a key component of the OpenDMAP system is the definition of semantic rules characterizing the expression of concepts in text. As in our previous publications on OpenDMAP, we refer to our semantic rules as patterns. These patterns are encoded as linguistic forms of concepts directly in the knowledge base. That is, rather than being constructed with a lexicon and a knowledge representation as two separate components, in OpenDMAP, concepts and their associated potential linguistic manifestations are jointly encoded. This association works out differently in the case of different types of ontologies. In the case of an ontology such as the Gene Ontology Cellular Component hierarchy (GO-CC), which has an extensive set of concepts that have associated terms, the terms themselves serve as the patterns. In the case of an ontology such as the Sequence Ontology, a different mechanism is used. In the Sequence Ontology, we have a protein concept (SO:000358). Unlike concepts in GO-CC, which have a high likelihood in many cases of showing up in the same form as their associated terms, or with minor variations that can be captured using linguistic rules (at least for cellular components with which we are most likely to be concerned, the more general concepts in the upper levels of GO-CC), there are many proteins that will be mentioned by a name that does not appear in the Sequence Ontology because it is not intended to be a catalog of all proteins or genes, but rather a specification of various sequence-related concepts and entity types. That is, it is not designed to represent specific entities, but rather classes of entities. In this case, we use a named entity recognizer (or, in the case of this shared task, the given annotations) to recognize members of the appropriate semantic class, and then map them to the appropriate element in the Sequence Ontology. In neither case is there a separate lexicon and knowledge model; rather, either linguistic patterns are directly associated with their concepts, as in the case of the Gene Ontology, or entities of a particular type are identified in free text through an alternative strategy such as a third-party named entity recognizer and their semantic type is mapped directly to the appropriate element in the ontology, as in the case of many Sequence Ontology elements.
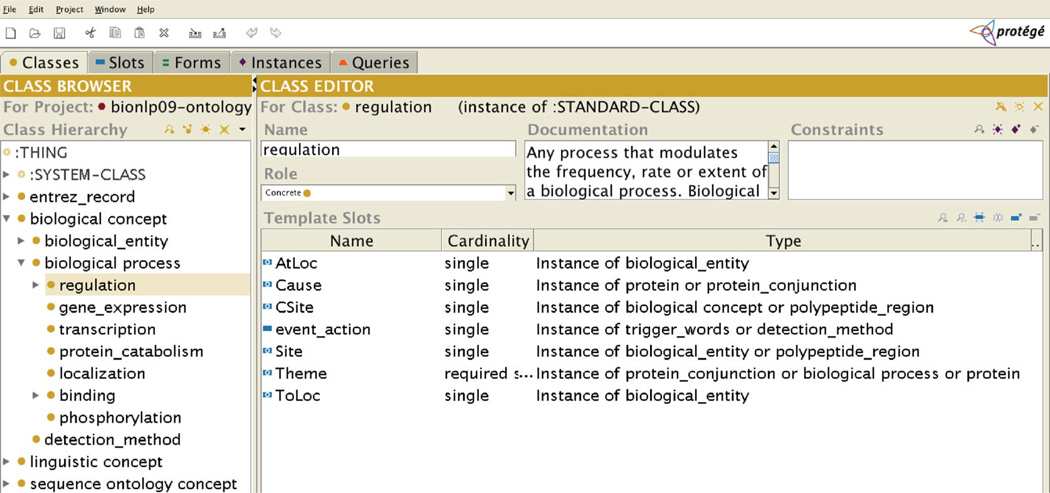
The OpenDMAP pattern language has a number of features that allow for semantic typing of arguments and for flexible specification of ordering. Because the semantic typing is a major distinctive feature of the system, we discuss it first. Semantic typing works in conjunction with an ontology and a frame specified in a Protégé project. Figure 2 shows the representation of a representative frame from our implementation. Note that each slot is constrained to belong to a particular semantic class (labeled Type in the Protégé interface). For example, the Site slot is constrained to be a member of the class biological_entity or polypeptide_region. These classes correspond directly to other ontology elements.
Figure 2.
Representation of the regulation frame in Protégé, showing semantic constraints on slot-fillers.
Patterns are then written for each concept, referencing the names of the slots to take advantage of their semantic types. For example, in the following simplified pattern for the regulation concept:
regulation := [regulation_trigger_word] of [Theme] (by [Cause])?
use of the word Theme in square brackets constrains the text matching that part of the pattern to be a protein, a protein conjunction, or a biological process, as specified by the ontology. These constraints can seen in the frame specification in Figure 2. The parenthesized (by [Cause])? is made optional by the following question mark.
For the BioNLP’09 task, each event pattern has at a minimum a Theme argument and an event-specific trigger word. For example, {phosphorylation} := [phosphorylation_nominalization] [Theme], where [phosphorylization_nominalization] represents a trigger word. Both elements are defined semantically.
The pattern language syntax is of context-free power. One unusual operator allows for flexible ordering of strings. It is illustrated here:
Protein_transport := [Transported-Entity] translocation @(from [Transport-Origin])@(to [Transport-Destination])
Here, the Transported-Entity must precede the text literal translocation. However, the @-sign allows for flexible ordering of the from-phrase and the to-phrase. They may occur in any position relative to the ordered parts of the rule. Thus, that single rule 1 will allow for matching of both Bax translocation to mitochondria from the cytosol and Bax translocation from the cytosol to the mitochondria.
Additionally, regular expression operators are included. These include a wildcard character (although see below for its effect on our rules’ performance), Kleene star, Kleene plus, ? for zero or 1, and a conjunction operator. The various operators can be combined to build arbitrarily complex rules. We gave examples of some simplified rules above. An example of a full rule set for the protein localization event type, follows. These are relatively straight-forward rules. They achieved precision of 0.6543, recall of 0.2046, and F-measure of 0.3118 on the devtest data, and precision of 1.0, recall of 0.1034, and F-measure of 0.1875 on the official test data:
{localization_trig_word} := secretion;
{localization_trig_word} := release;
{localization_trig_word} := localization;
{localization_trig_word_translocation} := translocation;
{localization_trig_word_secretion_release} := secretion, release;
{localization} := [ToLoc] [event_action localization_trig_word] of _ [Theme];
{localization} := [event_action localization_trig_word_secretion_release] of [Theme];
{localization} := [Theme ] [event_action localization_trig_word_secretion_release];
{localization} := ([ToLoc])? [event_action localization_trig_word] of [Theme];
{localization} := ([ToLoc])? [event_action localization_trig_word_translocation] of _[Theme];
Some things to notice about these rules are that although the := operator is reminiscent of Prolog, these are not Prolog productions; the first five rules function purely to define patterns for relational concept trigger words; and comments can be included in the rule files. More specifically to the shared task, note that each rule minimally includes (1) a typed slot (indicated by square brackets) for the trigger word, which the rules required us to return, and (2) another typed slot (again, indicated by square brackets) for the theme.
Another example of a full rule set follows, this time for gene expression events. This rule set contrasts with the previous one by making extensive use of regular expression operators to deal with inflectional and derivational morphology when defining the trigger words. It achieved a precision of 0.8595, recall of 0.3643, and F-measure of 0.5117 on the official test data:
{expression_trig_word} := r’express.*’, r’coexpress.*’, r’co-express.*’, r’overexpress.*’;
{expression_trig_word} := r’produc.*’, r’resynthes.*’, r’synthes.*’;
{expression_trig_word} := r’nonproduc.*’, r’non-produc.*’, r’generat.*’, r’nonexpress.*’, r’non-express.*’;
{gene_expression} := [Theme] (gene—protein)? adv? be? adv? [event_action expression_trig_word];
{gene_expression} := [event_action expression_trig_word]prep det? [Theme];
{gene_expression} := [event_action expression_trig_word] and nominalization prep det?[Theme];
{gene_expression} := [Theme] [event_action expression_trig_word];
The rule elements prep and det represent prepositions and determiners and are defined in a separate file; the curly braces indicate that an element is defined within the ontology, rather than being text literals.
2.3. OpenDMAP Pattern Match Scoring
Previous versions of our OpenDMAP system utilized a simple scoring algorithm for ranking competingmatches beginning at a given span of text. This algorithm prefers matches that cover every word of the span to those that have intervening words (the span score). For BioNLP’09, the algorithm was refined to include two other factors, a pattern score that penalizes matches to patterns which have optional pattern elements that are uninstantiated in the match, and a concept score that penalizes matches that do not fill all slots associated with a concept in the ontology. The final score is then calculated based on a weighted average of the three component scores. This adjusted algorithm was found to improve the selection of the correct pattern match in cases where there were multiple patterns that matched a span of text.
2.4. Named Entity Recognition
For proteins, we used the gold standard annotations provided by the organizers. For other semantic classes, we constructed a compound named entity recognition system which consists of a LingPipe GENIA tagging module (LingPipe,2 Alias-i (2008)), and several dictionary lookup modules. The dictionary lookup was done using a component from the UIMA (IBM 2009); Ferrucci and Lally 2004) sandbox called the ConceptMapper.
We loaded the ConceptMapper with dictionaries derived from the relevant ontologies identified in Section 2.1. The dictionaries contained the names and name variants for each concept in each ontology, and matches in the input documents were annotated with the relevant concept ID for the match. The only modifications that we made to these community-consensus ontologies were to remove the single concept cell from the Cell Type Ontology and to add the synonym nuclear to the Gene Ontology Cell Component concept nucleus.
The protein annotations were used to constrain the text entities that could satisfy the theme role in the events of interest. The other named entities were added for the identification of noncore event participants for Task 2, generally to characterize varying semantic constraints for Site arguments of the different event types as indicated in Table 1, based on input from a biological expert.
2.5. Handling of Coordination
Coordination was handled using the OpenNLP 3 constituent parser along with the UIMA wrappers that they provide via their code repository. We chose OpenNLP because it is easy to train a model, it integrates easily into a UIMA pipeline, and because of competitive parsing results as reported by Buyko (Buyko et al. 2006). We built a new model for the parser using 500 abstracts from the beta version of the GENIA treebank and 10 full-text articles from the CRAFT corpus (Verspoor, Cohen, and Hunter 2009) as training data. From the constituent parse, we extracted coordination structures into a simplified data structure that captures each conjunction along with its conjuncts. These were provided to downstream components. The coordination component achieves an F-score of 74.6% at the token level and an F-score of 57.5% at the conjunct level when evaluated against GENIA. For both measures the recall was higher than the precision by 4% and 8%, respectively.
We utilized the coordination analysis to identify events in which the THEME argument was expressed as a conjoined noun phrase. These were assumed to have a distributed reading and were postprocessed to create an individual event involving each conjunct, and further filtered to only include given (A1) protein references. So, for instance, analysis of the sentence in the example below should result in the detection of three separate gene expression events, involving the proteins HLA-DR, CD86, and CD40 respectively.
NAC was shown to down-regulate the production of cytokines by DC as well as their surface expression of HLA-DR, CD86 (B7-2), and CD40 molecules . . . (PMID 10072497)
2.6. Software Infrastructure
2.6.1. Core Text Processing Infrastructure
We took advantage of our existing infrastructure based on UIMA (The Unstructured Information Management Architecture, IBM (2009); Ferrucci and Lally (2004)) to support text processing and data analysis. Our tools are available from http://bionlp.sourceforge.org.
2.6.2. Development Tools
We developed a visualization tool to enable the linguistic pattern writers to better analyze the training data. This tool shows the source text one sentence at a time with the annotated words highlighted. A list following each sentence shows details of the annotations.
The tool is implemented as a UIMA Analysis Engine and reads annotations inserted into the CAS (Common Analysis Structure) data structure for a document within UIMA, which in turn was created based on a UIMA module for loading the provided A1 (protein) and A2 (gold standard event) annotations. The tool generates static HTML files to leverage the graphics capabilities of a browser.
For in-house testing, we were able to take advantage of a previously developed UIMA annotation comparator which would compare the annotations loaded from the A2 files with the annotations added in our processing and output an analysis of Precision/Recall/F-score. We similarly utilized an annotation printer for outputting and reviewing annotations in the CAS.
3. PATTERN DEVELOPMENT STRATEGIES
3.1. Corpus Analysis
Using the tool that we developed for visualizing the training data (described in Section 2.6.2), a subset of the gold-standard annotations were grouped by event type and by trigger word type (nominalization, passive verb, active verb, or multiword phrase). This organization helped to suggest the argument structures of the event predicates and also highlighted the variation within argument structures. It also showed the nature of more extensive intervening text that would need to be handled for the patterns to achieve higher recall.
Based on this corpus analysis, patterns were developed manually using an iterative process in which individual patterns or groups of patterns were tested on the training data to determine their impact on performance. Pattern writers started with the most frequent trigger words and argument structures.
3.2. Trigger Words
In the training data, we were provided annotations of all relevant event types occurring in the training documents. These annotations included a trigger word specifying the specific word in the input text which indicated the occurrence of each event. We utilized the trigger words in the training set as anchors for our linguistic patterns. We built patterns around the generic concept of, e.g., an expression trigger word and then varied the actual strings that were allowed to satisfy that concept. We then ran experiments with our patterns and these varying sets of trigger words for each event type, discarding those that degraded system performance when evaluated with respect to the gold standard annotations.
Most often a trigger word was removed from an event type trigger list because it was also a trigger word for another event type and therefore reduced performance by increasing the false positive rate. For example, the trigger words “level” and “levels” appear in the training data trigger word lists of gene expression, transcription, and all three regulation event types.
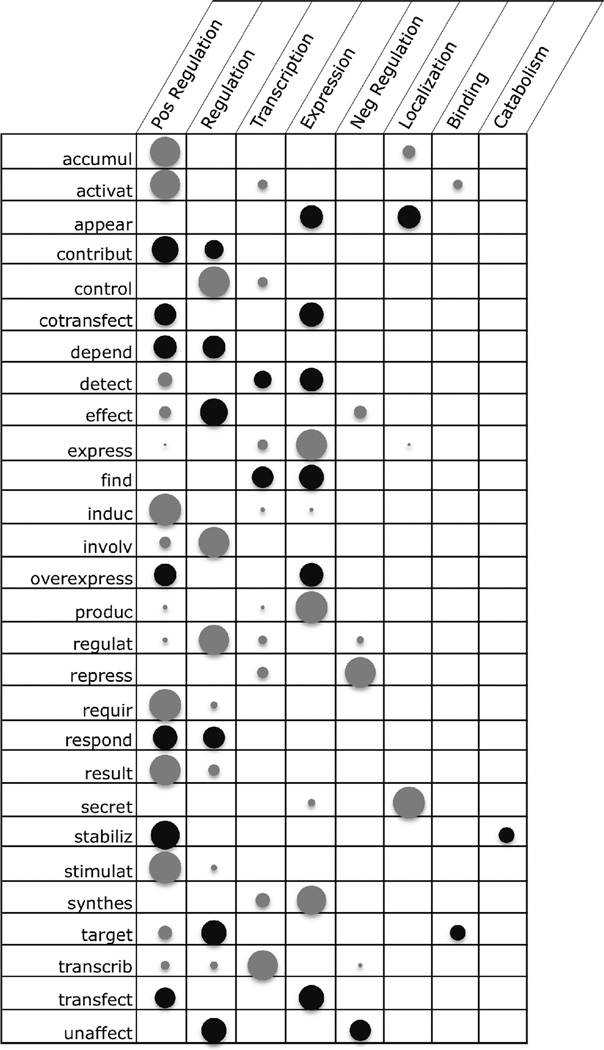
An analysis of the trigger words in the training data was performed to determine the extent of trigger word ambiguity. For each event type, trigger words were collected from the gold standard data files, lemmatized, and counted, removing any triggers that occurred less than three times for a single event type. The counts for each trigger word were normalized to a value between 0 and 1, which represents the relative frequency of a trigger word across event types. The frequencies were then compared across event types. Table 3 shows this comparison. The larger bubbles represent higher frequency of occurrence of a trigger word with an event type. The darker bubbles represent those trigger words whose relative frequency falls within the range 22–78%. These medium-range trigger words will be more challenging to disambiguate because their frequency is more evenly distributed across event types. Another outcome of this analysis to note is that some event types rely on many ambiguous trigger words, e.g., Positive Regulation.
Table 3.
Updated Results on Test Data for Tasks 1–3, with Important Bug Fixes in the Code Base. Key above.
| Tasks 1 and 3 |
Task 2 |
|||||||
|---|---|---|---|---|---|---|---|---|
| Event class | GS | answer | R | P | F | R | P | F |
| Localization | 174 (33) | 41 (33) | 18.97 | 80.49 | 30.70 | 16.67 | 69.05 | 26.85 |
| Binding | 347 (62) | 152 (62) | 17.87 | 40.79 | 24.85 | 17.48 | 40.13 | 24.35 |
| Gene expression | 722 (290) | 344 (290) | 40.17 | 84.30 | 54.41 | 40.17 | 84.30 | 54.41 |
| Transcription | 137 (28) | 31 (28) | 20.44 | 90.32 | 33.33 | 20.44 | 90.32 | 33.33 |
| Protein catabolism | 14 (4) | 6 (4) | 28.57 | 66.67 | 40.00 | 28.57 | 66.67 | 40.00 |
| Phosphorylation | 135 (47) | 48 (47) | 34.81 | 97.92 | 51.37 | 32.37 | 84.91 | 46.88 |
| Event Total | 1,529 (464) | 622 (464) | 30.35 | 74.60 | 43.14 | 29.77 | 72.77 | 42.26 |
| Regulation | 291 (11) | 31 (11) | 3.78 | 35.48 | 6.83 | 3.77 | 35.48 | 6.81 |
| Positive regulation | 983 (60) | 129 (60) | 6.10 | 46.51 | 10.79 | 6.08 | 46.51 | 10.75 |
| Negative regulation | 379 (18) | 41 (18) | 4.75 | 43.90 | 8.57 | 4.49 | 41.46 | 8.10 |
| Regulation Total | 1,653 (89) | 201 (89) | 5.38 | 44.28 | 9.60 | 5.31 | 43.78 | 9.47 |
| Negation | 227 (6) | 129 (6) | 2.64 | 4.65 | 3.37 | |||
| Speculation | 208 (25) | 165 (25) | 12.02 | 15.15 | 13.40 | |||
| Modification Total | 435 (31) | 294 (31) | 7.13 | 10.54 | 8.50 | |||
| All Total | 3,182 (553) | 823 (553) | 17.38 | 67.19 | 27.62 | 17.07 | 65.74 | 27.10 |
The analysis of ambiguity among trigger words is important for future disambiguation efforts. There is an interplay between the lexical and the semantic components of a sentence. A trigger word of little or no ambiguity can signal an event type even if the arguments of the event are ambiguous, underspecified, or not explicitly stated in the sentence, and can, subsequently, help to disambiguate said arguments using the event definition in the ontology. On the other hand, a trigger word that signals multiple event types will require stated arguments that are semantically specific enough to disambiguate the event type selection. In the case of the shared task events, because the semantic constraints on themes for all event types include proteins, the event types with patterns that used ambiguous trigger words were effectively indistinguishable. For example, OpenDMAP cannot discriminate between a gene expression event and a positive regulation event if the only clue it has is the phrase “elevated IgE levels,” which satisfies a pattern constraint for both event types. Patterns that use the high ambiguity trigger words discovered in this analysis will need further specification, whether that is in the form of tighter semantic restrictions on the arguments, or more contextual clues matched in text for disambiguation.
The selection of trigger words was guided by this frequency analysis. In a post hoc analysis, we find that a different proportion of the set of trigger words in the training data was finally chosen for inclusion in the list of trigger words used in the runtime system for each different event type. Between 10% and 20% of the top frequency-ranked trigger words were used for simple event types, with the exception that phosphorylation trigger words were chosen from the top 30%. For instance, for gene expression all of the top 15 most frequent trigger words were used (corresponding to the top 16%). For complex event types (the regulations) better performance was achieved by limiting the list to between 5% and 10% of the most frequent trigger words. This difference is explicable through the data in Figure 3; the most ambiguous trigger words are the least discriminating clues for the various event types, and the complex event types are characterized by the more ambiguous trigger words, as well as having substantially more variability in the relevant trigger words.
Figure 3.
Columns are event types. Rows represent ambiguous lemmata with frequency greater than three. Circle size represents relative frequency of the lemma for the given event type. Black circles represent lemmata that are especially difficult to disambiguate because their frequencies are distributed relatively evenly across different event types.
In addition, variants of frequent trigger words were identified and included. For instance, the nominalization “expression” is the most frequent gene expression trigger word and the verbal inflections “expressed” and “express” are also in the top 20%. The verbal inflection “expresses” is ranked lower than the top 30%, but was nonetheless included as a trigger word in the gene expression patterns.
3.3. Patterns for Complex Events
The methodology for creating complex event patterns such as regulation was the same as for simple events, with the exception that the Themes were defined in the ontology to also include biological processes to allow for the second-order relations. Iterative pattern writing and testing was a little more arduous because these patterns relied on the success of the simple event patterns, and hence more in-depth analysis was required to perform performance-increasing pattern adjustments.
3.4. Nominalizations
Nominalizationswere very frequent in the training data; for seven out of nine event types, the most common trigger word was a nominalization. In writing our patterns, we focused on these nominalizations. To write patterns for nominalizations, we capitalized on some of the insights from Cohen, Palmer, and Hunter (2008). Realized arguments of nominalizations can occur in three basic positions:
Within the noun phrase, after the nominalization; typically in a prepositional phrase
Within the noun phrase, immediately preceding the nominalization
External to the noun phrase
The first of these is the most straightforward to handle in a rule-based approach. This is particularly true in the case of a task definition like that of BioNLP’09, which focused on themes, because an examination of the training data showed that when themes were postnominal in a prepositional phrase, then that phrase was most commonly headed by of.
The second of these is somewhat more challenging. This is because both agents and themes can occur immediately before the nominalization, e.g., phenobarbital induction (induction by phenobarbital) and trkA expression (expression of trkA). To decide how to handle pre-nominal arguments, we made use of the data on semantic roles and syntactic position found in Cohen et al. (2008). That study found that themes outnumbered agents in the prenominal position by a ratio of 2.5 to 1. Based on this observation, we assigned pre-nominal arguments to the theme role.
Noun-phrase-external arguments are the most challenging, both for automatic processing and for human interpreters; one of the major problems is to differentiate between situations where they are present but outside of the noun phrase, and situations where they are entirely absent. An example of a phrase with a difficult structure is “EWS/FLI-1 antagonists induce growth inhibition of Ewing tumor cells.” This phrase could be paraphrased as EWS/FLI-1 antagonists inhibit growth of Ewing tumor cells, but the addition of the verb “induce” increases the complexity of the sentence by pushing the agent outside of the primary event noun phrase. Because the current implementation of OpenDMAP does not have robust access to syntactic structure, our only recourse for handling these arguments was through wildcards, and because they mostly decreased precision without a corresponding increase in recall, we were not able to capture these external arguments.
3.5. Negation and Speculation
Corpus analysis of the training set revealed two broad categories each for negation and speculation modifications, all of which can be described in terms of the scope of modification.
3.5.1. Negation
Broadly speaking, an event itself can be negated or some aspect of an event can be negated. In other words, the scope of a negation modification can be over the existence of an event (first example below), or over an argument of an existing event (second example).
This failure to degrade IkappaBalpha . . . (PMID 10087185)
AP-1 but not NF-IL-6 DNA binding activity . . . (PMID 10233875)
Patterns were written to handle both types of negation. The negation phrases “but not” and “but neither” were included within event patterns to catch those events that were negated as a result of a negated argument. For event negation, a more extensive list of trigger words was used that included verbal phrases such as “failure to” and “absence of.” In this case, the Theme of a negation event is defined to be a biological event, and the negation of the event can be recognized with a simple pattern such as negation := [Theme] {negation_trigger_word}.
The search for negated events was conducted in two passes. Events for which negation cues fall outside the span of text that stretches from argument to event trigger word were handled concurrently with the search for events. A second search was conducted on extracted events for negation cues that fell within the argument to event trigger word span, such as
...IL-2 does not induce I kappa B alpha degradation (PMID 10092783)
This second pass allowed us to capture one additional negation (6 rather than 5) on the test data.
3.5.2. Speculation
The two types of speculation in the training data can be described by the distinction, originally made by Frege, between “de dicto” and “de re” assertions. The “de dicto” assertions of speculation in the training data are modifications that call into question the degree of known truth of an event, as in
. . . CTLA-4 ligation did not appear to affect the CD28-mediated stabilization (PMID 10029815)
The “de re” speculation address the potential existence of an event rather that its degree of truth. In these cases, the event is often being introduced in text by a statement of intention to study the event, as in
. . .we investigated CTCF expression . . . [10037138]
To address these distinct types of speculation, two sets of trigger words were developed. One set consisted largely of verbs denoting research activities, e.g., ‘research,’ ‘study,’ ‘examine,’ ‘investigate,’ etc. The other set consisted of verbs and adverbs that denote uncertainty, and included trigger words such as ‘suggests,’ ‘unknown,’ and ‘seems.’
3.6. Errors in the Training Data
In some cases, there were discrepancies between the training data and the official problem definitions. This was a source of problems in the pattern development phase. For example, phosphorylation events are defined in the task definition as having only a theme and a site. However, there were instances in the training data that included both a theme and a cause argument. When those events were identified by our system and the cause was labeled, they were rejected during a syntactic error check by the test server.
4. RESULTS
4.1. Official Results
We participated in the challenge as Team 13. Table 2 shows our results on the official metrics. Our precision was the highest achieved by any group for Task 1 and Task 2, at 71.81 for Task 1 and 70.97 for Task 2. Our recalls were much lower and adversely impacted our F-measure; ranked by F-measure, we ranked 19th of 24 groups.
Table 2.
Official Scores for Tasks 1 and 2, and Modification Scores Only for Task 3, from the Approximate Span Matching/Approximate Recursive Matching Table.
| Tasks 1 and 3 |
Task 2 |
|||||||
|---|---|---|---|---|---|---|---|---|
| Event class | GS | answer | R | P | F | R | P | F |
| Localization | 174 (18) | 18 (18) | 10.34 | 100.00 | 18.75 | 9.77 | 94.44 | 17.71 |
| Binding | 347 (44) | 110 (44) | 12.68 | 40.00 | 19.26 | 12.32 | 39.09 | 18.74 |
| Gene expression | 722 (263) | 306 (263) | 36.43 | 85.95 | 51.17 | 36.43 | 85.95 | 51.17 |
| Transcription | 137 (18) | 20 (18) | 13.14 | 90.00 | 22.93 | 13.14 | 90.00 | 22.93 |
| Protein catabolism | 14 (4) | 6 (4) | 28.57 | 66.67 | 40.00 | 28.57 | 66.67 | 40.00 |
| Phosphorylation | 135 (30) | 30 (30) | 22.22 | 100.00 | 36.36 | 20.14 | 93.33 | 33.14 |
| Event Total | 1,529 (377) | 490 (377) | 24.66 | 76.94 | 37.35 | 24.30 | 76.12 | 36.84 |
| Regulation | 291 (9) | 19 (9) | 3.09 | 47.37 | 5.81 | 3.08 | 47.37 | 5.79 |
| Positive regulation | 983 (32) | 65 (32) | 3.26 | 49.23 | 6.11 | 3.24 | 49.23 | 6.08 |
| Negative regulation | 379 (10) | 22 (10) | 2.64 | 45.45 | 4.99 | 2.37 | 40.91 | 4.49 |
| Regulation Total | 1,653 (51) | 106 (51) | 3.09 | 48.11 | 5.80 | 3.02 | 47.17 | 5.67 |
| Negation | 227 (4) | 76 (4) | 1.76 | 5.26 | 2.64 | |||
| Speculation | 208 (14) | 105 (14) | 6.73 | 13.33 | 8.95 | |||
| Modification Total | 435 (18) | 181 (18) | 4.14 | 9.94 | 5.84 | |||
| All Total | 3,182 (428) | 596 (428) | 13.45 | 71.81 | 22.66 | 13.25 | 70.97 | 22.33 |
GS = gold standard (true positives) (given for Tasks 1/3 only), answer = all responses (true positives) (given for tasks 1/3 only), R = recall, P = precision, F = F-measure. All results are as calculated by the official scoring application.
In the evaluation, several different matching metrics were utilized for comparing submitted results to the gold standard data. These took into consideration various aspects of the predicted events: the event type, the identified event triggers, the event participants, and in turn the correctness of the entities and events that these participants refer to. In the exact matching (or strict equality), all of these aspects need to be identical for a prediction to count as a true positive. For approximate boundary matching, the text spans of the identified trigger words and entity participants are allowed to vary from the gold standard spans by one word to the right and/or left of the gold standard span. An additional variant, called approximate recursive matching, is like exact matching but relaxes the event participant match constraint to only consider Themes, allowing non-Theme arguments to differ. Note that the official system results presented in Table 2 use both approximate boundary matching and approximate recursive matching.
We noted that our results for the exact match metric and for the approximate boundary match metric were very close, suggesting that our techniques for named entity recognition and for recognizing trigger words are doing a good job of capturing the appropriate spans.
4.2. Bug Fixes and Coordination Handling
In addition to our official results, we also report in Table 3 the results of a run in which we fixed a number of bugs. This represents our current best estimate of our performance. The precision drops from 71.81 for Task 1 to 67.19, and from 70.97 for Task 2 to 65.74, but these precisions are still well above the second-highest precisions of 62.21 for Task 1 and 56.87 for Task 2. As the table shows, we had corresponding small increases in our recall to 17.38 and in our F-measure to 27.62 for Task 1, and in our recall to 17.07 and F-measure to 27.10 for Task 2.
We evaluated the effects of coordination handling by doing separate runs with and without this element of the processing pipeline. Compared to our unofficial results, which had an overall F-measure for Task 1 of 27.62 and for Task 2 of 27.10, a version of the system without handling of coordination had an overall F-measure for Task 1 of 24.72 and for Task 2 of 24.21.
4.3. Impact of Cascading Errors on Higher-Order Events
To assess the performance of our system on higher-order events (the regulation, negation, and speculation events) under the assumption of perfect recognition of the basic event types, we constructed a test case for our system in which all events corresponding to the basic event types (localization, binding, gene expression, transcription, protein catabolism, and phosphorylation)were extracted from the gold standard data and used as input to the detection of the complex event types. This allows us to get a sense of the impact of the limited recall of our system on the basic event types for the extraction of the more complex events that depend on their recognition.
The results of this analysis on the devtest data are shown in Table 4, with the system results under normal circumstances in the top section, and the system results using the gold standard data for the basic event types in the bottom section. As we might expect, we see a significant increase in the performance of the system on the negation and speculation events, from an F-score of 6.93 to 25.13 for negation and from 5.03 to 11.95 for speculation. The smaller improvement for speculation is a result of the more limited number of speculation patterns that we wrote for the shared task; the coverage of our negation patterns is simply broader.
Table 4.
Results on devtest Data on Complex Event Types without (Top Section) and with (Bottom Section) Use of Gold Standard Events for Basic Event Type.
| Event class | GS | Answer | R | P | F |
|---|---|---|---|---|---|
| Regulation | 173 (8) | 27 (8) | 4.62 | 29.63 | 8.00 |
| Positive regulation | 618 (30) | 79 (30) | 4.85 | 37.97 | 8.61 |
| Negative regulation | 196 (6) | 19 (6) | 3.06 | 31.58 | 5.58 |
| Negation | 107 (7) | 95 (7) | 6.54 | 7.37 | 6.93 |
| Speculation | 95 (5) | 104 (5) | 5.26 | 4.81 | 5.03 |
| Regulation | 173 (4) | 21 (4) | 2.31 | 19.05 | 4.12 |
| Positive regulation | 618 (12) | 52 (12) | 1.94 | 23.08 | 3.58 |
| Negative regulation | 196 (2) | 11 (2) | 1.02 | 18.18 | 1.93 |
| Negation | 107 (25) | 92 (25) | 23.36 | 27.17 | 25.13 |
| Speculation | 95 (15) | 156 (15) | 15.79 | 9.62 | 11.95 |
Interestingly, for all three regulation event types, we see a drop in performance when using the gold standard basic events. This can be attributed to the tension between regulation events that have proteins as their THEME, and those that have a basic event as their Theme. When both analyses are available, for instance in a clause such as regulation of CAT expression which would be matched by OpenDMAP as both regulation of [CAT] and regulation of [CAT expression], the system will make a choice between the two analyses. The inclusion of the gold standard events means that the system will be faced with this ambiguity more often; the performance on this test suggests that the system is making the incorrect choice in many of those instances.
4.4. Error Analysis
4.4.1. False Negatives
To better understand the causes of our low recall, we performed a detailed error analysis of false negatives using the devtest data. (Note that this section includes a very small number of examples from the devtest data.) We found four major causes of false negatives:
Intervening material between trigger words and arguments
Coordination that was not handled by our coordination component
Low coverage of trigger words
Anaphora and coreference
Intervening material For reasons that we detail in the Discussion section, we avoided the use of wildcards. This, and the lack of syntactic analysis in the version of the system that we used (note that syntactic analyses can be incorporated into an OpenDMAP workflow), meant that if there was text intervening between a trigger word and an argument, e.g., in to efficiently [express] in developing thymocytes a mutant form of the [NF-kappa B inhibitor] (PMID 10092801), where the bracketed strings are the trigger word and the argument, respectively, our pattern would not match.
Unhandled coordination Our coordination system only handled coordinated protein names. Thus, in cases where other important elements of the utterance, such as the trigger word transcription in transcription and subsequent synthesis and secretion of galectin-3 (PMID 8623933) were in coordinated structures, we missed the relevant events.
Low coverage of trigger words As we discuss in the Methods section, we did not attempt to cover all trigger words, in part because some less-frequent trigger words were involved in multiple event types, in part because some of them were extremely low-frequency and we did not want to overfit to the training data, and in part due to the time constraints of the shared task.
Anaphora and coreference Recognition of some events in the data would require the ability to do anaphora and coreference resolution. For example, in Although 2 early lytic transcripts, [BZLF1] and [BHRF1], were also detected in 13 and 10 cases, respectively, the lack of ZEBRA staining in any case indicates that these lytic transcripts are most likely [expressed] by rare cells in the biopsies entering lytic cycle (PMID 8903467), where the bracketed text is the arguments and the trigger word, the syntactic object of the verb is the anaphoric noun phrase these lytic transcripts, so even with the addition of a syntactic component to our system, we still would not have recognized the appropriate arguments without the ability to do anaphora resolution.
We return to a discussion of recall and its implications for systems like ours in the Discussion section.
4.4.2. False Positives
Although our overall rate of false positives was low, we sampled 90 false positive events from the devtest data, out of a total of 412, distributed across the nine event types. These were reviewed with a biologist to better understand the causes of this type of error.
We randomly selected 10 documents for each event type from the training data; the biologist then examined all events scored as false positives in those documents. An interesting finding was that the biologist judged 17% of them (15/90) as actually being true positives. We give the breakdown of true positives and false positives in these putative false positives in Table 5.
Table 5.
Analysis of Events Marked as False Positive in the Training Data.
| Event class | Analyzed | TP | FP |
|---|---|---|---|
| Localization | 4 | 1 | 3 |
| Binding | 21 | 1 | 20 |
| Gene expression | 11 | 7 | 4 |
| Transcription | 4 | 0 | 4 |
| Protein catabolism | 1 | 0 | 1 |
| Phosphorylation | 3 | 1 | 2 |
| Regulation | 14 | 1 | 13 |
| Positive regulation | 19 | 3 | 16 |
| Negative regulation | 13 | 1 | 12 |
| Totals | 90 | 15 | 75 |
TP is the number of events that were false positives according to the gold standard but that our biologist judged as true positive. FP is the number of events that were judged as false positives according to the gold standard and that our biologist agreed were false positives.
We found a number of cases in which a protein was listed in the A1 file, but a trigger word was apparently missed by the annotators, causing a false positive to be assigned when our system did find the trigger word. Examples of this are Tax-expressing (10090947), expression of CAT (10229275), RelA protein expression (10415057), fos gene expression (1712226), interleukin 2 production (7506531), and expression of the GM-CSF (7605990). In other cases, both the protein and the trigger word were in the gold standard, but there was no annotated event linking the two together. Examples of this include expression of vascular cell adhesion molecule 1 (10403270), where the expression trigger word and the protein name were both in the gold standard, but the expression event was not.
Because we had a relatively small number of actual false positives compared to other groups, we give examples of some of them.
We noted two main causes of false positive errors. The most common was that we misidentified a slot filler or were missing a slot filler completely for an actual event. The other main reason for false positives was when we erroneously identified a (non)event. For example, in coexpression of NF-kappa B/Rel and Sp1 transcription factors (PMID 7479915), we mistakenly identified Sp1 transcription as an event.
Failing to recognize embedded events while recognizing protein themes was one contributor to our poor results for the regulation event types. For example, where the intended output was [induction] of [I kappa B alpha phosphorylation] (7499266), we recognized only the protein portion of the theme and output [induction] of [I kappa B alpha] phosphorylation. Another cause of false positives was recognizing a theme, but not the associated cause, for regulation events. For example, where the intended output was [induction] of [IL-10 production] by [gp41] (10089566), we output [induction] of [IL-10 production] by gp41. Another cause of false positives was misrecognition of trigger words. In some cases this was due to words that were trigger words for events in some contexts, but merely parts of complex nominals in others, e.g. our incorrect output upstream of the [GM-CSF] [transcription] initiation site (7478534). In other cases, we failed to resolve cases of polysemy, such as the general English use of the word association, which can be a trigger word for binding in its biomedical use, in effects of [IL-11] were [associated] with reduced [NF-kappaB] activation (10411003). This also occurred when tokenization errors caused us to confuse regulation types, as in our incorrect output up-[regulation] of [CD80 Ag] (8690900). Finally, sentence segmentation errors from the incorporated OpenNLP sentence detector were an occasional contributor of false positives, as in our incorrect output for ...was suppressed by [alpha B2]. [Coexpression] of . . . (7605990).
5. DISCUSSION
Our results demonstrate that it is possible to achieve state-of-the art precision over a broad range of tasks and event types using our approach of manually constructed, ontologically typed rules—our precision of 71.81 on Task 1 was ten points higher than the second-highest precision (62.21), and our precision of 70.97 on Task 2 was 14 points higher than the second-highest precision (56.87). It remains the case that our recall was low enough to drop our F-measure considerably. Will it be the case that a system like ours can scale to practical performance levels nonetheless? Four factors suggest that it can.
The first is that there is considerable redundancy in the data; although we have not quantified it for this data set, we note that the same event is often mentioned repeatedly, but for knowledge base building and other uses of the extracted information, it is only strictly necessary to recognize an event once (although multiple recognition of the same assertion may increase our confidence in its correctness).
The second is that there is often redundancy across the literature; the best-supported assertions will be reported as initial findings and then repeated as background information.
The third is that these recall results reflect an approach that made no use of syntactic analysis beyond handling coordination. There is often text present in the input that cannot be disregarded without either using wildcards, which generally decreased precision in our experiments and which we generally eschewed, or making use of syntactic information to isolate phrasal heads. Syntactic analysis, particularly when combined with analysis of predicate-argument structure, has recently been shown to be an effective tool in biomedical information extraction (Miyao et al. 2009). There is broad need for this—for example, of the thirty localization events in the training data whose trigger word was translocation, a full eighteen had intervening textual material that made it impossible for simple patterns like translocation of [Theme] or [ToLoc] translocation to match.
Finally, our recall numbers reflect a very short development cycle, with as few as four patterns written for many event types. A less time-constrained pattern-writing effort would almost certainly result in increased recall. Some evidence for this comes from recent work by Hakenberg et al. (2009), who used our OpenDMAP parser with a set of 4,774 automatically learnt rules and produced the winning entry in the BioCreative II.5 protein-protein interaction information extraction competition.
We also note that there are users and use cases for which high precision is more important than high recall (Alex et al. 2008). For example, the analyst whose work on the Hanalyzer system is reported inLeach et al. (2009) never wanted to see any incorrect assertions in the system. Other such users may be model organism database curators, as suggested by Alex et al. (2008). Nonetheless, we continue to pursue methods to increase recall.
6. CONCLUSION
The results from the shared task indicate that systems like OpenDMAP, which takes advantage of semantic constraints defined in a background ontology coupled with linguistic patterns to identify text that corresponds to particular event types and identify the event participants, are reasonable approaches to problems like those defined by the BioNLP’09 challenge. In this case, the approach resulted in high-precision performance, as compared to other systems that participated in the challenge. It was found that “higher-order” events, in which one (or more) participant in the event is itself an event, are handled seamlessly in the OpenDMAP approach, but are harder to recognize due to their dependency on the performance of the base event recognition. Several challenges for the linguistic rules of our system were identified; we intend to improve on our handling of them in future versions of our system, including improved handling of syntactic structure (both to identify syntactic dependencies and to handle coordination more effectively), and treatment of coreference.
ACKNOWLEDGMENTS
We gratefully acknowledge Mike Bada’s help in loading the Sequence Ontology into Protégé.
This work was supported by NIH grants R01LM009254, R01GM083649, and R01LM008111 to Lawrence Hunter and T15LM009451 to Philip Ogren.
Footnotes
Determiners have been omitted from the rule for succinctness.
REFERENCES
- Alex B, Grover C, Haddow B, Kabadjov M, Klein E, Matthews M, Roebuck S, Tobin R, Wang X. Assisted curation: Does text mining really help? Pac Symp Biocomput. 2008 [PubMed] [Google Scholar]
- ALIAS-I. LingPipe 3.1.2. 2008. http://alias-i.com/lingpipe/ [Google Scholar]
- Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biology. 2005;6(2) doi: 10.1186/gb-2005-6-2-r21. Retrieved file cell.obo (versison 1.24 25:05:2007 09:56) from http://obofoundry.org/cgi-bin/detail.cgi?cell on June 14, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer S, Yesilada Y, Horan B, Goble C. Using ontologies and vocabularies for dynamic linking. IEEE Internet Computing. 2008;12(3):32–39. [Google Scholar]
- BjöRNE J, Heimonen J, Ginter F, Airola A, Pahikkala T, Salakoski T. Extracting complex biological events with rich graph-based feature sets. Proceedings of the Workshop on BioNLP: Shared Task. 2009;10:18. [Google Scholar]
- Blaschke C, Valencia A. The potential use of SUISEKI as a protein interaction discovery tool. Genome Inform. 2001;12:123–134. [PubMed] [Google Scholar]
- Blaschke C, Valencia A. The frame-based module of the SUISEKI information extraction system. IEEE Intelligent Systems. 2002;17:14–20. [Google Scholar]
- Buyko E, Wermter J, Poprat M, Hahn U. Automatically mapping an NLP core engine to the biology domain. Proceedings of the ISMB 2006 Joint BioLINK/Bio-Ontologies Meeting. 2006 [Google Scholar]
- Ciravegna F, Wilks Y. Designing adaptive information extraction for the semantic web in Amilcare. In: Handschuh S, Staab S, editors. Annotation for the SemanticWeb. Amsterdam, The Netherlands: IOS Press; 2003. [Google Scholar]
- Cohen KB, Tanabe L, Kinoshita S, Hunter L. A resource for constructing customized test suites for molecular biology entity identification systems. BioLINK. 2004:1–8. 2004. [Google Scholar]
- Cohen KB, Palmer M, Hunter L. Nominalization and alternations in biomedical language. PLoS ONE. 2008;3(9) doi: 10.1371/journal.pone.0003158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen KB, Verspoor K, Johnson HL, Roeder C, Ogren PV, Baumgartner WA, Jr, White E, Tipney H, Hunter L. BioNLP 2009 Companion Volume: Shared Task on Entity Extraction. 2009. High-precision biological event extraction with a concept recognizer; pp. 50–58. [Google Scholar]
- Consortium, T. G. O. Creating the Gene Ontology resource: Design and implementation. Genome Research. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daraselia N, Yuryev A, Egorov S, Novichkova S, Nikitin A, Mazo I. Extracting human protein interactions from MEDLINE using a full-sentence parser. Bioinformatics. 2004;20:604–611. doi: 10.1093/bioinformatics/btg452. [DOI] [PubMed] [Google Scholar]
- Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The sequence ontology: A tool for the unification of genome annotations. Genome Biology. 2005;6(5) doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci D, Lally A. Building an example application with the unstructured information management architecture. IBM Systems Journal. 2004;43(3):455–475. [Google Scholar]
- Hakenberg J, Leaman RJ, Vo NH, Jonnalagadda S, Sullivan R, Miller C, Tari L, Baral C, Gonzalez G. Online protein interaction extraction and normalization at Arizona State University. Proceedings of the BioCreative II.5 Workshop 2009 on Digital Annotations. 2009 [Google Scholar]
- Handschuh S, Maedche A, Staab S, Maedche E. CREAM—Creating relational metadata with a component-based, ontology-driven annotation framework. Proceedings of the First International Conference on Knowledge Capture (K-CAP) 2001 http://siegfried-handschuh.net/pub/2001/annotate_kcap2001.pdf. [Google Scholar]
- Hunter L, Lu Z, Firby J, Jr, B WA, Johnson HL, Ogren PV, Cohen KB. OpenDMAP: An open-source, ontology-driven concept analysis engine, with applications to capturing knowledge regarding protein transport, protein interactions and cell-specific gene expression. BMC Bioinformatics. 2008;9(78) doi: 10.1186/1471-2105-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IBM. UIMA Java framework. 2009. http://uima-framework.sourceforge.net/ [Google Scholar]
- Kim J-D, Ohta T, Pyysalo S, Kano Y, Tsujii J. Overview of BioNLP’09 shared task on event extraction. BioNLP 2009 Companion Volume: Shared Task on Entity Extraction. 2009:1–9. [Google Scholar]
- Leach SM, Tipney H, Feng W, Jr, B WA, Kasliwal P, Schuyler RP, Williams T, Spritz RA, Hunter L. Biomedical discovery acceleration, with applications to craniofacial development. PLoS Computational Biology. 2009;5(3) doi: 10.1371/journal.pcbi.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao Y, Sagae K, Saetre R, Matsuzaki T, Tsujii J. Evaluating contributions of natural language parsers to protein-protein interaction extraction. Bioinformatics. 2009;25(3):394–400. doi: 10.1093/bioinformatics/btn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: The Foundational Model of Anatomy. Journal of Biomedical Informatics. 2003;36(6):478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Saetre R, Miwa M, Yoshida K, Tsujii J. From protein-protein interaction to molecular event extraction. Natural language processing in biomedicine 2009. 2009:103–106. [Google Scholar]
- Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. BRENDA, the enzyme database: Updates and major new developments. Nucleic Acids Res. 2004;32(Database issue) doi: 10.1093/nar/gkh081. Retrieved file BrendaTissue.obo from http://obofoundry.org/cgi-bin/detail.cgi?brenda on June 14, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Gene Ontology: Tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Vera M, Motta E, Domingue J, Lanzoni M, Stutt A, Ciravegna F. MNM: Ontology Driven Semi-automatic and Automatic Support for Semantic Markup. Berlin, Germany: Springer Verlag; 2002. pp. 379–391. [Google Scholar]
- Verspoor K, Cohen KB, Hunter L. The textual characteristics of traditional and open access scientific journals are similar. BMC Bioinformatics. 2009;10 doi: 10.1186/1471-2105-10-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushiji A, Tateisi Y, Miyao Y, Tsujii J. Event extraction from biomedical papers using a full parser. Proceedings of the Pac Symp Biocomput. 2001:408–419. doi: 10.1142/9789814447362_0040. [DOI] [PubMed] [Google Scholar]