Abstract
Background
Carbohydrates are involved in a variety of fundamental biological processes and pathological situations. They therefore have a large pharmaceutical and diagnostic potential. Knowledge of the 3D structure of glycans is a prerequisite for a complete understanding of their biological functions. The largest source of biomolecular 3D structures is the Protein Data Bank. However, about 30% of all 1663 PDB entries (version September 2003) containing carbohydrates comprise errors in glycan description. Unfortunately, no software is currently available which aligns the 3D information with the reported assignments. It is the aim of this work to fill this gap.
Results
The pdb-care program http://www.glycosciences.de/tools/pdb-care/ is able to identify and assign carbohydrate structures using only atom types and their 3D atom coordinates given in PDB-files. Looking up a translation table where systematic names and the respective PDB residue codes are listed, both assignments are compared and inconsistencies are reported. Additionally, the reliability of reported and calculated connectivities for molecules listed within the HETATOM records is checked and unusual values are reported.
Conclusion
Frequent use of pdb-care will help to improve the quality of carbohydrate data contained in the PDB. Automatic assignment of carbohydrate structures contained in PDB entries will enable the cross-linking of glycobiology resources with genomic and proteomic data collections.
Background
Carbohydrates are involved in a variety of fundamental biological processes (cellular differentiation, embryonic development, fertilization) and pathological situations (bacterial and viral infections, inflammatory diseases, cancer) [1-3]. They therefore have a large pharmaceutical and diagnostic potential. Protein-carbohydrate interactions are intensively investigated using a variety of experimental methods. Among these, X-ray and NMR measurements provide a detailed 3D picture of the spatial location of the ligand as well as the protein.
About 70% of all proteins deposited in sequence databases show potential N-glycosylation sites, which can be identified by the presence of the Asn-X-Ser/Thr sequon [4,5]. For reasons that are still unclear, not all such sequons are glycosylated. It is estimated that more than half of all the proteins in the human body have carbohydrate molecules attached (see Fig. 1a). Unfortunately, extensive analytical procedures are required to determine which of these potential sites are occupied, even if the protein in question is known to be glycosylated. Due to several reasons, the absence of carbohydrate data in 3D structural data taken from X-ray crystallography does not necessarily mean that a potential glycosylation site is unoccupied; but the presence of carbohydrate residues in the 3D structures provides direct and unambiguous evidence for the occupancy of a glycosylation site. Therefore, these data have been intensively used to gain deeper insights into factors that regulate glycan attachment to a N-glycosylation site [6].
Figure 1.
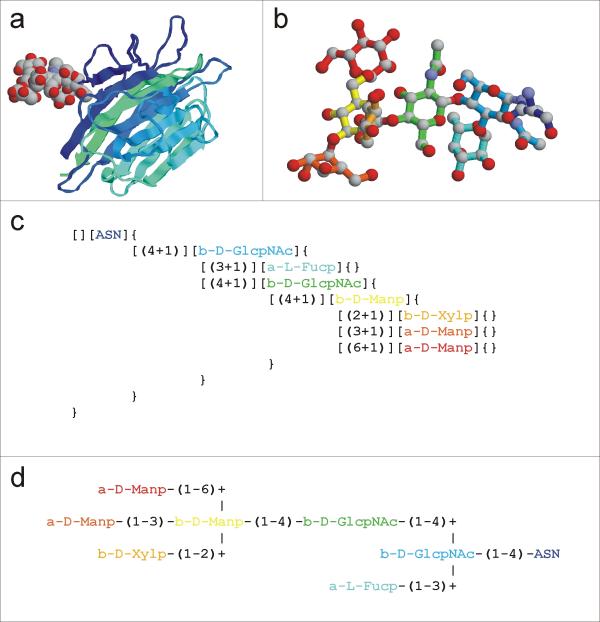
Description of carbohydrate structures (a) Typical PDB-entry (PDB-code: 1axy) with an attached N-Glycan given in spacefilling representation. (b) Ball-and-Stick 3D-representation of the same N-Glycan. For a simple comparison of the different representations, the same colour code for each residue is used for Figure 1b-d. (c) LINUCS representation of the same N-Glycan structure. (d) IUPAC-like description of the same N-Glycan.
Among the 25667 PDB entries (May 2004) [7], 1995 structures were detected which contain a total of 6714 carbohydrate chains. About half of them are covalently attached, the other half belongs to non-covalently bound ligands. In a recent study, however, it was found that about 30% of all PDB entries containing carbohydrates comprise one or several errors in glycan description, which are mainly due to wrong assignment of saccharide units [8]. The reason for this unacceptable high rate of errors is obvious. Sequences for complex carbohydrates differ significantly from the simple linear one-letter code used to describe genes and proteins: a) the number of naturally occurring residues is much larger for glycans, b) each pair of monosaccharide residues can be linked in several ways, and c) a residue can be connected to three or four others (branching). Unfortunately, no simple representation of complex carbohydrates does exist that is accepted by scientists from all disciplines such as the one-letter code of amino acid sequences for proteins. Sugar resides present in the PDB are defined in the so-called HET Group Dictionary [9]. Since a three-letter code is used to uniquely assign carbohydrates, a new residue name is required for each stereochemically different sugar unit and each substitution. This procedure makes correct assignment of sugar units tedious, complicated and obviously error-prone. Unfortunately, no software is currently available which automatically aligns the 3D information contained in PDB and the given assignments. It is the focus of pdb-care [10] to fill this gap.
Implementation
The pdb-care [10] program is based on the carbohydrate detection software pdb2linucs [8] that is able to identify and assign carbohydrate structures using only the reported atom types and their 3D atom coordinates. The LINUCS-notation [11], which is closely related to the IUPAC nomenclature recommendation for carbohydrates [12], is used to normalise complex carbohydrate structures (Fig. 1c and 1d). To be able to compare the detected carbohydrate structures in LINUCS notation to the residue assignments as reported in the PDB HET Group Dictionary [9] a translation table in XML format was created, where both descriptions are confronted. Three types of residues are included: monosaccharides, oligosaccharides and combined residues consisting of a carbohydrate moiety and a non-carbohydrate part, e.g. BOG standing for octyl-β-D-Glcp (see table 1 for definitions of PDB residue names mentioned in this article). The translation table actually contains 141 monosaccharides, 31 oligosaccharides and 77 combined residues and is still growing. The pdb-care [10] protocol reports the type of problems, inconsistencies and errors detected. The messages are classified as info, describing the type of checks performed, warnings when non-resolvable discrepancies are detected and errors when obviously wrong assignments have been found.
Table 1.
PDB carbohydrate residues (examples): Definitions of carbohydrate residues used in the Protein Data Bank. PDB residue names are defined using a three-letter encoding. There are more than 200 different carbohydrate residue names used in PDB entries. This table lists those mentioned in this article and some further examples in alphabetic order.
| Name | Definition |
| AFL | alpha-L-Fucose |
| AGC | alpha-D-Glucopyranose |
| BGC | beta-D-Glucopyranose |
| BOG | octyl-beta-D-Glucopyranose |
| FCA | alpha-D-Fucose |
| FCB | beta-D-Fucose |
| FMF | 2-deoxy-2-fluoro alpha-D-Mannopyranose |
| FUC | Fucose |
| FUL | beta-L-Fucose |
| G4S | D-Galactose-4-sulphate |
| GAL | D-Galactopyranose |
| GLA | alpha-D-Galactopyranose |
| GLB | beta-D-Galactopyranose |
| GLC | D-Glucopyranose |
| GLS | beta-D-Glucopyranose spirohydantoin [also used for D-Galactopyranose-6-sulphate] |
| GSA | D-Galactose-4-sulphate |
| LAK | Allolactose [b-D-Galp-(1-6)-b-D-Glcp] |
| LAT | Lactose [b-D-Galp-(1-4)-b-D-Glcp] |
| MAF | 2-deoxy-2-fluoro alpha-D-Mannopyranose |
| MAL | Maltose [a-D-Glcp-(1-4)-a-D-Glcp] |
| NAG | N-acetyl D-Glucosamin |
| NAN | 5-N-acetyl alpha-D-Neuraminic Acid |
| NGA | N-acetyl D-Galactosamin |
| SIA | 5-N-acetyl D-Neuraminic Acid (Sialic acid) |
| SLB | 5-N-acetyl beta-D-Neuraminic Acid |
pdb-care is written in C. Interaction with the user is done through a web-interface, which is implemented in PHP. The pdb-care [10] service is hosted at the central spectroscopic department of the German Cancer Research Centre in Heidelberg, Germany.
Results and discussion
The pdb-care [10] web interface allows either to analyse a file obtained directly from PDB using the PDB-ID, or to provide a pdb-file located on the local computer by upload or by copy / paste into the provided input window. Since carbohydrate structures are described as so-called hetero atoms within the HETATM records, all data assigned to the ATOM records (amino acids, nucleotides) are neglected.
Connectivity check
The examination of information given in the CONECT records of the pdb-files comprises two types of checks, which can be separately activated by the user. The first check analyses the reported connections. If a bond length differs for more than the user-definable tolerance from the normal length or if the number of connections for an atom exceeds the maximally allowed number for the respective element, a warning is displayed (Fig. 2a and 2b).
Figure 2.
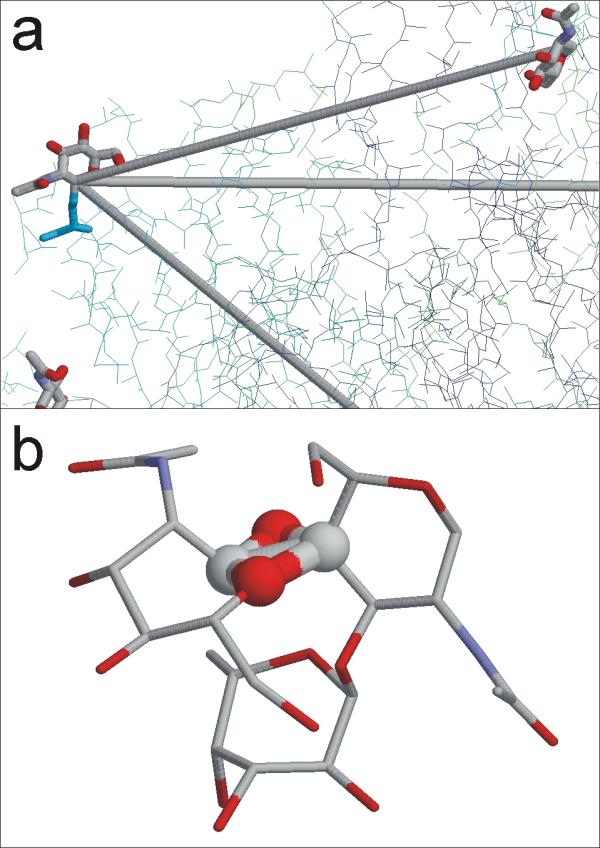
Erroneous connections in PDB entries. Besides missing connection information, some entries contain surplus connections. (a) In case the wrongly connected atoms are far distant from each other, these errors can be observed on the first view (PDB entry 1qoo, residue NAG401A). In this example, the spuriously assigned connections result in a hexavalent carbon atom. (b) Surplus connections ranging on short distances are much more difficult to discover by visual inspection (PDB entry 1bcs, residues NAG1051, NAG1052).
The second check analyses the reported data for completeness. For each hetero-atom its distances to all other atoms are determined. In case this distance is within the usual bond length range, and the connection is not listed in the CONECT records, a warning is displayed. In some pdb-files, there are overlapping residues, e.g. due to the fact that the crystal was soaked with both the alpha- and the beta-anomer of a ligand, like the residues FCA307 and FCB308 of PDB entry 1abf. In this case, the determination of bonds by atom distances may generate wrong positive warnings. This may also happen when there are atom pairs in the structure that lie close together but are not covalently bound. Therefore, bond length inconsistencies are reported as warnings and not as errors. It is up to the user to finally decide if the reported problems are in fact owing to incorrect information within the CONECT records.
Since the coordination of ions depends on various electronic factors, pdb-care can be configured in such a way that all bonds to ions are ignored. Both connectivity checks analyse the entire data given in the HETATM records and are therefore not limited to carbohydrate residues.
Nomenclature check
Mismatches between residue nomenclature and determined structure are the most common type of errors found within carbohydrate residues in the PDB [8]. This type of error probably results from the facts that the number of available carbohydrate residues by far exceeds the number of amino acids or nucleotides. Additionally, the individual residues often differ from each other only through the orientation of hydroxyl groups attached to ring carbons (Fig. 1b). These small structural differences make it difficult to correctly assign the stereochemistry of sugar units.
pdb-care generates carbohydrate nomenclature based only on the given atom types and their 3D atom coordinates using the pdb2linucs algorithm [8].
Subsequently, looking up the translation table where systematic names and the respective PDB residue codes are listed, both independently derived assignments are compared. In case they do not coincide, an error message is reported. The correct PDB residue code corresponding to the detected monosaccharide structure will be provided on demand. This option facilitates subsequent corrections of assignments.
Residue nomenclature in pdb-files is complicated by the fact that there is a large amount of ambiguities and redundancies. On the one hand there are many residues that stand for both the alpha- and the beta-anomer of one monosaccharide type, e.g. GAL for D-Galactose or GLC for Glucose. In an older version of the PDB HET Group Dictionary (March 2002), GLC was defined as 'alpha-D-Glucose', but since there was no residue name for the beta-anomer defined, it was often used for beta-D-Glucose residues as well. In the actual HET Group Dictionary, there are two further residues specifying the two Glucose anomers: AGC for α-D-Glcp and BGC for β-D-Glcp. Some residue names are even used for two entirely different residues. GLS, for example, is defined as 'beta-D-Glucopyranose spirohydantoin', but in entry 1kes, it was defined as 'D-Galactose-6-sulphate'.
On the other hand, there are several residues for which more than one PDB residue name exists. 'D-Galactose-4-sulphate', e.g., is encoded by both GSA and G4S; and MAF as well as FMF encode for '2-deoxy-2-fluoro-alpha-D-Mannose'.
To reduce the amount of ambiguously defined residues, pdb-care offers an option to suggest unique residue names – provided they are available – when ambiguous ones are reported in the PDB entry.
As already discussed for the interpretation of conflicting connectivity information, again it is up to the experimentalist to judge if inconsistencies in nomenclature are caused just by selecting a wrong PDB residue name or if they are due to erroneous interpretation of the electron density maps. This decision can hardly be solved automatically by a software.
The experimentalist has to decide if, for example, a residue that is detected as 'D-GlcpNAc' (PDB residue code NAG), but is named as NGA (defined as 'D-GalpNAc') is in fact 'D-GlcpNAc' and was just wrongly named, or as said in the PDB residue code is a NGA, which would mean that there is a problem with the structure.
Linkage information
In most pdb-files, information on how the single monosaccharides are linked to each other, to the proteins or to further, non-carbohydrate residues is entirely missing or present in a non-standardised way within the REMARK sections. Therefore, pdb-care verifies linkages only for PDB residues encoding for oligosaccharides, where linkage data is given implicitly within the residue definition. However, the pdb2linucs algorithm is able to generate a complete structural description of the complete glycan, which can be easily transformed to the IUPAC nomenclature (Fig. 1c,1d).
Conclusions
The currently available check software for molecular 3D structures in PDB like WhatCheck [13] or ProCheck [14,15] is focussed on the protein part of pdb-files. Intensive use of these programs has led to a high quality of the annotation of protein structures deposited in the PDB. The lack of a corresponding software for carbohydrate residues results in a high rate of assignment errors for this part of PDB information. It can be anticipated, that frequent use of pdb-care will improve the quality of carbohydrate data contained in the PDB. This enhancement of the glyco-related information will make it more reliable for the realm of glycobiology.
The automatic assignment of carbohydrate structures contained in PDB entries will improve the cross-linking of glycobiology resources with genomic and proteomic data collections, which will be an important issue of the upcoming glycomics projects. Due to the current high rate of errors in the carbohydrate parts of PDB structures, however, it is not possible to extract this data from pdb-files and include it into carbohydrate databases without any quality control. The existence of a check program makes it feasible to update GlycosciencesDB – the former SweetDB [16] – automatically with data obtained from PDB in case there are no inconsistencies detected. For entries that are not included automatically, the software aids the database administrator in judging if a structure should be accepted or not.
To further improve the quality as well as the accessibility of glyco-related data contained in PDB entries, a complete structural description including stereochemistry as well as linkage information of a glycan should be reported. With pdb-care and pdb2linucs two software tools are now available which produce such a description automatically based on the data already contained in PDB entries.
Availability and requirements
• Project name: PDB CArbohydrate REsidue check (pdb-care)
• Project home page: http://www.glycosciences.de/tools/pdb-care/
• Operating systems: Platform independent
• Programming language: C, PHP
• Other requirements: none
• Any restrictions to use by non-academics: none
Abbreviations
PDB: Protein Data Bank
XML: extensible markup language
PHP: PHP hypertext preprocessor
Authors' contributions
TL was responsible for the software design and implementation. CWvdL contributed to aspects of design and oversaw and managed contributions to the project. Both authors contributed equally drafting the manuscript and read and approved the final manuscript.
Acknowledgments
Acknowledgements
The development of 'pdb-care' is funded by a grant from the German Research Council (Deutsche Forschungsgemeinschaft: DFG) within the digital library program.
Contributor Information
Thomas Lütteke, Email: t.luetteke@dkfz-heidelberg.de.
Claus-W von der Lieth, Email: w.vonderlieth@dkfz-heidelberg.de.
References
- Rademacher TW, Parekh RB, Dwek RA. Glycobiology. Annu Rev Biochem. 1998;57:785–838. doi: 10.1146/annurev.bi.57.070188.004033. [DOI] [PubMed] [Google Scholar]
- Taylor ME, Drickamer K. Introduction to Glycobiology. Oxford University Press; 2002. p. 224. [Google Scholar]
- Varki A, Esko J, Freeze H, G. Hart, J. Marth. Essential of Glycobiology. New York, Cold Spring Habor Laboratory Press; 1999. [PubMed] [Google Scholar]
- Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/S0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- Ben-Dor S, Esterman N, Rubin E, Sharon N. Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology. 2004;14:95–101. doi: 10.1093/glycob/cwh004. [DOI] [PubMed] [Google Scholar]
- Petrescu AJ, Milac AL, Petrescu SM, Dwek RA, Wormald MR. Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology. 2004;14:103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütteke T, Frank M, von der Lieth C-W. Data mining the protein data bank: automatic detection and assignment of carbohydrate structures. Carbohydr Res. 2004;339:1010–1020. doi: 10.1016/j.carres.2003.09.038. [DOI] [PubMed] [Google Scholar]
- PDB HET Group Dictionary http://deposit.pdb.org/het_dictionary.txt
- PDB-Care Web interface http://www.glycosciences.de/tools/pdb-care/
- Bohne-Lang A, Lang E, Forster T, von der Lieth CW. LINUCS: linear notation for unique description of carbohydrate sequences. Carbohydr Res. 2001;336:1–11. doi: 10.1016/S0008-6215(01)00230-0. [DOI] [PubMed] [Google Scholar]
- McNaught AD. Nomenclature of carbohydrates (recommendations 1996) Adv Carbohydr Chem Biochem. 1997;52:43–177. [PubMed] [Google Scholar]
- Hooft RWW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272–272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- Loss A, Bunsmann P, Bohne A, Schwarzer E, Lang E, von der Lieth CW. SWEET-DB: an attempt to create annotated data collections for carbohydrates. Nucleic Acids Res. 2002;30:405–408. doi: 10.1093/nar/30.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]


