RNA beacon technology is a promising method to detect and separate cells expressing a particular gene, but developing a successful, specific beacon can take months and in some cases is impossible. This study reports an off-the-shelf universal beacon that decreases the time and cost of applying beacon technology to select any living cell population transfected with an exogenous gene.
Keywords: RNA beacons, Stem cell separation, Stem cell selection, Fluorescence-activated cell sorting analysis, Gene expression
Abstract
Stem cell therapy requires a nontoxic and high-throughput method to achieve a pure cell population to prevent teratomas that can occur if even one cell in the implant has not been transformed. A promising method to detect and separate cells expressing a particular gene is RNA beacon technology. However, developing a successful, specific beacon to a particular transfected gene can take months to develop and in some cases is impossible. Here, we report on an off-the-shelf universal beacon that decreases the time and cost of applying beacon technology to select any living cell population transfected with an exogenous gene.
Significance
Stem cell therapy requires pure cell populations. RNA beacons have a high potential to select and separate differentiated cells from undifferentiated cells. However, development of a beacon for a specific gene can be difficult and time-consuming. An RNA tag was developed that can be attached to any gene without affecting the protein product. The tagged gene is detected by an off-the-shelf RNA beacon that can be used for cell selection.
Introduction
Although stem cell therapies offer promise to alleviate many human diseases, they require an exceedingly pure population because teratomas may result from as little as one remaining pluripotent cell in the implanted tissue. Presently, identifying and removing a very small population of cells from tissue is a major roadblock in stem cell therapy. Whereas some cells can be detected and separated by extracellular epitopes of specific biomarkers, such markers do not exist for intracellular proteins such as the transcription factors used for in stem cell transformation. RNA beacons have the potential to detect mRNAs that code for any protein, despite their ultimate localization. RNA beacons can be used alone without the use of a reporter or other accessory molecules. Importantly, RNA beacons can be used to separate cell populations in a high-throughput manner based on the mRNA for any gene of interest.
RNA beacons are stem-loop structures [1–3] with a fluorescent dye and quencher attached to the 5′ and 3′ ends of a base-paired stem. The ∼10 nucleotides (nt) in the loop are complementary to a target mRNA so that hybridization with mRNA opens the stem, resulting in a large increase in fluorescence. The nature of the fluorescence pair is variable, and so RNA beacons can be made in many colors that can be optimized for detection by flow cytometry and separation by fluorescence-activated cell sorting (FACS).
Although RNA beacons can be developed to detect genes associated with pluripotency (e.g., Oct4, nanog, Sox2) [1, 4], many stem cell applications create induced pluripotent stem cells and induce them to differentiate toward specific lineages [5] by introducing specific genes (often transcription factors). Current selection methods require a distinctive surface marker that can detect target cells using antibodies or require introduction of a reporter gene such as green fluorescent protein (GFP). The clear advantage of RNA beacons is that they allow for the selection of live cells independent of a reporter gene and without the requirement of a surface marker. This technology may be valuable for applications in which the stem cell is used as a suitcase to deliver a therapeutic gene, such as for biological pacemakers in which human mesenchymal stem cells (hMSCs) are transfected with an HCN gene that underlies the pacemaker current (IF) in cardiac cells [6–8]. Promising studies of this technology were recently reported by Ban et al. [9] who used five molecular beacons to separate cardiomyocytes to 99% purity. The implanted tissue did not show tumors for up to 4 weeks [10].
Most stem cell applications introduce a gene of interest to promote a cell line with the desired properties. Detection of these cells using RNA beacons would be simpler if a tested, off-the-shelf beacon were available, because developing a beacon for a specific gene requires finding an accessible and specific target on the mRNA to allow the beacon to bind, and constructing a beacon with a hybridization energy high enough to unwind the stem and allow opening but low enough so that the beacon does not open prematurely. This procedure can be time-consuming and for some genes is impossible.
Here, we report on the use of a universal beacon (UB) that targets a nonsense sequence that can be placed in an untranslated region of any gene of interest and allows detection of cells expressing a specific exogenous gene. We present a side-by-side comparison of UB with a beacon that specifically targets the stem cell marker, nanog, and then with a gene that encodes for human HCN2 channels used to develop biological pacemakers. We find the UB is a safe and nontoxic sensor of genes coding for intracellular proteins that preserves the function and localization of the protein product and allows for high-throughput separation of pure cell populations.
Materials and Methods
Cell Culture and Isolation
hMSCs, human normal adult dermal fibroblasts (NHDF), and human aortic adventitial fibroblast (AOAF) were purchased from Lonza Group Ltd. (Basel, Switzerland) and cultured in MSCGM BulletKit (Lonza), FGM BulletKit (Lonza), and SCGM BulletKit (Lonza), respectively. In all cases, passages 2–5 were used. HEK 293 cells, macrophage cells, and Chinese hamster ovary (CHO) cells were purchased from ATCC. HEK293 and macrophage cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; catalog no. 11965; Gibco, Grand Island, NY, http://www.invitrogen.com) supplemented with 10% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin sulfate. Fischer rat thyroid (FRT) cells were a gift from Dr. Deborah Brown (Stony Brook University). FRT and CHO cells were cultured in Ham’s F12 medium with 10% fetal bovine serum, 50 units/ml penicillin, and 50 μg/ml streptomycin sulfates. All cells were maintained at 37°C in a humidified atmosphere of 5% CO2.
Canine mesenchymal stem cells (cMSCs) were isolated from 1-year-old dogs according the protocol [8]. Briefly, cMSCs were isolated by Ficoll-Paque Plus density gradients centrifugation from aspirated bone marrow. Primary cultures of cMSCs were maintained at 37°C in 5% CO2, 95% air with an initial medium for 48 hours. The medium was changed every 3–4 days. Cell colonies with spindle-like morphology were transferred 7 days after initial plating. After confluence, cells were harvested with 0.25% trypsin-EDTA and replaced. Isolated cells were characterized at passages 2–4 by flow cytometric analysis of specific surface antigens with fluorescein isothiocyanate (FITC)-conjugated rat anti-canine CD44, FITC-conjugated rat anti-canine CD45 unconjugated rat anti-canine CD90, and phycoerythrin (PE)-conjugated mouse anti-canine CD34. A large majority of the cells were CD44-positive (99.61%) and CD90-positive (93%) and CD34-negative or CD45-negative (98%), suggesting that a significant majority were MSCs.
To further validate the cMSC properties of the isolated cells, we subjected subsets of the cells to osteogenic, adipogenic, and chondrogenic differentiation protocols. For adipogenic and osteogenic differentiation, the cells were plated in 6-well or 12-well plates. Adipogenic and osteogenic induction was initiated using kits available from Lonza. For adipogenesis, three to five cycles of the following medium changes were performed: 2–3 days of exposure to adipogenic induction medium followed by 2–3 days of exposure to maintenance medium. Osteogenic induction was carried by feeding the cells with osteogenic induction medium every 3–4 days for 2–3 weeks. Chondrogenic induction was performed by pelleting 2.5 × 105 cells in chondrogenic induction medium containing transforming growth factor β3. Complete medium changes were performed every 2–3 days for 3–4 weeks. At the end of the induction protocols, the cells were rinsed with phosphate-buffered saline (PBS) and fixed with 10% formalin. Adipogenesis was assayed using Oil Red O staining. Osteogenesis was assayed by staining for calcium deposition using alizarin red staining. Chondrogenic pellets were embedded in cryogenic cutting medium and sectioned for histology, and glycosaminoglycans were stained by Safranin O.
Universal Beacon Design
A universal beacon sequence was prepared based on sequences that do not match any human mRNA transcript and genomic DNA sequence as described in the text. These “nonsense” sequences were identified using the Bioinformatics Sequence Manipulation Suite [11] and were checked by the NIH BLAST program [12] to determine whether they matched any human transcript mRNA and genomic DNA sequences.
We took the most promising nonsense sequence and attached CCACC and GGtGG on the 5′ and 3′ ends to form the stems and enclose the candidate sequence. We then attached the fluorophore 6-Carboxyfluorescein (6-FAM) and its corresponding quencher BHQ1 to the 5′ and 3′ ends. The fluorophore Tide Fluor 5 (TF5) (ATT Bioquest, Sunnyvale, CA, http://aatbio.com/prods.html) was inserted at the end of the nonsense sequence and at the start of a stem, which will be used as beacon delivery indicator (e.g., 5′-6-FAM-ccaccCGTACGCGTCGGAGATTF5gggtgg-BHQ1-3′). The beacon was synthesized by Eurofins MWG Operon (Alameda, CA, http://www.operon.com).
Introducing the Nonsense Sequence Into nanog and hHCN2
We placed the nonsense sequence at the 5′-untranslated region (UTR) of the nanog and hHCN2 genes to avoid any influence on mRNA transcription or protein function, the sequence located at 5′-UTR. We verified that the insertion site does not disrupt regulatory regions such as CAAT, CCAAT, TATA boxes, etc. Briefly, the antisense nonsense sequence (5′-CTAGCATCTCCGACGCGTACGGC-3′) was inserted to Nanog/pcDAN3.1 before the start codon by NheI and NotI restriction enzyme sites (named N5JR1). We introduced the sequence into hHCN2/pcDNA3.1 before or after the stop codon and avoided potential small interfering RNA (siRNA) binding sites using the RNAi Consortium Public TRC Portal [13]. The nonsense sequence (AGCTTATCTCCGACGCGTACGG) was inserted to hHCN2/pcDNA3 by HindIII and BamHI restriction enzymes sits before start codon (named H5JR1) or after the stop codon of hHCN2/pcDNA 3.1 XhoI restriction enzyme site (TCGAGATCTCCGACGCGTACGC, named H3JR1). The nonsense sequence was also doubly inserted to hHCN2/pcDNA3 by HindIII/BamHI and XhoI (named H5, 3JR1).
The nanog or hHCN2 plasmids were transfected into HMSCs or NHDF cell by electroporation according to the protocol of Lonza (Walkersville, MD, http://www.lonza.com); 2 μg of plasmid DNA was transfected to 4–5 × 105 cells by Nucleofector program C-17. Ten micrograms of plasmid DNA was transfected into HEK 293 cells by calcium phosphate coprecipitation. Usually, universal beacon selection was done after 48 hours of transfection.
Delivering Beacons Into Cells
Microinjection
Cells were cultured in glass-bottomed MatTek wells for 24 hours. Before microinjection, the medium was changed to phenol-free Leibovitz-15. (For primary cardiomyocytes, the medium was first changed to phenol-free Leibovitz-15 with 14 mM EGTA.) The beacon concentration in the needle was 50 μM. The control cells were injected with the dye tracer alone. Injection was carried out using self-pulled needles using an InjectMan NI2 with a FemtoJet pump from Eppendorf mounted on Axiovert 200 M (Carl Zeiss) equipped with a long working distance ×40 phase 2 objective. Solutions were microinjected into the cytoplasm. We typically set the injection pressure (Pi) at 40 hPa, and kept the compensation pressure Pc at 20 hPa (for all cells expect cardiomyocytes. For cardiomyocytes, the injection pressure was 90 hPa, and the compensation pressure was kept at 45 hPa. The injection time was t = 0.7 s. Typically, we injected ∼10–25 cells within a 10- to 20-minute period. We examined the microinjected cells under the phase microscope to select viable cells.
Lipofectamine
Beacon transfections were carried out using Lipofectamine (Invitrogen) according to the manufacturer’s instructions. A final concentration of 25 nM beacon was used for 0.5 × 106 cells in suspension in serum-free medium. Cells were incubated at 37°C with 5% CO2 before performing fluorescence microscopy imaging, flow cytometric analysis, or fluorescence-activated cell sorting.
Calcium Phosphate Precipitation
Starting with ∼50% confluent cells in a 100-mm dish, the cells were fed 7 ml of fresh medium. The reagent (4–10 μg) was incubated with 120 μM CaCl2 and HEPES buffer containing NaCl, NaHPO4, and HEPES, pH 7.1, for 10 minutes on ice. The mixture was immediately added to the surface of the cultured cells, swirled gently, and returned to the incubator at 37°C with 5% CO2 until needed.
FuGENE 6
Transfections were according to the manufacturer’s instructions (Roche Diagnostics, Basel, Switzerland, http://www.roche-applied-science.com). Briefly, cells were grown to 50–80% confluent in 35-mm culture dishes. FuGENE 6 transfection reagent was added in a 3:1 serum-free medium. After 3–8 hours, serum was added to the medium, or the medium was changed to one containing serum.
Electroporation
Beacons were introduced into cells by electroporation using a protocol adapted from Lonza cell culture. Briefly, the medium was aspirated, and cells were harvested by adding 1 ml of trypsin/EDTA. Cells were then spun down for 5 minutes at 1,500g, resuspended with 2–4 μg of beacon, and mixed. For electroporation of hMSCs, beacons were mixed with 100 μl of hMSC nucleofector obtained from Lonza. The mixture was transferred into a electroporation cuvette by a 200-μl pipette being careful to avoid generating air bubbles. Electroporation was carried out in a Bio-Rad (Hercules, CA, http://www.bio-rad.com) Xcell electroporation instrument using the programs C-17 and U-23.
Confocal Imaging
Images of fluorescent cells were collected 1 hour after microinjection on an Olympus (Center Valley, PA, http://www.olympusamerica.com) Fluoview 1000 confocal microscope equipped with a ×40 1.4 numerical aperture oil immersion objective. The tracer fluorophore TF5 was excited by the 633-nm line of a He:Ne laser, and images were recorded using a bandpass 650–750-nm emission filter. The fluorophore 6-FAM was excited with the 488-nm argon-ion laser line, and images were recorded using a bandpass 505–605-nm emission filter. The two fluorophores were excited in a sequential manner using Multi Track acquisition. This procedure minimizes channel crosstalk.
Western Blotting
Cell lysates from HEK293 cells-HEK 293 cells transfected with wild-type (wt) nanog, and HEK293 cells transfected with the nanog gene also containing the complementary universal beacon sequence (UB-nanog) were subjected to gel electrophoresis followed by the transfer of the samples to a nitrocellulose membrane. The membrane was incubated for 1 hour with 1% bovine serum albumin to block nonspecific binding. nanog protein was detected by immunoblotting with mouse anti-nanog (Santa Cruz Biotechnology, Santa Cruz, CA, http://www.scbt.com) antibody. The primary antibody was diluted according to the manufacturer's instructions in Tris-buffered saline with 0.5% Tween, added to the membrane, and incubated for 1 hour. This procedure was followed by three 15-minute washes in 25 mM Tris, 150 mM NaCl, 0.2% Tween 20, and pH 7.6 (TBST). Afterward, horseradish peroxidase-conjugated secondary antibody was added and incubated for 1 hour. The membrane was washed 3 times for 15 minutes in TBST and exposed to a photographic film after a chemiluminescence reaction. The housekeeping protein Hsp90 was detected on the same membrane as a loading control.
Flow Cytometry Analysis and FACS
Cells were dissociated with trypsin-EDTA solution (Lonza). Twenty-five nM UB was delivered into 0.5 × 106 cells in suspension and incubated for 1 hour in an incubator at 37°C with 5% CO2. Cells were washed twice with PBS to remove the extracellular beacon, and the cells were suspended in 0.5 ml of DMEM (GIBCO). Negative control samples included wild type cells transfected with UB and positive controls cells stably transfected with the UB tag gene.
FACS experiments were carried out on a BD Biosciences (San Diego, CA, http://www.bdbiosciences.com) FACSAria at the Stony Brook University flow cytometry facility. Cells were sorted using 3–5 × 106 cells per milliliter in serum-free medium for which the sorting pressure was 40 ψ, and the nozzle diameter was 85 μm. The sorting rate was set at 1,600 events/second. Detection filters used for the 488-nm laser excitation were FITC (530-nm peak, 30-nm half-width) and PE (575-nm peak, 26-nm half-width). For the 633-nm laser excitation, the detection filter was APC (660-nm peak, 20-nm half-width).
Patch-Clamp Studies of Pacemaker Current in cMSCs
We used whole-cell patch clamp to study membrane currents in wild-type cMSCs and those transfected with hHCN2-UB (cMSCs hHCN2-UB). The IF values of expressed hHCN2 channels were measured under voltage clamp by an Axopatch-1B (Axon Instruments, Union City, CA, http://www.moleculardevices.com) amplifier. Patch electrode resistance was 4–6 MΩ before sealing. The cells were constantly superfused using a gravitational perfusion system with a complete change of the chamber solutions in approximately 0.5 minutes. The temperature of the bath as well as of the perfusion solution was kept constant at 22 ± 1°C. The pipette solution was filled with 50 mmol/l KCl, 80 mmol/l potassium aspartate, 1 mmol/l MgCl2, 3 mmol/l Mg-ATP, 10 mmol/l EGTA, and 10 mmol/l HEPES (pH adjusted to 7.2 with KOH). The external solution contained 137.7 mmol/l NaCl, 5.4 mmol/l KCl, 2.3 mmol/l NaOH, 1.8 mmol/l CaCl2, 1 mmol/l MgCl2, 10 mmol/l glucose, 5 mmol/l HEPES, and 2–5 mM BaCl2, for which the pH was adjusted to 7.4 with NaOH. To measure the IF activation curve, a standard two-step protocol was used. Hyperpolarizing steps from −40 to −130 mV at 10-mV increments were applied from a holding potential of −40 mV, followed by a depolarizing step to +50 mV to measure the tail current. For each cell, the normalized plot of tail current of IF versus test voltage was fit with a Boltzmann function, and then the voltage of half-maximum activation (Vh) and slope factor (K) were defined from the fitting. No detectable IF was expressed in the control cells (data not shown). Robust expression of IF was detected in cMSCs transfected with hHCN2-UB with a Vh at 79.0 ± 0.3 mV and a K at 6.7 ± 0.3 mV (n = 6 cells), respectively [6, 10].
Results
Preparing a Successful Beacon
Before describing UB technology, we illustrate the procedure used to develop and optimize a beacon to detect the mRNA for an intracellular protein. We choose the intracellular stem cell marker nanog, which has been successfully used by other investigators. We first identified a free loop in the predicted mRNA structure (217–237 nt). The beacon was prepared by synthesizing a 10-nt complimentary strand and placing 5 base-paired nucleotides on the 5′ and 3′ ends. The bases were chosen so that their pairing energy was less than the energy to hybridize the beacon loop to its target mRNA. We then tested several different probe/quencher pairs and found that FAM/BHQ1 pair yielded the best response with the lowest signal/noise ratio. In solution, the resulting beacon 5′-6′-FAM-ggtgc-gacaagctggatccacact-gcacc-BHQ1-3′, shows a ∼30-fold increase in fluorescence when digested with RNase.
We tested several transfection methods: electroporation, FuGENE, calcium phosphate precipitation, and Lipofectamine as described under Materials and Methods. By far, Lipofectamine gave the highest transfection efficiency (>95%) and the highest survival rate (80%) in all cell lines tested.
We tested the nanog beacon in several cell lines. We could not detect a beacon response in human aortic adventitial fibroblasts, which do not express nanog, but found a robust response when transfected with nanog. Additionally, we could not detect beacon fluorescence in normal human adult dermal fibroblasts, Chinese hamster ovary cells, Fischer rat thyroid cells, canine MSCs, and mouse macrophages but detected beacon fluorescence in hMSCs and HEK293 cells (Fig. 1A). These studies validate the specificity of the human nanog beacon.
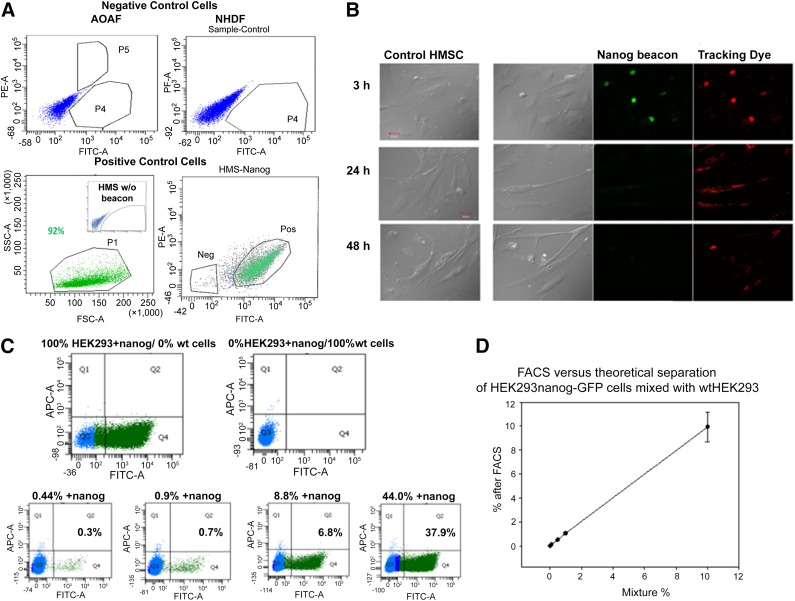
Figure 1.
FACS separation. (A): FACS separation of cultured cell lines and human mesenchymal stem cells (hMSCs) with nanog molecular beacon delivered by Lipofectamine. (B): Phase contrast (left) of control hMSCs and hMSCs transfected with nanog beacon. Green fluorescence indicates hybridization of the nanog beacon to nanog mRNA. Red fluorescence is caused by the tracking dye on the beacon and shows that the beacon has been delivered into the cell. We find that the beacon begins to degrade after 3 hours, and after 48 hours only a small amount of tracer dye can be detected. (C): FACS separation of wtHEK293 from HEK293 cells stably transfected with nanog using the nanog beacon. The top panels are for 100% nanog-transfected HEK293 (100% HEK293+nanog) and wild-type HEK293 (0% HEK293+nanog), whereas the bottom panels indicate the theoretical percentages in the heading of the HEK293+nanog. The actual values are indicated in the plots. (D): Results showing the amount of HEK293-nanog-GFP cells separated from nonfluorescent wtHEK293 cells versus the theoretical amount. We find an exact match between experimental and theoretical values until 0.01% theoretical with the experimental reading 0.03%. The results show the separation of different mixtures of HEK293 cells expressing GFP-nanog from unlabeled cells by FACS, as a function of their theoretical value. As can be seen, the experimental values are identical to the theoretical ones. All cell images were taken using a ×60 objective. Scale bars = 30 μm. Abbreviations: AOAF, aortic adventitial fibroblast; FACS, fluorescence-activated cell sorting; FITC, fluorescein isothiocyanate; GFP, green fluorescent protein; HMS, human mesenchymal stem; HMSC, human mesenchymal stem cell; Neg, negative; NHDF, normal adult dermal fibroblasts; PE, phycoerythrin; Pos, positive; SSC, side scatter corresponding to the amount of scattered light from the laser; wt, wild type.
We monitored beacon degradation (Fig. 1B). Beginning ∼3 hours post-transfection, we observed dispersion and weakening of the signal that was complete after 12 hours in all cell types. No signal is seen after 24 hours in a fast growing cells and 48 hours in slow growing cells (i.e., SK-H-SH versus AOAF cells overexpressing nanog).
We then optimized the use of beacon for cell separation. To isolate cells that are not expressing the gene of interest, we needed to determine whether a lack of beacon response was due to the absence of the target gene or because the cell was not transfected with beacon. To resolve this problem, we designed a beacon that incorporated a tracking fluorophore at the base of the stem: 5′-FAM-ggtgc-gacaagctggatccacact-TF5-gcacc-BHQ1-3′, where TF5 is the red probe Tide Fluor 5. By simultaneously sorting cells by the red tracking dye and the green fluorescence from a positive response, we separated cells that lacked nanog from ones that express it.
We then determined the limit of FACS sensitivity by mixing wild-type HEK293 cells with ones stably transfected with nanog. In Figure 1C we show that the nanog beacon is able to separate 0.3% of nanog-positive cells from a mixture in which the theoretical value is 0.44%, thus giving an ∼70% separation accuracy at values less than 1/100 for positive selection. This selection accuracy increases to ∼85% as the amount of nanog-positive cells increases. We can compare this accuracy of separation using beacon detection to the experimental accuracy of cell separation independent of beacon in which we simply mixed HEK293-nanog-GFP with wtHEK293 cells (Fig. 1D). These studies show that separation is accurate to at least 0.01%, and thus the differences seen between the theoretical and actual values seen in Figure 1C are due to the beacon response rather than errors in the experimental procedure. Nevertheless, these data show that the yield for positive selection of cells using beacon is quite high.
Development of a Universal Beacon
First, we randomly generated 20-nt DNA sequences and searched for them in the human genome. We found that CGTACGCGTCGGAGAT does not match any transcript mRNA and has only a 75% match to 6 genomic DNA sequences and an 81% match to 3 genomic DNA sequences. We inserted this sequence before the start codon of nanog after verifying that it does not disrupt regulatory regions (e.g., CAAT or TATA boxes) and avoided potential siRNA sites. Neither the UB tag nor UB (5′-6-FAM-ccaccCGTACGCGTCGGAGAT-TF5-gggtgg-BHQ1) affected nanog protein expression, cell growth, or cell morphology (Fig. 2A). We directly compared cell separation by FACS of cMSCs transiently transfected with UB-nanog using the nanog beacon described above versus the UB-nanog. We find an identical 99.1% recovery of nanog-positive cells using both the nanog beacon and the UB after correcting for the transfection efficiency of UB-nanog (25.9%).
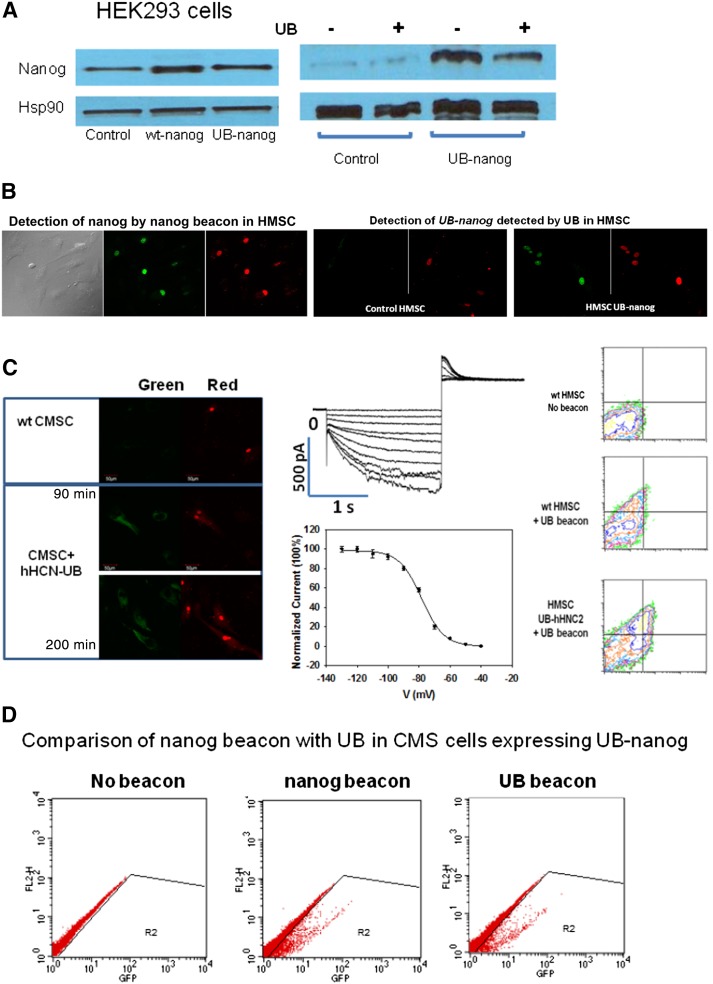
Figure 2.
Fluorescence-activated cell sorting (FACS) and Western blotting. (A): The leftmost set of Western blots shows that expression of both wt and UB-nanog are similar in HEK293 cells as compared with the housekeeping gene Hsp90. For this study, cells were grown under conditions so that the level of endogenous nanog is clearly visible and can be compared with the transfected cells. We find that transfection of UB-nanog at the levels needed to elicit beacon response results in similar levels of wild-type nanog and UB-nanog; thus, the UB tag does not alter nanog expression. The rightmost set of Western blots shows that under conditions in which endogenous nanog is low, the presence of the UB tag does not affect the expression of endogenous nanog or UB-nanog. (B): Comparison of the detection of nanog by the nanog beacon and UB-nanog by UB in HMSCs. (C): Wild-type canine MSCs (top) and cells transfected with UB tag-hHNC2, where the red fluorescence is caused by tracking dye and the green fluorescence is due to beacon opening (left). Middle: patch clamp studies showing that the cells expressing UB tag-hHNC2 as separated by FACS using UB forms functional channels. Right: FACS results of wild-type HMSCswithout (top) and with (middle) UB (negative control) and of hMSCs transfected with UB tag-hHNC2 and UB (bottom), where the red fluorescence is from the TF5 tracking dye, whereas the green fluorescence corresponds to UB opening. Scale bars = 50 mm. Magnification, ×360. (D): Comparison of FACS results of cMSCs transiently transfected with UB tag-nanog gene (N5JR1) detected by nanog beacon versus UB-nanog. Although the transfection efficiency was low, both MB-nanog and UB selected the same percentage of cells. Abbreviations: CMSC, canine mesenchymal stem cell; GFP, green fluorescent protein; HSMC, human mesenchymal stem cell; UB, universal beacon; wt, wild type.
We applied UB technology to the pacemaker channel gene hHCN2 in a project with the ultimate goal of using stem cell technology to replace electrical pacemakers. Unlike with the nanog beacon, we were unable to construct a sensitive and specific traditional beacon to this gene because of its high G/C content. Additionally, the hHCN2 channel is a transmembrane protein and does not have a distinguishable epitope for antibody recognition. Therefore, only the UB approach would allow us to separate cells expressing this gene.
We inserted the UB tag before the start and/or after the codon in the 5′-UTR of hHCN2. We transfected UB-hHCN2 or hHNCN2 into cMSCs followed by transfection with the UB. Red UB fluorescence was seen in all cells, but only cells transfected with UB tag-hHCN2 showed green fluorescence. The highest level of beacon response is seen ∼60–90 minute after transfection followed by degradation (Fig. 2B). We then separated cells transfected with UB tag-hHCM2 by FACS. All cells that showed green fluorescence had channel activity (Fig. 2C). Thus, UB tag-hHNC2 is expressed, and the protein properly localizes to the plasma membrane to form functional channels that are identical to wild type (Fig. 2B, 2C). FACS studies indicate a small background that we find is due to the 11.1% impurity of the beacon preparation, as determined by mass spectrometry. Using purer beacon preparations diminished this background to give a separation accuracy on par with those in Figure 1C. Additionally, both the nanog beacon and UB gave identical selection and separation of cMSCs transiently transfected with UB-nanog (Fig. 2D). Thus, our studies suggest that UB can be applied to positive as well as negative selection.
Discussion
RNA beacons have the potential to deliver pure cell preparations needed for stem cell therapy, but developing a specific and responsive beacon can be difficult if not impossible. Here, we report on an off-the-shelf universal beacon that targets a nonsense tag placed in the untranslated region of a functional protein or even a biomarker. We show that UB technology allows for detection and high-throughput separation of any exogenous gene without altering the properties of the protein product.
Conclusion
Here we have shown that separation of UB-positive cells using UB yields a pure population of cells expressing the gene of interest. This technology should greatly enhance the ability to prepare pure populations of transformed stem cells for therapeutic use and can be used in other cell types that allow transfection or infection as well.
Acknowledgments
This work was supported by NIH Grant GM053132 (to S.S.) and NIH Grant HL-111401 (to I.C.).
Author Contributions
Y.G.: collection and/or assembly of data, data analysis and interpretation, final approval of manuscript; Z.L.: collection and/or assembly of data; I.S.C.: conception and design, financial support, final approval of manuscript; S.S.: conception and design, data analysis and interpretation, financial support, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.Rhee WJ, Bao G. Simultaneous detection of mRNA and protein stem cell markers in live cells. BMC Biotechnol. 2009;9:30. doi: 10.1186/1472-6750-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhee WJ, Santangelo PJ, Jo H, et al. Target accessibility and signal specificity in live-cell detection of BMP-4 mRNA using molecular beacons. Nucleic Acids Res. 2008;36:e30. doi: 10.1093/nar/gkn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ban K, Wile B, Kim S, et al. Purification of cardiomyocytes from differentiating pluripotent stem cells using molecular beacons that target cardiomyocyte-specific mRNA. Circulation. 2013;128:1897–1909. doi: 10.1161/CIRCULATIONAHA.113.004228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King FW, Liszewski W, Ritner C, et al. High-throughput tracking of pluripotent human embryonic stem cells with dual fluorescence resonance energy transfer molecular beacons. Stem Cells Dev. 2011;20:475–484. doi: 10.1089/scd.2010.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itzhaki I, Maizels L, Huber I, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 6.Plotnikov AN, Shlapakova I, Szabolcs MJ, et al. Xenografted adult human mesenchymal stem cells provide a platform for sustained biological pacemaker function in canine heart. Circulation. 2007;116:706–713. doi: 10.1161/CIRCULATIONAHA.107.703231. [DOI] [PubMed] [Google Scholar]
- 7.Potapova I, Plotnikov A, Lu Z, et al. Human mesenchymal stem cells as a gene delivery system to create cardiac pacemakers. Circ Res. 2004;94:952–959. doi: 10.1161/01.RES.0000123827.60210.72. [DOI] [PubMed] [Google Scholar]
- 8.Potapova IA, Doronin SV, Kelly DJ, et al. Enhanced recovery of mechanical function in the canine heart by seeding an extracellular matrix patch with mesenchymal stem cells committed to a cardiac lineage. Am J Physiol Heart Circ Physiol. 2008;295:H2257–H2263. doi: 10.1152/ajpheart.00219.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ban K, Wile B, Kim S, et al. Purification of cardiomyocytes from differentiating pluripotent stem cells using molecular beacons that target cardiomyocyte-specific mRNA. Circulation. 2013;128:1897–1909. doi: 10.1161/CIRCULATIONAHA.113.004228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryazantsev DY, Kvach MV, Tsybulsky DA, et al. Design of molecular beacons: 3′ couple quenchers improve fluorogenic properties of a probe in real-time PCR assay. Analyst (Lond) 2014;139:2867–2872. doi: 10.1039/c4an00081a. [DOI] [PubMed] [Google Scholar]
- 11. Bioinformatics Sequence Manipulation Suite. Available at http://www.bioinformatics.org/sms2/random_dna.html. Accessed February 20, 2015.
- 12. BLAST. Available at http://blast.ncbi.nlm.nih.gov/Blast.cgi. Accessed February 20, 2015.
- 13. RNAi Consortium Public TRC Portal. Available at http://www.broadinstitute.org/rnai/public/trans/candidates. Accessed February 20, 2015.