Abstract
Background
CD8+ T cell responses are known to be important to the control of HIV-1 infection. While responses to reverse transcriptase and most structural and accessory proteins have been extensively studied, CD8 T cell responses specifically directed to the HIV-1 enzymes Protease and Integrase have not been well characterized, and few epitopes have been described in detail.
Methods
We assessed comprehensively the CD8 T cell responses to synthetic peptides spanning Protease and Integrase in 56 HIV-1 infected subjects with acute, chronic, or controlled infection using IFN-γ-Elispot assays and intracellular cytokine staining. Fine-characterization of novel CTL epitopes was performed on peptide-specific CTL lines in Elispot and 51Chromium-release assays.
Results
Thirteen (23%) and 38 (68%) of the 56 subjects had detectable responses to Protease and Integrase, respectively, and together these targeted most regions within both proteins. Sequence variability analysis confirmed that responses cluster largely around conserved regions of Integrase, but responses against a large, highly conserved region of the N-terminal DNA-binding domain of Integrase were not readily detected. CD8 T cell responses targeted regions of Protease that contain known Protease inhibitor mutation residues, but strong Protease-specific CD8 T cell responses were rare. Fine-mapping of targeted epitopes allowed the identification of three novel, HLA class I-restricted, frequently-targeted optimal epitopes. There were no significant correlations between CD8 T cell responses to Protease and Integrase and clinical disease category in the study subjects, nor was there a correlation with viral load.
Conclusions
These findings confirm that CD8 T cell responses directed against HIV-1 include potentially important functional regions of Protease and Integrase, and that pharmacologic targeting of these enzymes will place them under both drug and immune selection pressure.
Introduction
In HIV-1 infection, virus-specific CD8 T cell responses are readily detected in peripheral blood and lymph nodes, but HIV-1 replication typically persists in the face of an exuberant CD8 response [1-3]. Although ineffective at eradicating virus, HIV-specific CD8 T cells nonetheless play an important role in decreasing viremia. In SIV-infected macaques, depletion of CD8 cells results in uncontrolled infection [4,5]. In human studies, partial control of viremia during acute infection correlates with the appearance of HIV-specific CD8 T cells [6,7], and some reports have suggested that there is an inverse correlation between the CD8 response and HIV-1 viral load, although this remains controversial [8-11]. Escape from CTL recognition has been linked to disease progression in some studies [12-14], and recent population-based studies have confirmed that immune selection pressure mediated through HLA class I-restricted responses influence viral evolution, providing additional evidence that immune selection pressure persists in the chronic phase of HIV-1 infection [15]. Thus, although the specific relationship between CD8 T cells and viral control in HIV-1 infection remains unclear, CD8 responses appear to be a critical component of an effective HIV-1-specific immune response [16,17].
Significant efforts have been made to identify HLA-restricted CTL epitopes important for the control of HIV-1 infection, but this analysis remains incomplete. More than 300 peptides containing CD8 T cell epitopes have been reported to the HIV-1 Molecular Immunology Database, of which approximately 150 have been optimally defined [18]. This work has largely focused on the HIV-1 proteins Gag p17, p24, Nef, Env and Reverse Transcriptase (RT). The distribution of epitopes targeted within these proteins is highly variable, with clustering in relatively conserved regions of the virus [19,20]. Recently, studies have also identified CD8 T cell responses to several HIV-1 accessory proteins, including Tat, Rev, Vpr, Vpu and Vif, and shown that they comprise a significant percentage of the overall CTL response [21,22].
In contrast, studies of CD8 T cell responses to two enzymes within the Pol gene, Protease and Integrase, have been limited. These proteins are relatively highly conserved, and also are targets for drug development that place them under pharmacologic selection pressure. Moreover, since both proteins are relatively highly conserved, they may be valuable targets for vaccine development. The potential dual selective pressures on these genes may have important clinical implications [23]. Here, we describe the comprehensive assessment of the CD8 T cell response directed against Protease and Integrase in a large, diverse cohort of HIV-1 infected subjects, show that they are frequently targeted by HIV-specific CD8 T cell, and identify novel optimal epitopes that are frequently targeted.
Materials and methods
Subjects
Fifty-six subjects with documented HIV-1 infection based on serologic criteria who are followed clinically at the Massachusetts General Hospital, the Brigham and Women's Hospital, the Fenway Community Health Center or the Lemuel Shattuck Hospital in Boston were recruited and divided into three groups based on disease characteristics. Twenty-eight subjects were identified, and began effective treatment, during acute HIV-1 infection, defined as within 180 days of seroconversion ("acute cohort"). Twenty-two subjects with chronic HIV-1 infection followed for routine longitudinal care were also studied ("chronic" cohort). Of these, thirteen were receiving effective antiretroviral treatment and nine were not receiving treatment at the time of study. Finally, six individuals who control HIV-1 infection without treatment, defined as repeated HIV-1 RNA measurements below 1000 copies/ml in the absence of antiretroviral medications, were studied ("HIV-1 controller" cohort). Clinical and immunologic aspects of several of these patients have previously been described [21,24]. The study was approved by the Institutional Review Boards of the respective institutions, and all subjects gave informed consent for their participation. A subset of subjects in the acute cohort was studied while they were enrolled simultaneously in a study of structured treatment interruption in acute HIV-1 infection [25]; thus, for some acutely infected subjects, data both on and off therapy were obtained.
Synthetic HIV-1 peptides
Overlapping peptides 15 to 18 amino acids in length spanning the complete clade B consensus amino acid sequence of HIV-1 Protease (13 peptides) and Integrase (37 peptides) were synthesized on an automated peptide synthesizer (MBS 396, Advanced Chem Tech, Louisville, Kentucky, USA) by fluorenylmethoxycarbonyl chemistry. The algorithm used to design overlapping peptides has been described [11]. Briefly, consecutive 18-mers overlapping by 10 amino acids served as the basic template. If the terminal amino acid was not a defined class I MHC anchor residue, a shortened 15-, 16- or 17-mer with a compatible anchor residue was synthesized instead. A ten amino acid overlap was maintained for all peptides. Truncated peptides (8- to 11-mers) used to map novel optimal CTL epitopes were obtained from Research Genetics (Birmingham, Alabama, USA).
ELISPOT assay
Protease- and Integrase-specific CD8 T cell responses were quantified by IFN-γ ELISPOT assay as previously described [21,26]. Briefly, fresh peripheral blood mononuclear cells (PBMC) were isolated by Ficoll-Hypaque (Sigma, St. Louis, Missouri, USA) density gradient centrifugation. 100 μL of complete RPMI/10% fetal calf serum containing 0.5–1 × 105 PBMC were plated in each well of a 96-well polyvinylidene plate (MAIP S45; Millipore, Bedford, Massachusetts, USA) pre-coated with 0.5 μg/ml of the anti-IFN-γ MAb 1-DIK (Mabtech, Stockholm, Sweden). Individual peptides were added to wells at a final concentration of 1 × 10-5 M; wells without peptide served as a negative control, and phytohemagglutinin (PHA) was used as a non-specific activator of IFN-γ production to serve as a positive control. Plates were incubated overnight at 37°C. After washing with PBS, biotinylated anti-IFN-γ MAb 7-B6-1 was added at 0.5 μg/ml and incubated for 60–90 minutes at room temperature. After washing, 100 μl of 1:20,000 streptavidin-conjugated alkaline phosphatase (Mabtech) was added to each well, and individual IFN-γ secreting cells were visualized as dark spots after reacting with 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium (Bio Rad Labs, Hercules, California, USA). Specific IFN-γ producing cells (spot-forming cells, or SFC) were counted by direct visualization. Responses of greater than 40 SFC/million PBMC after subtracting the negative control value were considered positive; negative control values in all cases were less than 30 SFC/million PBMC.
Flow cytometric detection of peptide-stimulated IFN-γ production
Intracellular cytokine staining assays were performed as described previously [27]. Briefly, 0.5–1 × 106 PBMC were incubated with 4 μM peptide and 1 μg/ml each of anti-CD28 and anti-CD49 MAbs (Becton Dickinson, San Jose, California, USA) for one hour, followed by the addition of 10 ug/ml of brefeldin A (Sigma). Cells were incubated at 37°C for 6 hours, and then at 4°C overnight. Cells were then washed, stained with fluorescent-labeled CD4 and CD8 antibodies (Becton Dickinson), and then fixed and permeabilized using the Caltag Fixation/Permeabilization Kit according to the manufacturer's instructions (Caltag, Burlingame, California, USA). Fixed and permeabilized cells were then stained with anti-IFN-γ-fluoresceine isothiocyanate antibody (Becton Dickinson), washed and analyzed on a FACSCalibur flow cytometer (Becton Dickinson). In all but one detected T cell response, IFN-γ producing cells were exclusively CD8+.
Generation of peptide-specific CD8 CTL lines and HLA restriction of responses
PBMC were expanded with a bispecific CD3/CD4b monoclonal antibody [22] for 10 to 14 days in R10 medium [RPMI 1640 medium supplemented with 10 mM HEPES, 2 mM L-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin and 10% heat-inactivated fetal calf serum (Sigma)] supplemented with 50 U/ml recombinant interleukin-2 (Hoffman LaRoche, Nutley, New Jersey, USA). Peptide-specific CD8 T cell lines were isolated from expanded PBMC as previously described, using 20 μM peptide in an IFN-γ catching assay [22]. Peptide specificity of CD8 CTL lines was confirmed by flow cytometry, and lines were further expanded for an additional 7–10 days in the presence of irradiated feeder cells before use in epitope mapping and HLA restriction studies. HLA-restriction assays were performed using extensively washed, peptide-pulsed B-LCL as the peptide-presenting cell. HIV-1-specific cytotoxicity was assessed by 51chromium-release assay using an E:T ratio of 10:1. HLA-restriction of CTL epitopes was determined using a panel of target cells matched through only one of the HLA-A, HLA-B or HLA-C class I alleles expressed by the effector cells [28]. HLA tissue typing was performed at the MGH Tissue Typing Laboratory using sequence-specific primer PCR.
Fine mapping of CTL epitopes
In some cases, putative CTL responses to overlapping 15–18 mer peptides were further fine mapped to define the optimal, HLA-restricted epitope, as previously described [21,29]. Briefly, 8-, 9-, 10- and 11-mer truncations of the parent peptide were obtained (Research Genetics), and serial dilutions from 1 × 10-4 to 1 × 10-11 M were used in an ELISPOT assay. The optimal epitope was defined as the peptide that induced 50% maximal SFC at the lowest peptide concentration [29].
Comparison of CD8 T cell responses with amino acid sequence variability
To correlate CD8 T cell responses with conserved and non-conserved regions of Protease and Integrase, two calculations were performed. First, primary sequence data for individual Protease and Integrase protein sequences (n = 155) were obtained from the HIV-1 Molecular Immunology Database [30]. All subtypes were represented, and all clade B sequences with known dates of isolation were prior to 1997, so that Protease sequence variability would not have been influenced by Protease inhibitor-selected variations. Normalized Shannon entropy scores for each amino acid position were calculated using the general formulae:
(1) Cent =  log2 pa/log2(min(N, K)) and
log2 pa/log2(min(N, K)) and
(2) pa = na/N
where na is the number of amino acid residues of type a, N is the number of residues in the sequence database, and K is the number of residue types. In the subsequent analysis, N was set equal to155 (the number of sequences analyzed) and K was set equal to 21, representing the 20 amino acids and an extra symbol for any gaps in the sequence. The program Scorecons http://www.biochem.ucl.ac.uk/cgi-bin/valdar/scorecons_server.pl was used for all calculations. Second, because few optimal epitopes have been mapped in Protease or Integrase, there are insufficient data to develop a score based on known CTL epitopes directed against each amino acid in the two proteins. Thus, for each amino acid position, the number of subjects in the current study with detectable responses against peptides containing that amino acid were summed and used as a measure of CD8 responses to that amino acid residue. Raw normalized entropy scores were then correlated with the amount of CD8 T cell responses for each amino acid residue in both Protease and Integrase. Entropy scores were also smoothed over nine amino acids (corresponding to the size of a typical CD8 T cell epitope) and correlated with CD8 T cell responses. Correlations were made using the Spearman's rank-order correlation test [20].
Comparison of CTL responses against HIV-1 proteins by size
The HIV-1 Molecular Immunology Database was reviewed for reports describing CTL epitopes [18,30]. Published reports of cohorts in whom subjects were comprehensively screened against peptides spanning the entire length of one or more HIV-1 proteins were identified [8-10,24], and data on CTL frequency against individual HIV-1 proteins extracted for the comparison plot presented as Figure 5.
Figure 5.
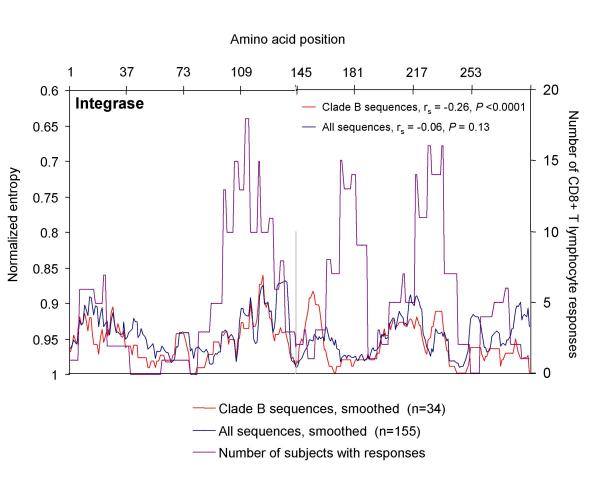
Correlation of amino acid sequence variability with frequency of CD8 T cell responses targeting Integrase. For Integrase, amino acid sequences were obtained from at the HIV-1 Molecular Immunology Database (27), and aligned relative to the HIV-1 clade B consensus sequence. Entropy scores for each amino acid residue were calculated based on this alignment, smoothed over nine amino acids, and plotted for all sequences (n = 155, blue line, left axis) and clade B sequences only (n = 34, red line, left axis). Entropy scores of 1 correspond to 100% conserved residues, while lower scores (plotted here on an inverse scale) correspond to increasing sequence variability. The number of responses in the 56 study subjects against peptides containing each amino acid was also plotted (purple line, right axis) to correlate regions with high sequence variability with regions targeted by CD8 T cells. Spearman's rank-order correlation coefficient was calculated to correlate CD8 T cell responses against sequence variability for each protein.
Results
Characteristics of study subjects
A total of 56 HIV-infected subjects were studied, including cohorts with acute, chronic, and controlled HIV-1 infection, as depicted in Table 1. Cohorts were similar with respect to basic demographics and ethnic background, as well as CD4 cell counts. The expected differences in viral load between controllers and the other cohorts were seen. Mean log10HIV-1 RNA level in the controller cohort was 2.03 ± 2.15 copies/ml; mean value in the untreated chronic cohort was 4.52 ± 4.56 copies/ml. Acute cohort subjects had been infected for a mean 23 months (range, 1 to 49 months) at the time of study. Although all but one of the 28 subjects enrolled in the acute cohort began effective antiretroviral treatment at the time of enrollment, 12 subjects were subsequently enrolled in a supervised treatment interruption trial [25], and thus had CD8 responses measured while off therapy.
Table 1.
Baseline characteristics of the 56 HIV-infected study subjects
| Acute (n = 28) | Chronic (n = 22) | Controller (n = 6) | |
| Age, years | 40.6 ± 9.1 | 43.7 ± 8.1 | 35.8 ± 5.0 |
| Male/Female | 27/1 | 18/4 | 5/1 |
| Race/Ethnicity | |||
| African-American | 3 | 3 | 2 |
| Caucasian | 23 | 16 | 3 |
| Haitian | 0 | 1 | 0 |
| Latino/Hispanic | 2 | 2 | 1 |
| Duration of infection, monthsa | 23 ± 13 | 122 ± 58 | 137 ± 83 |
| [range] | [1 – 49] | [30 – 191] | [16 – 241] |
| CD4+ T cells, cells × 106/mla | 675 ± 239 | 546 ± 236 | 615 ± 285 |
| [range] | [253 – 1252] | [89 – 828] | [176 – 1000] |
| Mean plasma HIV-1 RNA, log10 copies/ml | 3.99 | 4.13 | 2.03 |
| [range] | [1.70 – 5.27] | [1.70 – 4.99] | [1.70 – 2.60] |
| Receiving antiretroviral therapyb | |||
| Yes (%) | 57 % | 59 % | 0 % |
| No (%) | 43 % | 41 % | 100 % |
a Values are mean +/- S.D. b Two patients in the acute cohort were evaluated at two different time points, once on therapy, once off therapy.
CD8 T cell responses against HIV-1 Protease
We generated a series of 13 overlapping peptides (15 to 18 amino acids in length) spanning the complete HIV-1 Protease sequence, using the clade B consensus sequence [30] as a template (see Figure 1A for peptide sequences). Of the 13 peptides spanning Protease, a total of 10 (77%) were recognized by at least one study subject. Eight of these ten responses were confirmed as CD8-mediated by either CD4 cell depletion or intracellular cytokine staining; the remaining two responses could not be further evaluated due to sample availability. Thirteen of 56 subjects (23%) recognized at least one Protease peptide, with magnitudes ranging from 50 to 750 spot-forming cells (SFC) per million PBMC. The mean Protease-specific response in those recognizing this protein was 212 ± 202 SFC per million PBMC (Figures 1B and 1C and Table 2). These values are similar to reported CD8 T cell responses against other HIV-1 proteins [8-10,24]. There were not statistically significant differences in the percentage of subjects responding to Protease peptides among the three cohorts, or in the magnitude of the responses. Of the 13 subjects with identifiable Protease-specific responses, most targeted only one peptide, although the single peptide targeted varied among the persons tested. The broadest Protease-specific responses were in two subjects, both in the acute cohort, each of whom recognized four of the 13 Protease peptides, two of which were overlapping and therefore suggested recognition of the overlap region common to both peptides.
Figure 1.
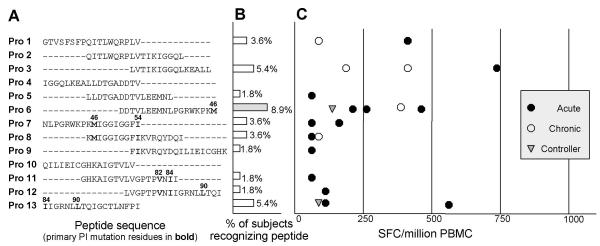
CD8 T cell responses to HIV-1 Protease. PBMC were stimulated with the indicated peptide in an overnight IFN-γ ELISPOT assay. Each row represents an individual peptide. (A) Amino acid sequence, using standard single-letter amino acid abbreviations. Numbers above peptide sequences refer to the amino acid position within Protease, with key Protease inhibitor mutation residues indicated in bold. (B) Bars represent the percentage of 56 study subjects who responded to the peptide in an ELISPOT assay. Peptides with the highest number of responses are shaded gray. (C) The magnitude of every CTL response detected in the study cohort. Each symbol represents a single CTL response against that peptide by one individual. Magnitudes of responses are shown after subtraction of background, which in all cases was <30 SFC/million PBMC. Closed circle (●): acute cohort. Open circle (○): chronic cohort. Shaded triangle ( ): controllers.
): controllers.
Table 2.
Number and percentage of subjects with detectable CTL responses, by group
| Group | Protease | Integrase | |||
| Number of subjects with any response (%) | Mean responsea | Number of subjects with any response (%) | Mean responsea | ||
| Acute | Total | 7/28 (25) | 221 | 18/28 (64) | 378 |
| On therapyb | 3/17 (18) | --- | 9/17 (53) | --- | |
| Off therapyb | 4/13 (31) | --- | 10/13 (77) | --- | |
| Chronic | Total | 4/22 (18) | 204 | 15/22 (68) | 281 |
| On therapy | 2/13 (15) | --- | 9/13 (69) | --- | |
| Off therapy | 2/9 (22) | --- | 6/9 (67) | --- | |
| Controller | 2/6 (33) | 125 | 5/6 (83) | 215 | |
| Overall | 13/56 (23) | 212 | 38/56 (68) | 320 | |
a Values are SFC/million PBMC. b Two patients in the acute cohort were evaluated at two different time points, once on therapy, once off therapy.
Although CD8 responses directed against the majority of Protease peptides were found, most of the individual Protease peptides were infrequently targeted by CTL. Only three peptides, Protease 3, Protease 6, and Protease 13 were recognized by more than two subjects; these were also the only peptides against which the mean magnitude of the CD8 response was greater than 250 SFC/million PBMC (Figure 1B and 1C). Protease 6 was the most frequently recognized peptide, targeted by five subjects (9%), and was thus chosen for further analysis and optimal epitope fine-mapping.
CD8 T cell responses against HIV-1 Integrase
Thirty-seven overlapping peptides spanning the complete HIV-1 Integrase sequence were used to assess CD8 responses in the same cohorts, also using the HIV-1 clade B consensus sequence as a template (Figure 2A). Twenty-six of the 37 Integrase peptides (70%) were recognized by at least one subject. Thirteen responses were confirmed as CD8-mediated by either CD4 cell depletion or intracellular cytokine staining. One response, against Integrase 29, was found to be CD4+ T cell mediated in one subject, and CD8-mediated in another subject, which suggests that this overlapping peptide contains both a CD4 and a CD8 T cell epitope. Unlike Protease, where a fairly uniform distribution of responses was seen across the entire protein, there were large regions in the Integrase sequence that were nearly devoid of CD8 responses. A stretch of nine peptides, Integrase 3 to Integrase 11, spanning 58 amino acids at the N-terminus of Integrase in the DNA-binding domain, were targeted by only three responses in the entire cohort of 56 study subjects (Figure 2C). Poorly immunogenic regions of Integrase were also seen at Integrase 18 to 22, Integrase 25 to 29, and at the C-terminus (Integrase 32 to 37).
Figure 2.
CD8 T cell responses to HIV-1 Integrase. PBMC were stimulated with the indicated peptide in an overnight IFN-γ ELISPOT assay. Each row represents an individual peptide. (A) Amino acid sequence, using standard single-letter amino acid abbreviations. Numbers above peptide sequence indicate amino acid position within Integrase. The conserved HHCC zinc finger-like domain and DDE element are indicated in bold. (B) Bars represent the percentage of 56 study subjects who responded to the peptide in an ELISPOT assay. Peptides with the highest number of responses are shaded gray. (C) The magnitude of every CTL response against Integrase peptides detected in the study cohort. Each symbol represents a single CTL response against that peptide by one individual. Magnitude of responses are after subtraction of background, which in all cases was <30 SFC/million PBMC. Symbols are the same as in Figure 1: Closed circle (●): acute cohort. Open circle (○): chronic cohort. Shaded triangle ( ): controllers.
): controllers.
Thirty-eight of fifty-six subjects (68%) recognized epitopes within Integrase, with a magnitude of response ranging from 50 to 1500 SFC per million PBMC (Figures 2B and 2C, Table 2). The mean magnitude of the response was 320 ± 301 SFC per million PBMC. Four subjects recognized as many as five Integrase peptides; most subjects recognized a single peptide. Three Integrase peptides were each recognized by more than 10% of study subjects: Integrase 14, Integrase 24 and Integrase 30 (Figure 2B). The majority of the CD8 T cell responses against Integrase were clustered around these three peptides.
Identification of optimal CD8 T cell epitopes within Protease and Integrase
Most of the previously described epitopes in Protease and Integrase have been defined based on predicted HLA-binding motifs, and published data on optimally-defined epitopes within Protease and Integrase identified directly from HIV-1 infected subjects are scarce [31-36]. We characterized the minimal amino acid sequences required for optimal recognition of the dominant Protease and Integrase peptides in these study subjects, as well as the restricting HLA class I alleles. Finemapping of the three novel CTL epitopes described in figure 3 was performed with cells from one patient, respectively. For each epitope peptide titrations were repeated and confirmed in at least one other study subject with a response to the corresponding 15 mer and the matching HLA type. In addition, the novel 9 or 10 mer was tested in all study subjects for which additional specimen were available.
Figure 3.
Fine-mapping of one novel epitope within Protease and two within Integrase. Peptide-specific CD8 cell lines were generated for three peptides, Protease 6, Integrase 17, and Integrase 29/30. PBMC collected from subjects with strong responses by ELISPOT were expanded using a bispecific CD3/4 antibody. Following expansion, peptide-specific cells were collected using an IFN-γ catching assay after stimulation with the appropriate peptide. Peptide specificity was confirmed by flow cytometry. HLA-restriction was then determined using peptide-pulsed target cells matched at only one MHC class I allele in a 51Cr-releasse assay at an E:T ratio of 10:1; peptide-pulsed autologous cells were used as a positive control. The sequences of the optimal epitopes were also determined by testing peptide-specific cell lines against serial dilutions of truncations of the original peptide in an ELISPOT assay. Data are shown for three epitopes: (A, B) – Protease 6. (C, D) – Integrase 17. (E, F) – Integrase 29/30.
Five subjects had strong responses to Protease 6 (range 150 to 460 SFC/million PBMC). Using serial dilutions of truncated peptides, we identified the optimal epitope within Protease 6 as EEMNLPGRW (EW9, amino acids Protease 34–42), as shown in Figure 3A,3B. HLA restriction of EW9 by HLA-B44 was determined using a 51chromium release assay. Overall, 4 of the 8 subjects (50%) expressing the HLA-B44 allele and evaluated in our study responded to Protease 6 and the novel EW9 epitope. Although the optimal epitope EW9 does not include the primary Protease inhibitor mutation site M46, it does include residue M36, which is a known accessory mutation site in PI-treated patients.
Using a similar approach to the fine-mapping of optimal epitopes and their HLA restriction, two frequently targeted CTL epitopes within Integrase were further characterized in detail. The most frequently targeted Integrase peptide is Integrase 30, which was recognized by 16% of study subjects. Several persons had responses to the adjacent peptide (Integrase 29), suggesting the presence of an epitope within the overlapping region of these peptides. Fine mapping confirmed the optimal epitope to be in the overlap region shared by both peptides, KIQNFRVYY (KY9), which was restricted by the HLA-A30 allele (Figure 3E,3F), an allele recently associated with decreased viral set point in non-clade B infection [37]. Both individuals with a response to Integrase 29/30 recognized the 9 mer peptide. Sabbaj et al. also described this epitope in a recently published article [38]. A related epitope, KIQNFRVYYR (KR10), has been predicted based on the HLA-A2 binding supermotif, and this 10 amino acid peptide has been previously studied in HLA-A2-positive long-term non-progressors [35]. Interestingly several study subjects had concomitant responses to Integrase 30 and 31 and expressed HLA alleles different from HLA-A30, suggesting that an additional epitope is contained within the C-terminal half of Integrase 30. It further indicates that the region of the Integrase sequence spanned by overlapping peptides Int-29–30 may be an immunodominant region of the Integrase protein.
Fine mapping confirmed the epitope within Integrase 17, a peptide targeted by 9% of study subjects, as the epitope STTVKAACWW (SW10). This epitope was restricted by HLA-B57, an MHC class I allele known to be associated with HIV-1 long-term non-progression, as shown in Figure 3C,3D. All five HLA-B57 positive study subjects in this cohort were long-term non-progressors and recognized STTVKAACWW, suggesting high immunogenicity of this newly defined epitope.
Correlation of regions targeted by CD8 T cell responses with amino acid variability
The above data indicate that numerous regions of both Protease and Integrase are potential targets for CD8 T cell responses, and suggest regions of epitope clustering. We further evaluated epitope clustering in these proteins through an analysis of primary sequence diversity. As a measure of sequence variability, we calculated the average entropy at each of the 288 amino acid positions within Integrase, based on 155 protein sequences, including 34 clade B sequences, reported to the HIV-1 Molecular Immunology Database [30]. A similar analysis has recently been reported for other HIV-1 proteins [20]. This analysis confirms that within Integrase, a large stretch of highly-conserved sequence exists at amino acids 40 to 100, and three smaller highly-conserved regions exist centered at amino acids 145, 181 and 240 (Figure 4, blue and red lines).
Figure 4.
Correlation of amino acid sequence variability with frequency of CD8 T cell responses targeting Protease. For Protease, amino acid sequences were obtained from at the HIV-1 Molecular Immunology Database (27), and aligned relative to the HIV-1 clade B consensus sequence. Entropy scores for each amino acid residue were calculated based on this alignment, smoothed over nine amino acids, and plotted for all sequences (n=155, blue line, left axis) and clade B sequences only (n=34, red line, left axis). Entropy scores of 1 correspond to 100% conserved residues, while lower scores (plotted here on an inverse scale) correspond to increasing sequence variability. The number of responses in the 56 study subjects against peptides containing each amino acid was also plotted (purple line, right axis) to correlate regions with high sequence variability with regions targeted by CD8 T cells. Spearman’s rank-order correlation coefficient was calculated to correlate CD8 T cell responses against sequence variability for each protein.
We next compared the entropy at each position with the number of subjects targeting peptides containing that amino acid (Figure 4, purple line). CD8 T cell responses cluster around three regions of Integrase, centered around amino acids 110, 180 and 220. The two clusters at the C-terminal end of Integrase correspond to regions of low amino acid variability, while the N-terminal epitope cluster centered on amino acid 110 overlaps with a region of high amino acid variability. Somewhat surprisingly, the highly conserved region of Integrase in the N-terminal domain from amino acids 40–90 with low sequence variability was largely devoid of CD8 T cell responses. Spearman's rank-order correlation coefficient (rs) and the P value for the correlation between the number of responses and raw entropy was rs = -0.07 and P = 0.11 for all sequences, and rs = -0.61 and P < 0.0001 for clade B sequences. Smoothing entropy scores over nine amino acids did not significantly alter the correlation between entropy and response frequency; for smoothed entropy, rs = -0.07 and P = 0.13 for all sequences, and rs = -0.26 and P < 0.0001 for clade B sequences. Thus overall, there is a slight inverse correlation between clade B sequence variability and CD8 T cell responses for the entire Integrase protein.
A similar analysis was performed for Protease. Notably, Protease has more clearly defined domains of high and low amino acid variability (Figure 5, blue and red lines). As the sequences used to calculate amino acid variability predate Protease inhibitor therapy, this is not the result of drug-induced selection pressure. Confirming this, non-clade B sequences from regions of the world where drugs are unavailable also show three domains with high and three domains with low variability (data not shown). Figure 5 also reveals a slight inverse correlation between CD8 T cell responses against Protease (Figure 5, purple line) and Protease sequence variability for clade B sequences. Both graphically and statistically, this association is not as strong for Protease as it is for Integrase. For Protease, Spearman's rank-order correlation coefficient using raw entropy scores was rs = -0.16 and P = 0.054 for all sequences, and rs = -0.61 and P < 0.0001 for clade B sequences. The corresponding values using smoothed entropy scores were rs = -0.003 and P = 0.49 for all sequences, and rs = -0.20 and P = 0.02 for clade B sequences. It should also be noted that the data on CD8 T cell responses presented here were obtained from subjects many of who were receiving, or had previously received, Protease inhibitor therapy. Because autologous protein sequences from these patients were not readily available, we were not able to assess the impact of prior PI treatment on the subsequent correlation between Protease-specific responses and Protease sequence diversity. Nonetheless, as has been found for other HIV proteins [20], there appears to be an inverse correlation between clade B sequence diversity and CD8 T cell responses against both Integrase and Protease in the study subjects.
Discussion
We here report a unique and most detailed assessment of the CD8 T cell response against the HIV-1 enzymes Protease and Integrase in a large cohort of HIV-1-infected subjects representing both early and chronic disease. To date, responses directed against two of the three key HIV-1 enzymes encoded by the pol gene, Protease and Integrase, have received limited attention, and the breadth and specificity of responses to these proteins remain poorly defined [18]. We show that both Protease and Integrase are significant targets for HIV-1-specific CD8 T cell responses, recognized by 23% and 68% of subjects in our cohort, respectively. These values are consistent with the frequency of responses targeting other HIV-1 proteins, although lower per unit protein length than more immunogenic proteins such as Gag p17, Gag p24, and Nef. Moreover, we optimally define the three most frequently targeted discrete epitopes within these proteins, and show that peptides containing Protease epitopes overlap with regions expected to be under pharmacologic selection pressure in those persons fortunate to have access to Protease inhibitor therapy.
Although studies of CD8 responses to the pol gene product have been conducted [10,11,39], few Protease and Integrase epitopes had been described, even in the highly conserved active sites of the enzyme. No epitopes within Protease have been defined de novo in infected persons; those reported to the HIV-1 CTL database have been identified either on the basis of predicted HLA-binding motifs, or characterized only in HIV-exposed, seronegative individuals [31,35,40]. Optimal epitope mapping for these epitopes and analysis of the frequency and breadth in HIV-1 infected populations has not been done. Similarly, rigorous optimal epitope mapping in Integrase has not been reported; peptides targeted within Integrase have been identified based largely on predicted binding motifs, as well as studies of exposed seronegative subjects or populations with selected HLA alleles [32,33,35,36]. Our data indicate that both of these proteins serve as frequent targets for CD8 T cells.
Significant epitope clustering in Integrase was seen in our study, and these epitopes cluster largely around highly conserved residues in the C-terminal portion of the protein. Interestingly, highly conserved residues in the N-terminal zinc finger domain and the conserved "DDE" catalytic core are largely devoid of CD8 T cell responses [41]. Without sequencing autologous virus, we cannot rule out the possibility that the peptides used in our study to evaluate CD8 responses in the conserved regions of Integrase failed to pick up responses that were actually present in our study subjects. However, because these regions of Integrase are highly conserved, the clade B consensus sequence used to generate the peptides should be a close reflection of the viral sequence present in our study subjects. Identification of factors that might contribute to a paucity of immune responses against a highly conserved region of this protein, such as poor proteasome cleavage sites [42] or reduced affinity for HLA class I molecules [43]. A recent study actually indicates that the frequency of recognition of a peptide was indeed correlated with the presence of predicted immunoproteasomal cleavage sites within the C-terminal half of the peptide and a reduced frequency of amino acids that impair binding of optimal epitopes to the restricting class I molecules [11]. However, this issue will require further study.
The HIV-1 Protease gene is under considerable selection pressure through the use of Protease inhibitor therapy in populations fortunate enough to have access to these drugs. Given the potentially complex interactions between drug selection pressure and immune selection pressure, we determined whether the epitopes targeted by Protease-specific CD8 cells were contained within regions known to mutate in the presence of drug selection pressure. Protease 6, the most frequently targeted Protease peptide, contains the primary mutation site M46 and the accessory mutation site M36. Primary PI mutations at M46, I54, V82, I84, and L90 [44,45] are clustered in peptides Protease 6 through 9, and Protease 11 through 13 (Figure 1A), all of which contain CD8 T cell epitopes.
Because of their relative immunogenicity and highly conserved nature, both Protease and Integrase could be potential targets for vaccines and immunotherapeutic interventions. However, features of the CD8 T cell response directed against each protein should be noted in this context. First, the Protease-specific responses identified here were of relatively low magnitude, even in those who control viremia without treatment. Second, although the Integrase-specific responses described here were of high magnitude, they cluster around three regions of the Integrase molecule, at least one of which falls largely outside of the highly conserved active sites of the enzyme [46]. Further epitope mapping within Protease and Integrase will be necessary to determine the extent of epitopes throughout these proteins, and further delineate the relationship between sequence diversity and effective CD8 T cell responses.
The frequent targeting of the Protease and Integrase proteins raises the question as to how immunogenic these proteins are compared to other HIV proteins. Figure 6 compares the frequency of CD8 T cell responses versus protein amino acid length for Integrase and Protease, as well as for all HIV-1 proteins based on published data from large cohorts evaluating the responses against individual proteins [8-10,24,47]. Although Integrase peptides were targeted by CD8 T cells in HIV-1 infected subjects at three times the frequency of Protease peptides, comparison of CD8 T cell responses per unit protein length suggests that the relative targeting of the two proteins is similar (Figure 6). In addition, published data on CTL frequencies per unit protein length for other non-structural proteins, including Tat, Rev, and Vif, are similar to Protease and Integrase. Reverse transcriptase, Vpr, and the envelope glycoproteins exhibit proportionately lower frequencies of CTL induction relative to their number of amino acid residues; conversely, the frequency of CTL responses per unit length of protein to Nef, Vpr, and the Gag proteins appear to be over-represented in HIV-1-infected subjects. There are multiple factors that may influence immunogenicity. Levels of the protein available for epitope processing are affected by the stability of the mRNA, polyprotein or mature protein and the protein's relative cytoplasmic abundance [48]. CTL epitopes are also affected by the presence of proteasome cleavage sites within a protein, sequence variation, the stability of HLA binding and TCR recognition. The role of each of these factors can be difficult to measure.
Figure 6.
Frequency of CD8 T cell responses to HIV-1 proteins relative to protein size. The frequency of responses directed against Protease and Integrase in the study cohort are plotted against the size of the proteins, in number of amino acids. Published data from cohorts where the frequency of CD8 T cell responses against at least one HIV-1 protein are plotted for comparison (see text for references).
Finally, in our study we did not determine any significant differences in CD8 T cell responses directed against Protease and Integrase when comparing acute and chronic HIV infection. Previous studies have suggested that CTL responses that develop during acute infection may differ from those seen during chronic infection, and that these differences may be important in the ultimate failure of the immune response to control viremia [10,12,24,49]. Responses directed against nef and accessory proteins appear to develop early in HIV infection, until Gag p24-specific responses emerge and dominate the CTL response in chronic infection. The generation and persistence of Protease and Integrase-specific responses do not appear to differ in acute versus chronic infections, although the impact of drug selection pressure on this process remains to be determined.
Conclusions
We conclude that Protease and Integrase are frequently targeted by the CD8 T cell response in infected individuals. These responses may be particularly important to examine in relation to viral immunopathogenesis and specific selection pressures as treatment with Protease inhibitors expands and Integrase inhibitors commence. In treated patients, viral sequence within these epitopes will be under selective pressures from two sources, drug and the immune system. Recent data from Moore et al. strongly suggest that HIV-1 sequence variation in individual patients can be directly attributed to escape from CTL, and previous studies in humans and primate models have confirmed CTL escape and its functional consequences [15,50-53]. Similar analyses have been undertaken on the evolution of virus under selective pressure from Protease inhibitors alone [54,55]. The dynamics of viral escape during selective pressure from both CTL and from drugs will be critical to examine, and will likely require assessment of immune responses to the autologous virus variants present in vivo to provide further insights regarding HIV immunopathogenesis and vaccine development.
Competing interests
None declared.
Authors' contributions
WRR carried out the majority of the Elispot assays, statistical analysis of the HIV database sequences, participated in study design and drafted the manuscript. MMA established the HIV controller Cohort, lead the epitope mapping efforts, participated in the immunoassays, and the draft of the manuscript. AR, XY, CAF and BP performed the flowcytometry-based immunoassays, carried out the cytotoxicity assays and participated in the processing of study specimen and in the Elispot assays. MA participated in study design, data analysis and lent statistical support. ESR and BDW were responsible for the establishment of the primary infection cohort, conceived of the study and participated in the study design and coordination. All authors read and approved the final manuscript
Abbreviations used in this paper
RT, reverse transcriptase;
SFC, spot-forming cells;
BCL, B cell line;
PBMC, peripheral blood mononuclear cells;
IFN, Interferon
Acknowledgments
Acknowledgements
The authors would like to thank the study subjects for their participation. We are also indebted to the dedicated research staff at the various clinical sites for their recruitment efforts.
This research was funded by the National Institutes of Health (R37 AI128568-BDW, ROI AI50429-MA), the Doris Duke Charitable Foundation (MA), Concerned Parents for AIDS Research (CPFA) (MMA), the Partners/Fenway/Shattuck Center for AIDS Research (XGY) and Howard Hughes Medical Institute (BDW). B.D.W. is the recipient of a Doris Duke Distinguished Clinical Scientist Award.
Contributor Information
William R Rodriguez, Email: wrodriguez@partners.org.
Marylyn M Addo, Email: Maddo@partners.org.
Almas Rathod, Email: arathod@partners.org.
Cecily A Fitzpatrick, Email: cfitzpatrick@zonemail.net.
Xu G Yu, Email: Xyu@partners.org.
Beth Perkins, Email: Bperkins@partners.org.
Eric S Rosenberg, Email: erosenberg1@partners.org.
Marcus Altfeld, Email: Maltfeld@partners.org.
Bruce D Walker, Email: bwalker@partners.org.
References
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- McMichael AJ, Rowland-Jones SL. Cellular immune responses to HIV. Nature. 2001;410:980–987. doi: 10.1038/35073658. [DOI] [PubMed] [Google Scholar]
- Soudeyns H, Pantaleo G. The moving target: mechanisms of HIV persistence during primary infection. Immunol Today. 1999;20:446–450. doi: 10.1016/S0167-5699(99)01504-2. [DOI] [PubMed] [Google Scholar]
- Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, Kostrikis LG, Zhang L, Perelson AS, Ho DD. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MR, Ambrozak DR, Douek DC, Bonhoeffer S, Brenchley JM, Casazza JP, Koup RA, Picker LJ. Analysis of Total Human Immunodeficiency Virus (HIV)-Specific CD4(+) and CD8(+) T-Cell Responses: Relationship to Viral Load in Untreated HIV Infection. J Virol. 2001;75:11983–11991. doi: 10.1128/JVI.75.24.11983-11991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards BH, Bansal A, Sabbaj S, Bakari J, Mulligan MJ, Goepfert PA. Magnitude of functional CD8(+) T-cell responses to the gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J Virol. 2002;76:2298–2305. doi: 10.1128/jvi.76.5.2298-2305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RE,, Strick D, Johnston MN, Corcoran C, Wurcel A, Fitzpatrick CA, Feeney ME, Rodriguez WR, Basgoz N, Drä nert R, Stone DR, Brander C, Goulder PJR, Rosenberg ES, Altfeld M, Walker BD. Comprehensive analysis of Human Immunodeficiency Virus type 1 (HIV-1)-specific T cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77:2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frahm N, Korber BT, Adams CM, Szinger JJ, Draenert R, Addo MM, Feeney ME, Yusim K, Sango K, Brown NV, SenGupta D, Piechocka-Trocha A, Simonis T, Marincola FM, Wurcel AG, Stone DR, Russell CJ, Adolf P, Cohen D, Roach T, StJohn A, Khatri A, Davis K, Mullins J, Goulder PJ, Walker BD, Brander C. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J Virol. 2004;78:2187–2200. doi: 10.1128/JVI.78.5.2187-2200.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- Evans DT, O'Connor DH, Jing P, Dzuris JL, Sidney J, da Silva J, Allen TM, Horton H, Venham JE, Rudersdorf RA, Vogel T, Pauza CD, Bontrop RE, DeMars R, Sette A, Hughes AL, Watkins DI. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat Med. 1999;5:1270–1276. doi: 10.1038/15224. [DOI] [PubMed] [Google Scholar]
- Goulder PJ, Brander C, Tang Y, Tremblay C, Colbert RA, Addo MM, Rosenberg ES, Nguyen T, Allen R, Trocha A, Altfeld M, He S, Bunce M, Funkhouser R, Pelton SI, Burchett SK, McIntosh K, Korber BT, Walker BD. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–338. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
- Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- Brander C, Walker BD. T lymphocyte responses in HIV-1 infection: implications for vaccine development. Curr Opin Immunol. 1999;11:451–459. doi: 10.1016/S0952-7915(99)80076-4. [DOI] [PubMed] [Google Scholar]
- Goulder PJ, Rowland-Jones SL, McMichael AJ, Walker BD. Anti-HIV cellular immunity: recent advances towards vaccine design. Aids. 1999;13:S121–36. [PubMed] [Google Scholar]
- Brander C, Goulder PJ. The evolving field of HIV-CTL mapping: New approaches for the identification of novel epitopes. 2001.
- Walker BD, Korber BT. Immune control of HIV: the obstacles of HLA and viral diversity. Nat Immunol. 2001;2:473–475. doi: 10.1038/88656. [DOI] [PubMed] [Google Scholar]
- Yusim K, Kesmir C, Gaschen B, Addo MM, Altfeld M, Brunak S, Chigaev A, Detours V, Korber BT. Clustering Patterns of Cytotoxic T-Lymphocyte Epitopes in Human Immunodeficiency Virus Type 1 (HIV-1) Proteins Reveal Imprints of Immune Evasion on HIV-1 Global Variation. J Virol. 2002;76:8757–8768. doi: 10.1128/JVI.76.17.8757-8768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo MM, Altfeld M, Rosenberg ES, Eldridge RL, Philips MN, Habeeb K, Khatri A, Brander C, Robbins GK, Mazzara GP, Goulder PJ, Walker BD. The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals. Proc Natl Acad Sci U S A. 2001;98:1781–1786. doi: 10.1073/pnas.98.4.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altfeld M, Addo MM, Eldridge RL, Yu XG, Thomas S, Khatri A, Strick D, Phillips MN, Cohen GB, Islam SA, Kalams SA, Brander C, Goulder PJ, Rosenberg ES, Walker BD. Vpr is preferentially targeted by CTL during HIV-1 infection. J Immunol. 2001;167:2743–2752. doi: 10.4049/jimmunol.167.5.2743. [DOI] [PubMed] [Google Scholar]
- Karlsson AC, Deeks SG, Barbour JD, Heiken BD, Younger SR, Hoh R, Lane M, Sallberg M, Ortiz GM, Demarest JF, Liegler T, Grant RM, Martin JN, Nixon DF. Dual pressure from antiretroviral therapy and cell-mediated immune response on the human immunodeficiency virus type 1 protease gene. J Virol. 2003;77:6743–6752. doi: 10.1128/JVI.77.12.6743-6752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altfeld M, Rosenberg ES, Shankarappa R, Mukherjee JS, Hecht FM, Eldridge RL, Addo MM, Poon SH, Phillips MN, Robbins GK, Sax PE, Boswell S, Kahn JO, Brander C, Goulder PJ, Levy JA, Mullins JI, Walker BD. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J Exp Med. 2001;193:169–180. doi: 10.1084/jem.193.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg ES, Altfeld M, Poon SH, Phillips MN, Wilkes BM, Eldridge RL, Robbins GK, D'Aquila RT, Goulder PJ, Walker BD. Immune control of HIV-1 after early treatment of acute infection. Nature. 2000;407:523–526. doi: 10.1038/35035103. [DOI] [PubMed] [Google Scholar]
- Goulder PJ, Addo MM, Altfeld MA, Rosenberg ES, Tang Y, Govender U, Mngqundaniso N, Annamalai K, Vogel TU, Hammond M, Bunce M, Coovadia HM, Walker BD. Rapid definition of five novel HLA-A*3002-restricted human immunodeficiency virus-specific cytotoxic T-lymphocyte epitopes by elispot and intracellular cytokine staining assays. J Virol. 2001;75:1339–1347. doi: 10.1128/JVI.75.3.1339-1347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher CJ, Quittner C, Peterson DM, Connors M, Koup RA, Maino VC, Picker LJ. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat Med. 1999;5:518–525. doi: 10.1038/8400. [DOI] [PubMed] [Google Scholar]
- Walker BD, Flexner C, Birch-Limberger K, Fisher L, Paradis TJ, Aldovini A, Young R, Moss B, Schooley RT. Long-term culture and fine specificity of human cytotoxic T-lymphocyte clones reactive with human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1989;86:9514–9518. doi: 10.1073/pnas.86.23.9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altfeld MA, Trocha A, Eldridge RL, Rosenberg ES, Phillips MN, Addo MM, Sekaly RP, Kalams SA, Burchett SA, McIntosh K, Walker BD, Goulder PJ. Identification of dominant optimal HLA-B60- and HLA-B61-restricted cytotoxic T-lymphocyte (CTL) epitopes: rapid characterization of CTL responses by enzyme-linked immunospot assay. J Virol. 2000;74:8541–8549. doi: 10.1128/JVI.74.18.8541-8549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Brander C, Haynes B, Koup R, Kuiken C, Moore J, Walker BD, Watkins D. HIV Molecular Immunology Database 2001, http://hiv-web.lanl.gov/immunology. Los Alamas National Laboratory: Theoretical Biology and Biophysics, Los Alamos, NM, USA. 2001.
- Altfeld MA, Livingston B, Reshamwala N, Nguyen PT, Addo MM, Shea A, Newman M, Fikes J, Sidney J, Wentworth P, Chesnut R, Eldridge RL, Rosenberg ES, Robbins GK, Brander C, Sax PE, Boswell S, Flynn T, Buchbinder S, Goulder PJ, Walker BD, Sette A, Kalams SA. Identification of novel HLA-A2-restricted human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte epitopes predicted by the HLA-A2 supertype peptide-binding motif. J Virol. 2001;75:1301–1311. doi: 10.1128/JVI.75.3.1301-1311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond KB, Sriwanthana B, Hodge TW, De Groot AS, Mastro TD, Young NL, Promadej N, Altman JD, Limpakarnjanarat K, McNicholl JM. An HLA-directed molecular and bioinformatics approach identifies new HLA-A11 HIV-1 subtype E cytotoxic T lymphocyte epitopes in HIV-1- infected Thais. AIDS Res Hum Retroviruses. 2001;17:703–717. doi: 10.1089/088922201750236988. [DOI] [PubMed] [Google Scholar]
- Ikeda-Moore Y, Tomiyama H, Miwa K, Oka S, Iwamoto A, Kaneko Y, Takiguchi M. Identification and characterization of multiple HLA-A24-restricted HIV-1 CTL epitopes: strong epitopes are derived from V regions of HIV-1. J Immunol. 1997;159:6242–6252. [PubMed] [Google Scholar]
- Kaul R, Dong T, Plummer FA, Kimani J, Rostron T, Kiama P, Njagi E, Irungu E, Farah B, Oyugi J, Chakraborty R, MacDonald KS, Bwayo JJ, McMichael A, Rowland-Jones SL. CD8(+) lymphocytes respond to different HIV epitopes in seronegative and infected subjects. J Clin Invest. 2001;107:1303–1310. doi: 10.1172/JCI12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Propato A, Schiaffella E, Vicenzi E, Francavilla V, Baloni L, Paroli M, Finocchi L, Tanigaki N, Ghezzi S, Ferrara R, Chesnut R, Livingston B, Sette A, Paganelli R, Aiuti F, Poli G, Barnaba V. Spreading of HIV-specific CD8+ T-cell repertoire in long-term nonprogressors and its role in the control of viral load and disease activity. Hum Immunol. 2001;62:561–576. doi: 10.1016/S0198-8859(01)00245-2. [DOI] [PubMed] [Google Scholar]
- Tomiyama H, Sakaguchi T, Miwa K, Oka S, Iwamoto A, Kaneko Y, Takiguchi M. Identification of multiple HIV-1 CTL epitopes presented by HLA-B*5101 molecules. Hum Immunol. 1999;60:177–186. doi: 10.1016/S0198-8859(98)00113-X. [DOI] [PubMed] [Google Scholar]
- Tang J, Tang S, Lobashevsky E, Myracle AD, Fideli U, Aldrovandi G, Allen S, Musonda R, Kaslow RA. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J Virol. 2002;76:8276–8284. doi: 10.1128/JVI.76.16.8276-8284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbaj S, Bansal A, Ritter GD, Perkins C, Edwards BH, Gough E, Tang J, Szinger JJ, Korber B, Wilson CM, Kaslow RA, Mulligan MJ, Goepfert PA. Cross-Reactive CD8+ T Cell Epitopes Identified in US Adolescent Minorities. J Acquir Immune Defic Syndr. 2003;33:426–438. doi: 10.1097/00126334-200308010-00003. [DOI] [PubMed] [Google Scholar]
- Haas G, Samri A, Gomard E, Hosmalin A, Duntze J, Bouley JM, Ihlenfeldt HG, Katlama C, Autran B. Cytotoxic T-cell responses to HIV-1 reverse transcriptase, integrase and protease. Aids. 1998;12:1427–1436. doi: 10.1097/00002030-199812000-00004. [DOI] [PubMed] [Google Scholar]
- Kaul R, Rowland-Jones SL, Kimani J, Dong T, Yang HB, Kiama P, Rostron T, Njagi E, Bwayo JJ, MacDonald KS, McMichael AJ, Plummer FA. Late seroconversion in HIV-resistant Nairobi prostitutes despite pre-existing HIV-specific CD8+ responses. J Clin Invest. 2001;107:341–349. doi: 10.1172/JCI10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci U S A. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedermann G, Geier E, Lucchiari-Hartz M, Hitziger N, Ramsperger A, Eichmann K. The specificity of proteasomes: impact on MHC class I processing and presentation of antigens. Immunol Rev. 1999;172:29–48. doi: 10.1111/j.1600-065x.1999.tb01354.x. [DOI] [PubMed] [Google Scholar]
- Rubio-Godoy V, Dutoit V, Rimoldi D, Lienard D, Lejeune F, Speiser D, Guillaume P, Cerottini JC, Romero P, Valmori D. Discrepancy between ELISPOT IFN-gamma secretion and binding of A2/peptide multimers to TCR reveals interclonal dissociation of CTL effector function from TCR-peptide/MHC complexes half-life. Proc Natl Acad Sci U S A. 2001;98:10302–10307. doi: 10.1073/pnas.181348898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Society-USA International AIDS. Update on Drug Resistance Mutations - Special contribution. Topics HIV Med. 2001;9:21–23. [Google Scholar]
- Martinez-Picado J, DePasquale MP, Kartsonis N, Hanna GJ, Wong J, Finzi D, Rosenberg E, Gunthard HF, Sutton L, Savara A, Petropoulos CJ, Hellmann N, Walker BD, Richman DD, Siliciano R, D'Aquila RT. Antiretroviral resistance during successful therapy of HIV type 1 infection. Proc Natl Acad Sci U S A. 2000;97:10948–10953. doi: 10.1073/pnas.97.20.10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MS, McClure MA, Feng DF, Gray J, Doolittle RF. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986;83:7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere Y, McChesney MB, Porrot F, Tanneau-Salvadori F, Sansonetti P, Lopez O, Pialoux G, Feuillie V, Mollereau M, Chamaret S, et al. Gag-specific cytotoxic responses to HIV type 1 are associated with a decreased risk of progression to AIDS-related complex or AIDS. AIDS Res Hum Retroviruses. 1995;11:903–907. doi: 10.1089/aid.1995.11.903. [DOI] [PubMed] [Google Scholar]
- Yewdell JW, Norbury CC, Bennink JR. Mechanisms of exogenous antigen presentation by MHC class I molecules in vitro and in vivo: implications for generating CD8+ T cell responses to infectious agents, tumors, transplants, and vaccines. Adv Immunol. 1999;73:1–77. doi: 10.1016/s0065-2776(08)60785-3. [DOI] [PubMed] [Google Scholar]
- Yu XG, Addo MM, Rosenberg ES, Rodriguez WR, Lee PK, Fitzpatrick CA, Johnston MN, Strick D, Goulder PJ, Walker BD, Altfeld M. Consistent Patterns in the Development and Immunodominance of Human Immunodeficiency Virus Type 1 (HIV-1)-Specific CD8(+) T-Cell Responses following Acute HIV-1 Infection. J Virol. 2002;76:8690–8701. doi: 10.1128/JVI.76.17.8690-8701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TM, O'Connor DH, Jing P, Dzuris JL, Mothe BR, Vogel TU, Dunphy E, Liebl ME, Emerson C, Wilson N, Kunstman KJ, Wang X, Allison DB, Hughes AL, Desrosiers RC, Altman JD, Wolinsky SM, Sette A, Watkins DI. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35036559. [DOI] [PubMed] [Google Scholar]
- Barouch DH, Kunstman J, Kuroda MJ, Schmitz JE, Santra S, Peyerl FW, Krivulka GR, Beaudry K, Lifton MA, Gorgone DA, Montefiori DC, Lewis MG, Wolinsky SM, Letvin NL. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature. 2002;415:335–339. doi: 10.1038/415335a. [DOI] [PubMed] [Google Scholar]
- O'Connor DH, Allen TM, Vogel TU, Jing P, DeSouza IP, Dodds E, Dunphy EJ, Melsaether C, Mothe B, Yamamoto H, Horton H, Wilson N, Hughes AL, Watkins DI. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat Med. 2002;8:493–499. doi: 10.1038/nm0502-493. [DOI] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Wei X, Horwitz MS, Peffer N, Meyers H, Nelson JA, Gairin JE, Hahn BH, Oldstone MB, Shaw GM. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- Doukhan L, Delwart E. Population genetic analysis of the protease locus of human immunodeficiency virus type 1 quasispecies undergoing drug selection, using a denaturing gradient-heteroduplex tracking assay. J Virol. 2001;75:6729–6736. doi: 10.1128/JVI.75.14.6729-6736.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhuis M, Boucher CA, Schipper P, Leitner T, Schuurman R, Albert J. Stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy. Proc Natl Acad Sci U S A. 1998;95:14441–14446. doi: 10.1073/pnas.95.24.14441. [DOI] [PMC free article] [PubMed] [Google Scholar]