Figure 5. Cut repeats stimulate the glycosylase and AP-lyase activities of OGG1.
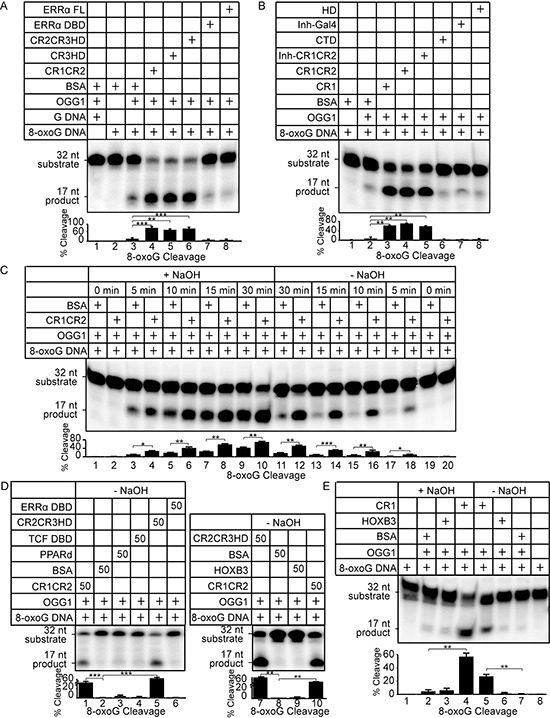
Double-stranded oligonucleotides containing an 8-oxoG or an unmodified G were radioactively end-labeled and used in cleavage and DNA binding assays. Sequences of probe A substrate and product are shown in Figure S1A. Percentages of cleaved product from 3 independent experiments are shown below each lane. Error bars represent standard error using Student's t-test, ***p < 0.001; **p < 0.01; *p < 0.05. (A and B) The 8-oxoG cleavage assay was performed for 30 min. using OGG1 (New England Biolabs, Ipswich, MA) and bacterially purified proteins as indicated. A diagrammatic representation of CUX1 proteins is shown in Figure 2A: CR1, CR1CR2, Inh-CR1CR2, CR2CR3HD, CR3HD, HD, CTD. Coomassie stains are presented in Figure S1B. ERRα FL is the full length estrogen-related receptor protein; ERRαDBD, the ERRα DNA binding domain; Inh-Gal4, the yeast Gal4 transcription factor fused to the CUX1 auto-inhibitory domain. (C) The 8-oxoG cleavage assay was performed using 50 nM OGG1 and bacterially purified CUX1 CR1CR2. After 0, 5, 10, 15, 30 min incubation at 37°C, reactions were stopped, and DNA was submitted to treatment with NaOH (+NaOH) or not (−NaOH). Reactions in the presence of NaOH monitor OGG1 glycosylase activity only, whereas reactions in the absence of NaOH reveal OGG1 glycosylase and AP-lyase activities. (D) The 8-oxoG cleavage assay was performed for 30 min. with OGG1 and the indicated proteins and was stopped without treatment with NaOH. (E) The 8-oxoG cleavage assay was performed using 50 nM OGG1 and bacterially purified CUX1 CR1. The reaction was incubated for 30 min at 37°C and the DNA was treated either NaOH (+NaOH) or not (−NaOH) prior to migration on the gel.
