Abstract
A multitude of RNA hairpins are directly implicated in human disease. Many of these RNAs are potentially valuable targets for drug discovery and basic research efforts. However, very little is known about the molecular requirements for achieving sequence-selective recognition of a particular RNA sequence and structure. While a relatively modest number of synthetic small to medium-sized RNA-binding molecules have been reported, rapid identification of sequence-selective disease-relevant RNA-binding molecules remains a daunting challenge. RNA Recognition Motif (RRM) domains may represent unique privileged scaffolds for the generation of synthetic proteins that selectively recognize disease-relevant RNAs, including RNA hairpins. As a demonstration of this potential, we mutated putative RNA-binding regions within the U1A RRM, and a variant thereof, and screened these synthetic proteins for affinity to HIV-1 Trans-Activation Response (TAR) element hairpin RNA. Impressively, some of these U1A-derived proteins bind TAR with single-digit micromolar dissociation constants, and do so preferentially over the native protein's original target RNA (U1hpII) and a DNA TAR variant. For two TAR-binding proteins, binding affinity is not appreciably diminished by addition of 10 molar equivalents of cellular tRNAs from Escherichia coli. Taken together, our findings represent the first synthetic RRMs that selectively bind a disease-relevant RNA hairpin, and may represent a general approach for achieving sequence-selective recognition of RNA hairpins, which are the focus of therapeutic discovery and basic research
Introduction
Ribonucleic acids (RNAs) play critical roles in functionally diverse cellular processes, including many of therapeutic significance.(1-3) Reagents capable of selectively recognizing disease-relevant RNAs, thus represent valuable starting points for drug discovery and basic research.(4-6) However, structural and electronic features associated with RNA can make the identification of such reagents a daunting challenge.(7) To date, only a modest number of synthetic small (<1 kDa) to medium-sized (∼1-2 kDa) RNA-binding molecules have been reported.(8-21) Of these, only a fraction exhibit moderate to good sequence selectivity. This limited success, coupled with sparse knowledge of the detailed requirements for specific recognition of a given folded RNA, suggests that traditional small molecule approaches will never provide a complete solution. We need a fundamentally different approach.
Nature has evolved a suite of RNA-binding proteins(22-24), which may serve as privileged scaffolds for the generation of synthetic proteins that bind disease-relevant RNAs.(22, 25) Some of these proteins have evolved to address precisely the challenge before us: Recognition of a particular folded RNA with good affinity and sequence selectivity. Converting them to our purpose requires deciphering and manipulating the elements that govern recognition specificity. Recently, exciting findings have been reported on the engineering of Pumilio/fem-3-binding factor (PUF) repeat domains and pentatricopeptide Repeat (PPR) proteins for sequence-selective recognition of single-stranded RNAs (ssRNAs).(26-30) Taken together, these efforts have resulted in the generation of a ssRNA “recognition code”, which enables thoughtful design of synthetic proteins capable of selectively binding virtually any ssRNA sequence. However, selective recognition of structured RNAs remains a serious challenge.
Here we report the first mutation of a known RNA Recognition Motif (RRM) to confer specificity for the HIV Trans-Activation Response (TAR) element hairpin, a significant target for therapeutic discovery and basic research. The best synthetic RRMs we identified bind to TAR with low micromolar affinity, and do so preferentially over the native protein's original target RNA, a DNA TAR variant, and in the presence of 10 molar equivalents of cellular tRNAs. Together these results establish a fundamentally new strategy for the sequence-specific recognition of high value RNA targets, and provide a potentially general framework for the development of new complexes between engineered or evolved RRMs and disease-relevant structured RNAs, including RNA hairpins.
The N-terminal RNA Recognition Motif (RRM) within the U1 small nuclear ribonucleoprotein (U1A, Figure 1A and 1B) selectively binds the RNA hairpin U1hpII (Figure 1B) with incredible affinity (dissociation constant, KD, ∼10-11 M).(31, 32) The U1A-U1hpII complex has attracted significant attention and frequently serves as a model for recognition of hairpin RNA by a protein.(33-39) Native U1A-U1hpII binding proceeds through a “lure and lock” mechanism.(40) Polyanionic RNA is first lured to U1A by strategically placed positively charged lysine residues.(41) The lure step is followed by sequence-selective interactions with the target RNA.(32, 34) Following an ion-paired lure step, the complex is additionally stabilized through π-π interactions that involve solvent-exposed loop nucleotides C5 and A6, and U1A residues Tyr13 and Phe56(42) (Figure 1B). In solution, the U1hpII loop exhibits a dynamic structure.(32) However, in the bound state, residues within the β2-β3 loop of U1A protrude into the U1hpII RNA loop, essentially locking this region of the RNA hairpin in place around the U1A β2-β3 loop.(32) (Figure 1C). This complex is further stabilized by RNA sequence-selective hydrogen bond networks, including well-defined interactions between U1A residues Asn15, Asn16, and Glu19 and U1hpII nucleotides U2, G3, and G4 (Figure 1D).(43)
Figure 1.
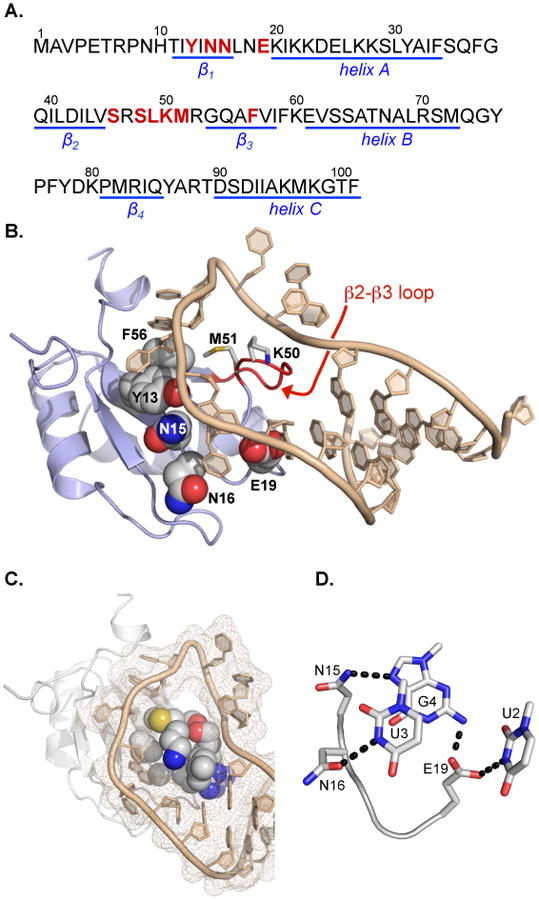
(A) Primary amino acid sequence of the wild-type N-terminal U1A RNA Recognition Motif (RRM). Structural features of U1A are highlighted in blue. Residues mutated in this work are highlighted in red. (B) U1A/U1hpII complex (PDB: 1URN). The b2-b3 loop and key residues that are mutated in this work are highlighted and annotated. (C) Space-filling model of U1A b2-b3 loop residues protruding through the U1hpII RNA loop. (D) Hydrogen bond network formed between U1A residues Asn15, Asn16, and Glu19 and U1hpII bases U2, U3, and G4. These contacts contribute to the source of sequence selective recognition of U1hpII by U1A. Hydrogen bonds are represented as black dashed lines.
The U1hpII RNA hairpin contains a relatively large 10-nucleotide loop (Figure 2A and 2B). However, a large number of disease-relevant RNA hairpins contain significantly smaller loops, which typically range four to eight nucleotides in length.(7) Well-studied features of the U1A-U1hpII interaction, however, suggest that variants of U1A and/or U1A-derived proteins could be made to recognize RNA hairpins with smaller loops. For example, U1A does not engage U1hpII loop nucleotides U9, C10, and C11, suggesting smaller RNA loops can be accommodated.(39, 44) U1A β2-β3 loop residues Lys50 and Met51 (highlighted in Figure 1B) form no interaction with U1hpII.(32) Laird-Offringa and coworkers have shown that while deletion of U1hpII loop nucleotides U8 and U9 reduces U1A affinity by ∼3000-fold, a compensatory mutation in U1A that removes residues Lys50 and Met51 (ΔK50ΔM51) results in a protein that binds either hairpin with similar affinity.(45) We have used fluorescence polarization to measure dissociation constants (KD) for numerous ΔK50ΔM51 mutants and U1hpII-derived RNA hairpins with eight or seven-nucleotide loops (ΔU8ΔC9 or ΔU8ΔC9ΔC10, respectively).(46) Our results indicate a similar binding mode for the wild-type complex and complexes involving ΔK50ΔM51 and U1hpII hairpins with shortened loops. Putative RNA-binding regions in the U1A-U1hpII context can play prominent roles in stabilizing interactions between ΔK50ΔM51 and U1hpII variants with shortened loops.
Figure 2.
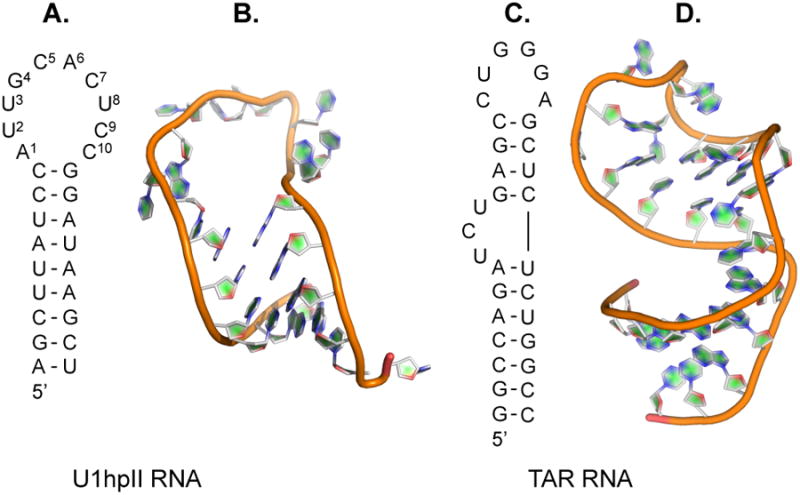
(A) Secondary sequence and structure of U1hpII. (B) Crystal structure of U1hpII (PDB: 1URN). (C) Secondary sequence and structure of HIV-1 Trans-Activation Response (TAR) hairpin RNA. (D) Crystal structure of TAR RNA (PDB: 1ARN).
Previous research has demonstrated a degree of plasticity within RNA-binding regions of U1A (and by relation, ΔK50ΔM51). Mutagenesis of residues within putative RNA recognition regions of U1A generally does not result in protein instability and unfolding.(32, 41-43, 46) In addition, certain mutations within the putative RNA-binding regions can lead to altered RNA binding selectivity. For example, researchers have shown that mutating residues within the b2-b3 loop lead to U1A-derived proteins with improved affinity for U1hpII.(47) Additionally, similar mutations have resulted in U1A variants capable of selectively binding RNA hairpins with high sequence homology to U1hpII.(47, 48) Finally, Baranger and coworkers have shown that mutating Glu19 can result in U1A-derived proteins that selectively bind a U1hpII G4U mutant RNA hairpin.(43) Taken together, these finding suggest that U1A is a potentially malleable protein scaffold from which new proteins capable of binding disease-relevant RNA hairpins can be generated. Thus far, however, researchers have only focused on the generation of U1A-derived proteins with improved affinity for U1hpII, or altered affinity for RNAs with high sequence homology to U1hpII. We questioned if U1A and/or ΔK50ΔM51 might serve as a starting point for the development of new synthetic proteins that selectively recognize a disease-relevant RNA hairpin that does not share significant sequence homology with U1hpII.
As an initial test of our ability to identify synthetic RRMs that recognize an RNA hairpin involved in human disease, we created a focused library of U1A- and ΔK50ΔM51-mutants and measured the affinity of these proteins to Trans-Activation Response (TAR) element from HIV-1 (Figures 2C and 2D). TAR RNA is a well-studied RNA hairpin with a six-nucleotide loop and three-nucleotide bulge within the stem.(49) TAR is required for trans-activation of the HIV-1 viral promoter, which ultimately results in the synthesis of proteins necessary for HIV-1 proliferation(50), and has therefore been the focus of therapeutic discovery and molecular recognition studies. In contrast to previous efforts to alter the sequence-selectivity of U1A-derived proteins, TAR shares little sequence and structural homology with U1hpII (Figure 2).
Results and Discussion
Alanine Scanning Identifies Mutations Within Putative RNA-Binding Regions of U1A that Stabilize Binding to TAR RNA
We used fluorescence polarization to measure the dissociation constants for complexes involving a 5′ fluorescein-labeled TAR RNA and U1A, ΔK50ΔM51, or mutants thereof. Both U1A and ΔK50ΔM51 bind TAR with low micromolar dissociation constants (KD = 19.1(±4.9) and 15.9 (±2.3) μM, respectively, Table 1, entries 1 and 10). These binding affinities are significantly better than recently reported RNA-binding molecules. Additionally, these data support our hypothesis that both U1A and ΔK50ΔM51 are privileged scaffolds for RNA hairpin recognition, and are valuable starting points for achieving good and selective affinity of disease-relevant RNA hairpins.
Table 1. Binding affinities for complexes involving TAR RNA and U1A or ΔK50ΔM51, and specific alanine mutants thereof.
| Entry | RRM scaffold | Mutationa | Nucleic acid | KD (μM)b | ΔG (kcal/mol)c | ΔΔG (kcal/mol)d |
|---|---|---|---|---|---|---|
| 1 | U1A | none | TAR RNA | 19.1 (±4.9) | -6.43 | ------ |
| 2 | U1A | Asn15Ala | TAR RNA | 11.7 (±0.9) | -6.72 | -0.29 |
| 3 | U1A | Asn16Ala | TAR RNA | 8.8 (±1.4) | -6.89 | -0.46 |
| 4 | U1A | Glu19Ala | TAR RNA | 5.9 (±0.8) | -7.13 | -0.70 |
| 5 | U1A | Ser46Ala | TAR RNA | 15.1 (±2.6) | -6.57 | -0.14 |
| 6 | U1A | Ser48Ala | TAR RNA | 14.2 (±1.3) | -6.61 | -0.18 |
| 7 | U1A | Leu49Ala | TAR RNA | 5.2 (±0.5) | -7.20 | -0.77 |
| 8 | U1A | Lys50Ala | TAR RNA | 39.6 (±12.2) | -6.00 | +0.43 |
| 9 | U1A | Met51Ala | TAR RNA | 4.9 (±0.6) | -7.24 | -0.81 |
| 10 | ΔK50ΔM51 | none | TAR RNA | 15.9 (±2.3) | -6.54 | ------ |
| 11 | ΔK50ΔM51 | Asn15Ala | TAR RNA | 91.1 (±8.8) | -5.51 | +1.03 |
| 12 | ΔK50ΔM51 | Asn16Ala | TAR RNA | 62.1 (±5.4) | -5.74 | +0.80 |
| 13 | ΔK50ΔM51 | Glu19Ala | TAR RNA | 10.2 (±2.2) | -6.80 | -0.26 |
| 14 | ΔK50ΔM51 | Ser46Ala | TAR RNA | 24.5 (±6.0) | -6.29 | +0.25 |
| 15 | ΔK50ΔM51 | Ser48Ala | TAR RNA | 33.9 (±15.9) | -6.09 | +0.45 |
| 16 | ΔK50ΔM51 | Leu49Ala | TAR RNA | 91.6 (±39.7) | -5.50 | +1.04 |
All point mutations are derived from native U1A (entries 2-9) or ΔK50ΔM51 (entries 11-16) scaffolds.
The error for each reported dissociation constant (KD) is the standard deviation of three separate experiments.
ΔG is the free energy of the protein in complex with TAR RNA calculated with the equation ΔG = -RTlnKD.
ΔΔG is the difference in binding free energy between the complexes in entry 1 (for U1A-derived mutants) or 10 (for ΔK50ΔM51-derived mutants), and indicated mutants thereof.
To identify residues within the U1A and ΔK50ΔM51 frameworks that specifically contact TAR RNA, as well as identify similarities in the mechanism of binding for these complexes and the native interaction, we mutated putative U1hpII RNA-binding residues to alanine and measured the effect of each mutation on affinity for TAR RNA. Given their well-established role in U1hpII recognition, we mutated residues Asn15, Asn16, Glu19, and residues within the b2-b3 loop (highlighted in red, Figures 1A and 1B). In addition to providing insight into the points of contact for this new protein-RNA interaction, we hypothesized that residues that affect TAR affinity could be optimized for affinity, and potentially selectivity. Previous studies have shown that Arg47 participates in U1hpII RNA recruitment through non-selective ion-paired interactions with the phosphate backbone of RNA.(41) Within both the U1A and ΔK50ΔM51 frameworks, Arg47Ala mutation dramatically lowered affinity for TAR RNA (KD >250 μM, data not shown). Of the eight remaining U1A-derived alanine mutants we tested, only the Lys50Ala mutant showed lower affinity for TAR RNA, compared to native U1A (Table 1, entry 8). The tightest binding U1A-derived alanine mutants, Glu19Ala, Leu49Ala and Met51Ala, bound TAR with 3.2-, 3.7- and 3.9-fold improved affinity, compared to native U1A (Table 1, entries 4, 7, and 9, respectively). Generally, these findings suggest that mutation of putative RNA-binding residues in the U1A-U1hpII complex can result in synthetic U1A proteins with good binding affinity for TAR RNA. Interestingly, while ΔK50ΔM51 binds TAR with slightly better affinity than native U1A, none of the ΔK50ΔM51 alanine mutants outperformed the analogous U1A alanine mutants. Of the six ΔK50ΔM51 alanine mutants we tested, only Glu19Ala showed significantly improved affinity for TAR RNA (1.6-fold improvement compared to ΔK50ΔM51).
Expanded Mutagenesis of TAR RNA-Binding Residues in U1A Improves Affinity
Expanding on these findings, we set out to optimize the three best-performing U1A-derived alanine mutants (U1A Glu19Ala, U1A Leu49Ala, and U1A Met51Ala) and the best ΔK50ΔM51-derived alanine mutant (ΔK50ΔM51 Glu19Ala). We measured the binding affinities for a focused library of U1A mutants with chemically diverse residues at positions 19, 49, and 51. This set of U1A variants was designed to test changes in complex stability as a result of incorporating negatively charged (glutamic acid, Glu), positively charged (lysine, Lys), hydrogen bond donating (serine, Ser), amide (asparagine, Asn), or aromatic (phenylalanine, Phe) amino acids at these important residues.
Alanine mutagenesis data indicated that the negatively charged carboxylic acid containing side chain at position 19 has a deleterious effect on the stability of a complex with TAR RNA. Therefore, we reasoned that U1A proteins containing chemically distinct side chains at this position might have improved affinity for TAR RNA. Indeed, U1A Glu19Lys, Glu19Ser, Glu19Asn and Glu19Phe mutants exhibited 3.2-, 4.7-, 1.6-, and 3.1-fold improved affinity for TAR, compared to native U1A (Table 2, 1-4, respectively).
Table 2. Binding affinities for complexes involving TAR RNA and specific U1A- and ΔK50ΔM51-derived mutants.
| Entry | RRM scaffold | Mutationa | Nucleic acid | KD (μM)b | ΔG (kcal/mol)c | ΔΔG (kcal/mol)d |
|---|---|---|---|---|---|---|
| 1 | U1A | Glu19Lys | TAR RNA | 5.9 (±0.4) | -7.13 | -0.70 |
| 2 | U1A | Glu19Ser | TAR RNA | 4.1 (±0.3) | -7.34 | -0.91 |
| 3 | U1A | Glu19Asn | TAR RNA | 12.0 (±0.9) | -6.71 | -0.28 |
| 4 | U1A | Glu19Phe | TAR RNA | 6.2 (±0.8) | -7.10 | -0.67 |
| 5 | U1A | Leu49Glu | TAR RNA | >250 | ------ | ------ |
| 6 | U1A | Leu49Lys | TAR RNA | 3.9 (±0.4) | -7.37 | -0.94 |
| 7 | U1A | Leu49Ser | TAR RNA | 12.4 (±2.3) | -6.69 | -0.26 |
| 8 | U1A | Leu49Asn | TAR RNA | 14.4 (±2.1) | -6.60 | -0.17 |
| 9 | U1A | Leu49Phe | TAR RNA | 24.1 (±4.3) | -6.30 | +0.13 |
| 10 | U1A | Met51Glu | TAR RNA | >250 | ------ | ------ |
| 11 | U1A | Met51Lys | TAR RNA | 7.7 (±0.5) | -6.97 | -0.54 |
| 12 | U1A | Met51Ser | TAR RNA | 29.9 (±5.7) | -6.17 | +0.26 |
| 13 | U1A | Met51Asn | TAR RNA | 13.8 (±1.6) | -6.62 | -0.19 |
| 14 | U1A | Met51Phe | TAR RNA | 16.4 (±2.3) | -6.52 | -0.09 |
| 15 | ΔK50ΔM51 | Glu19Lys | TAR RNA | 6.3 (±1.0) | -7.09 | -0.55 |
| 16 | ΔK50ΔM51 | Glu19Ser | TAR RNA | 7.0 (±1.9) | -7.03 | -0.47 |
| 17 | ΔK50ΔM51 | Glu19Asn | TAR RNA | 27.8 (±3.2) | -6.21 | +0.33 |
| 18 | ΔK50ΔM51 | Glu19Phe | TAR RNA | 12.6 (±1.8) | -6.68 | -0.14 |
Single residue mutants are derived from native U1A (entries 1-14) or ΔK50ΔM51 (entries 15-18) scaffolds.
The error for each reported dissociation constant (KD) is the standard deviation of three separate experiments. All point mutations are derived from native U1A (entries 2-9) or ΔK50ΔM51 (entries 11-16) scaffolds.
ΔG is the free energy of the protein in complex with RAR RNA calculated with the equation ΔG = -RTlnKD.
ΔΔG is the difference in binding free energy between the complexes with native U1A or ΔK50ΔM51, and indicated mutants thereof.
Table 4. Binding selectivity of U1A-derived double mutants for TAR RNA over U1hpII RNA or TAR DNA.
| Entry | RRM scaffold | TAR RNA - binding mutationa | U1hpII “off” mutation | KD (μM)b TAR RNA | KD (μM)c U1hpII RNA | Fold-selectivity for TAR RNAd |
|---|---|---|---|---|---|---|
| 1 | U1A | Glu19Ala | Tyr13Gln | 21.7 (±5.3) | NDe | ND |
| 2 | U1A | Glu19Ala | Asn15Val | 14.2 (±1.4) | 48.4 (±12.4) | 3.4 |
| 3 | U1A | Glu19Ala | Phe56Ala | 29.2 (±6.3) | ND | ND |
| 4 | U1A | Glu19Phe | Tyr13Gln | 58.0 (±24.1) | ND | ND |
| 5 | U1A | Glu19Phe | Asn15Val | 19.0 (±2.5) | ND | ND |
| 6 | U1A | Glu19Phe | Phe56Ala | 20.9 (±3.3) | ND | ND |
| 7 | U1A | Glu19Ser | Tyr13Gln | 10.3 (±2.8) | 63.6 (±8.9) | 6.2 |
| 8 | U1A | Glu19Ser | Asn15Val | 11.3 (±0.8) | 15.1 (±2.0) | 1.3 |
| 9 | U1A | Glu19Ser | Phe56Ala | 19.24 (±1.4) | ND | ND |
| 10 | U1A | Leu49Ala | Tyr13Gln | >1 mM | ND | ND |
| 11 | U1A | Leu49Ala | Asn15Val | 39.2 (±9.6) | ND | ND |
| 12 | U1A | Leu49Ala | Glu19Ala | 6.9 (±0.8) | 11.7(±2.1) | 1.7 |
| 13 | U1A | Leu49Ala | Phe56Ala | 123.2 (±38.7) | ND | ND |
| 14 | U1A | Met51Ala | Tyr13Gln | 209.8 (±84.3) | ND | ND |
| 15 | U1A | Met51Ala | Asn15Val | 46.0 (±13.4) | ND | ND |
| 16 | U1A | Met51Ala | Glu19Ala | 14.9 (±2.5) | 15.8(±2.7) | 1.1 |
| 17 | U1A | Met51Ala | Phe56Ala | 176.9 (±98.2) | ND | ND |
All double mutants are derived from native U1A.
The error for each reported dissociation constant (KD) is the standard deviation of three separate experiments.
Fold-selectivity for TAR RNA was calculated by (KD off-target nucleic acid/KD TAR RNA).
ND = not determined
Additional information on the molecular requirements for TAR RNA recognition by U1A-derived proteins was obtained by examining focused libraries of mutants at positions 49 and 51. None of the Leu49 mutants resulted in a higher affinity complex, compared to U1A Leu49Ala. However, the affinity of the Leu49Glu for TAR RNA was significantly diminished (KD > 250 μM), suggesting that a negative charge at this position is not tolerated (Table 2, entry 5). Lysine, serine, asparagine, and phenylalanine are tolerated at residue 49; however, these mutations resulted in modest to appreciably worse affinity for TAR RNA, compared to U1A Leu49Ala (Table 2, entries 6-9). Similar to the mutagenesis profile we observed for residue 49, the Met51Glu mutant exhibited significantly decreased affinity for TAR RNA (KD > 250 μM, Table 2, entry 10). Lysine, serine, asparagine, and phenylalanine were tolerated at amino acid 51; however, modest to appreciably worse affinity for TAR RNA was observed in each case, compared to Met51Ala (Table 2, entries 11-14). Taken together, these findings implicate putative RNA-binding residues in the native U1A-U1hpII complex as important contributors to binding TAR RNA, and suggest that mutation of these key residues may result in synthetc proteins with improved affinity for TAR RNA. These findings also suggest that various functionally diverse residues are tolerated at residues 49 and 51; however, decreasing the apparent size of the β2-β3 loop through Leu49Ala or Met51Ala mutations can result in improved affinity for TAR hairpin RNA. This is perhaps intuitive, since the loop in TAR is significantly smaller than the RNA loop in U1hpII.
To provide insight into complexes involving TAR RNA and ΔK50ΔM51-derived proteins, as well as potentially improve this binding interaction, we prepared a focused library of mutants at position 19 and measured the effect of each mutation on affinity for TAR RNA. Similar to our earlier observations for U1A-derived mutants, Glu19Lys, Glu19Ser and Glu19Phe exhibit modestly improved affinity for TAR RNA, compared to ΔK50ΔM51 (2.5-, 2.2-, and 1.3-fold, respectively; Table 2, entries 15, 16, and 18). Similar to the analogous U1A mutant, ΔK50ΔM51 Glu19Asn exhibited the worst affinity for TAR RNA of residue 19 mutants (Table 2, entry 17).
To summarize these initial results: Impressively, mutation of a single residue within the U1A or ΔK50ΔM51 framework can result in new synthetic RRMs capable of binding TAR RNA with good affinity (low to single-digit micromolar dissociation constants). These findings support our general hypothesis, that U1A and ΔK50ΔM51 are privileged RNA hairpin-binding scaffolds, and mutagenesis of these structures can result in new RNA-binding proteins with affinity for disease-relevant RNA hairpins. From a single mutation, we generated U1A- derived proteins that rival or exceed previously reported small molecule TAR-binding compounds(21, 51, 52), and are significantly tighter binders than very recently reported RNA-binding small molecules identified from high-throughput screening(53). While potentially higher affinity complexes may result from more extensive mutagenesis, we viewed these mutants as promising leads that warrant additional study.
Combining Beneficial Single Mutations Does Not Improve Affinity for TAR RNA
Synergistic effects can play important roles in a multitude of biological processes, including binding interactions.(54) Expanding on our preliminary findings, we next set out to determine if combinations of beneficial U1A mutations identified in Table 1 and Table 2 further stabilize binding to TAR hairpin RNA. We prepared a truncated combinatorial library of double mutants (derived from beneficial single mutants). Interestingly, none of these double mutants exhibited improved affinity for TAR RNA, compared to the single mutant U1A proteins from which they derived (Supporting Information, Figure S6). While the rationale for this observation is unknown, one hypothesis is that synthetic U1A proteins with multiple independently beneficial mutations bind TAR RNA in slightly different conformations, compared to single mutation versions from which they derive. If true, complexes between TAR RNA and double mutants may favor interactions between a beneficial mutation, while inhibiting (or significantly lessening) the other. Based on these findings, we focused our efforts on single residue mutants of U1A or ΔK50ΔM51 that exhibit good affinity (low micromolar dissociation constant) for TAR RNA.
Synthetic U1A Proteins Selectively Bind TAR RNA over an Analogous DNA Sequence
Perhaps foremost among the many challenges faced by researchers focused on RNA recognition is achieving selectivity for a particular folded RNA over other nucleotides. Unlike proteins, which are generated from 20 proteinogenic amino acids that can contain chemically diverse side chains, the functional group diversity of RNA originates from only four nucleobases, which have significant chemical and structural homology. An obvious assessment of RNA- selective recognition is to compare affinities between a molecule or protein for a target RNA and a sequence analogous DNA. While experimentally simple, this is a particularly significant challenge. Sequence analogous RNA and DNA hairpins only differ chemically by replacement of uracil (U) in RNA with a structurally related thymine (T) base, and addition of a stereodefined 2′-hydroxyl group within the ribose of RNA. Since relatively little is known about the molecular requirements for achieving RNA-selective interactions, thoughtful integration of RNA-selectivity into a nucleic acid-binding small molecule or protein a priori is daunting and an unsolved problem. Unsurprisingly, a number of previously reported RNA-binding small molecules also bind the sequence analogous DNA with very little selectivity.(55, 56) However, we hypothesized that since RRMs evolved function (recognition of a particular RNA sequence and structure) in a virtual sea of cellular nucleic acids, this scaffold might exhibit inherent selectivity for a target RNA hairpin over other nucleic acids such as DNA or tRNAs. Consistent with this hypothesis, native U1A bound TAR RNA with ∼7.6-fold selectivity over the sequence analogous DNA (referred to as TAR DNA herein, Table 3, entry 1). Among the newly identified TAR-RNA binding RRMs we tested (see Table 3), five of these proteins bound TAR RNA with ∼17-to ∼37-fold selectivity over TAR DNA. These proteins, and their selectivity for TAR RNA over TAR DNA (in parenthesis) are as follows: U1A Glu19Ala (17.0); U1A Glu19Phe (25.2); U1A Glu19Ser (22.4); U1A Leu49Ala (17.0); and, U1A Met51Ala (36.5), (Table 3, entries 2, 3, 5, 6, and 8, respectively). Unsurprisingly, perhaps, U1A mutants with a positively charged lysine (U1A Glu19Lys, U1A Leu49Lys, and U1A Met51Lys) have the lowest selectivity for TAR RNA over TAR DNA (2.1-, 5.0-, and 7.1-fold, respectively). While all of the U1A and ΔK50ΔM51 mutants we tested selectively bound TAR RNA over TAR DNA, ΔK50ΔM51 mutants exhibited lower selectivity compared to most of the U1A-derived synthetic RRMs. Based on these findings, we limited subsequent studies to the pool of U1A-derived RRMs.
Table 3. Binding selectivity of specific U1A- and ΔK50ΔM51-derived mutants for TAR RNA over TAR DNA or U1hpII RNA.
| Entry | RRM scaffold | Mutationa | Nucleic acid | KD (M)b | Fold-selectivity for TAR RNAc |
|---|---|---|---|---|---|
| 1 | U1A | ------ | TAR DNA | 145.5 (±46.0) × 10-6 | 7.6 |
| 2 | U1A | Glu19Ala | TAR DNA | 100.1 (±33.4) × 10-6 | 17.0 |
| 3 | U1A | Glu19Phe | TAR DNA | 156.4 (±80.0) × 10-6 | 25.2 |
| 4 | U1A | Glu19Lys | TAR DNA | 12.3 (±2.0) × 10-6 | 2.1 |
| 5 | U1A | Glu19Ser | TAR DNA | 91.9 (±33.4) × 10-6 | 22.4 |
| 6 | U1A | Leu49Ala | TAR DNA | 88.2 (±22.0) × 10-6 | 17.0 |
| 7 | U1A | Leu49Lys | TAR DNA | 19.5 (±2.9) × 10-6 | 5.0 |
| 8 | U1A | Met51Ala | TAR DNA | 178.7 (±37.5) × 10-6 | 36.5 |
| 9 | U1A | Met51Lys | TAR DNA | 54.3 (±7.9) × 10-6 | 7.1 |
| 10 | ΔK50ΔM51 | Glu19Lys | TAR DNA | 29.8 (±5.8) × 10-6 | 4.7 |
| 11 | ΔK50ΔM51 | Glu19Ser | TAR DNA | 59.9 (±14.2) × 10-6 | 8.6 |
| 12 | U1A | Glu19Ala | U1hpII RNA | d2.6 (±0.2) × 10-9 | <1 |
| 13 | U1A | Glu19Phe | U1hpII RNA | 36.1 (±6.8) × 10-6 | 5.8 |
| 14 | U1A | Glu19Ser | U1hpII RNA | 11.4 (±1.6) × 10-6 | 2.8 |
| 15 | U1A | Leu49Ala | U1hpII RNA | <1 × 10-8 | <1 |
| 16 | U1A | Met51Ala | U1hpII RNA | e2.3 (±0.2) × 10-10 | <1 |
| 17 | U1A | Glu19Ser:Tyr13Gln | TAR DNA | >250 | >24 |
Single residue mutants are derived from native U1A (entries 1-8 and 11-15) or ΔK50ΔM51 (entries 9-10 and 16-17) scaffolds.
The error for each reported dissociation constant (KD) is the standard deviation of three separate experiments.
Fold-selectivity for TAR RNA was calculated by (KD off-target nucleic acid/KD TAR RNA).
The binding affinity between this mutant and U1hpII RNA has previously been reported as shown in reference 41.
The binding affinity between this mutant and U1hpII RNA has previously been reported as shown in reference 43.
Synthetic U1A Proteins Selectively Bind TAR RNA Over U1hpII Hairpin RNA
Many groups have shown that U1A has exceptional affinity for U1hpII (KD ∼10-11 M). A serious challenge exists when attempting to repurpose the U1A scaffold to identify mutants with affinity for therapeutically relevant RNAs. Given the incredibly high affinity between native U1A and U1hpII, an obvious concern is that U1A mutants will still bind U1hpII with exceptional affinity. However, researchers have previously shown that a single mutation to U1A can dramatically lower affinity for U1hpII.(32, 43) Many of the single mutations capable of suppressing or abrogating affinity for U1hpII reside within the regions of U1A that are the focus of this work. When combined with our preliminary findings, these data suggest that a single mutation to U1A might result in affinity for a new RNA sequence (such as TAR RNA) and dramatically lowered affinity for U1hpII. Impressively, some of the TAR RNA-binding synthetic RRMs we identified preferentially recognize TAR RNA over U1hpII hairpin RNA. In particular, U1A Glu19Phe and Glu19Ser exhibit 5.8- and 2.8-fold selectivity for TAR RNA over U1hpII, respectively (Table 3, entries 13 and 14). In contrast, however, U1A Glu19Ala, U1A Leu49Ala, and U1A Met51Ala selectively bound U1hpII hairpin RNA over TAR RNA.
Researchers have previously shown that Asn15Val, Glu19Ala, Tyr13Gln or Phe56Ala single mutations to U1A dramatically lower affinity for U1hpII.(42, 43) We hypothesized that combining “TAR RNA binding” and “U1hpII turn off” mutations may result in U1A double mutants with improved selectivity for TAR RNA over U1hpII, compared to the single mutants from which there were generated. Starting from the five U1A single mutants with the highest TAR RNA:TAR DNA binding selectivity, we prepared seventeen double mutants (see Table 4). The binding affinity of each of these double mutants for TAR RNA was measured by fluorescence polarization. In order to maintain focus on the best TAR-RNA binding proteins, those with a TAR KD >15 μM were removed from the pool, and their selectivity over U1hpII was not determined. For the majority of proteins we tested, combining “TAR RNA binding” and “U1hpII turn off” mutations appreciably lowered affinity for TAR RNA (KD >15 μM). However, we identified 5 double mutants that selectively bound TAR RNA over U1hpII. These proteins, and their selectivity for TAR RNA over U1hpII RNA (in parenthesis) are as follows: U1A Glu19Ala:Asn15Val (3.4); U1A Glu19Ser:Tyr13Gln (6.2); U1A Glu19Ser:Asn15Val (1.3); U1A Leu49Ala:Glu19Ala (1.7); and, U1A Met51Ala:Glu19Ala (1.1) (Table 4, entries 2, 7, 8, 12, and 16, respectively). Taken together, these data show that while some residues critical to U1hpII binding can be mutated in such a way as to enable affinity for TAR RNA, mutations which are deleterious to the U1A-U1hpII complex may not dramatically effect the U1A-TAR RNA complex, presumably due to appreciable differences in the structures of these complexes.
From the single and double mutant data we have found that U1A Glu19Ser, U1A Glu19Phe and U1A Glu19Ser:Tyr13Gln exhibit the best TAR RNA binding affinity and selectivity profiles. These three proteins bind TAR RNA with a dissociation constant of 4.1, 6.2 and 10.3 μM, respectively (Table 2, entries 2 and 4, and Table 4, entry 7). In addition, these proteins were found to preferentially bind TAR RNA over TAR DNA with 22.9-, 25.2- and >24-fold selectivity, respectively (Table 3, entries 5, 3, and 17). Finally, in addition to identifying mutations that confer good affinity to TAR RNA, mutants we identified show significantly lowered affinity for the cognate RNA. These three proteins bind TAR RNA with 2.7, 5.8- and 6.2-fold selectivity over U1hpII (Table 3, entries 14 and 13; Table 4, entry 7, respectively).
Synthetic U1A Proteins Selectively Bind TAR RNA in 10 Molar Excess tRNA
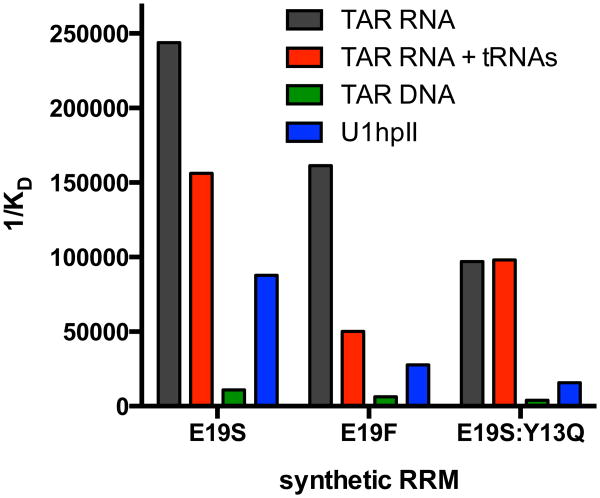
As a final assessment of selectivity, we measured binding affinities for complexes involving TAR RNA and U1A Glu19Ser, U1A Glu19Phe or U1A Glu19Ser:Tyr13Gln in the presence of 10-fold molar excess of cellular tRNAs from E.coli. In previously reported cases, addition of super-stoichiometric ratios of tRNAs has been shown to dramatically affect binding between folded RNAs and previously reported RNA-binding small molecules.(55) As previously mentioned, we hypothesized that since U1A evolved function (selective recognition of a particular RNA hairpin) in a virtual sea of nucleic acids, including a relatively high molarity of tRNAs, it might exhibit inherent selectivity for a particular RNA hairpin over these nucleic acids RNAs. Consistent with this hypothesis, addition of cellular tRNAs did not dramatically effect the affinity of these new protein – RNA interactions. Most impressively, the binding affinity between U1A Glu19Ser and TAR RNA was only lowered ∼1.6-fold in the presence of 10 molar equivalents of cellular tRNAs. Identical conditions did not appreciably affect binding between Glu19Ser:Tyr13Gln and TAR RNA. The binding affinities and selectivities for our three best mutants is summarized in Figure 3. Taken together, these findings demonstrate the relatively high level of selectivity for TAR RNA that is obtained by these synthetic RRMs.
Figure 3.
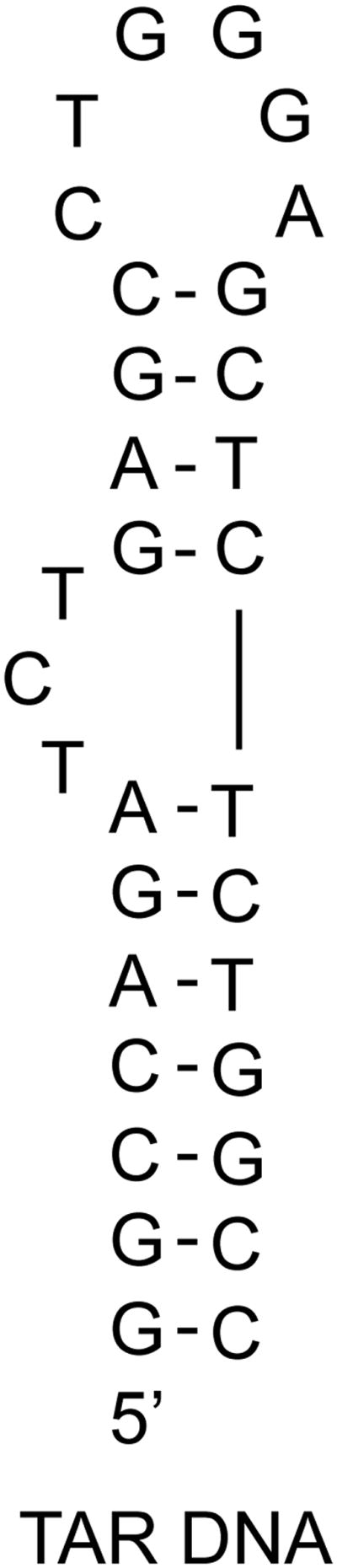
Sequence and proposed secondary structure of TAR DNA used in this work, which is analogous to TAR RNA.
Conclusions
Recent studies suggest that only a small percentage of the human proteome is susceptible to small molecule-dependent regulation.(57) In addition, the target diversity of current small molecule therapeutics is low. For example, ∼40% of current FDA approved drugs target a single class of protein target: G-protein coupled receptors (GPCRs).(57) The fundamental limitation of small-molecule reagents is encoded in the name itself: the molecules are small, and thus intrinsically challenged by extended contact surfaces found at many biologically important interfaces, including RNA. By virtue of this fact, there is a strong need to develop alternative approaches to recognition, and potentially therapeutic discovery. This includes those approaches that lie outside the paradigm in which proteins are the only molecular recognition and drug discovery targets. Additionally, the hyperfocus on a subset of protein targets highlights the need for new paradigms in target identification, especially those encompassing cellular components beyond the proteome.
The seemingly continuous emergence of complex and crucial new roles for RNA in cellular function and disease make it an attractive target for molecular recognition, therapeutic intervention, and biochemical probe development. Such vital efforts face a daunting bottleneck in the scarcity of methods for selective recognition of folded RNAs, a task generally beyond the scope of traditional small molecules. Inspired by selective recognition of a particular DNA sequence(58), which generally evades solution by small molecules, researchers have recently turned to medium-sized molecules (MW ∼1-5 kDa) for selective recognition of RNA. An alternative to this approach is the development of tailored RNA-binding proteins capable of selectively recognizing RNAs of therapeutic interest. Seminal studies on ssRNA-binding proteins such as Pumilio/fem-3-binding factor (PUF) repeat domains and pentatricopeptide repeat (PPR) proteins have resulted in the formation of a “recognition code” for ssRNAs.(25) However, most RNAs exist as folded structures. Therefore, an important consideration for the design of new RNA-binding proteins is recognition of folded RNAs.
The development of new RNA-binding small and medium-sized molecules is slowed by a modest understanding of the molecular requirements for achieving sequence-selective RNA recognition. In contrast, Nature has evolved a library of RNA-binding proteins, including the RNA Recognition Motif (RRM).(23) U1A is a well-studied RRM that binds U1hpII, an RNA hairpin with a 10-nucleotide loop. Mutation of putative RNA-binding regions within U1A can result in altered RNA selectivity.(32, 43, 47) Thus far, however, researchers have only examined RNAs with high sequence homology to U1hpII. We questioned if U1A and/or a U1A mutant we, and others, have studied (ΔK50ΔM51) might serve as a starting point for the generation of synthetic proteins that selectively bind a disease-relevant RNA hairpin that does not share significant sequence homology with U1hpII. In this initial report, we have shown that as few as 1-2 mutations to the putative RNA-binding regions within U1A or ΔK50ΔM51 can result in synthetic RRMs that bind TAR RNA, an HIV-relevant RNA hairpin that does not share appreciable sequence or structural homology with U1hpII. Impressively, some of these new synthetic RRMs bind TAR RNA with single-digit micromolar dissociation constants, and do so preferentially over the native protein's original target RNA (U1hpII) and a DNA TAR variant. For two TAR-binding proteins, binding affinity is not appreciably diminished by addition of 10 molar equivalents of cellular tRNAs from E. coli.
Taken together, our findings support the hypothesis that U1A and variants thereof can serve as privileged scaffolds for the generation of new RNA-binding proteins that selectively recognize a disease-relevant RNA hairpin, which does not share appreciable sequence or structural homology with U1hpII. Our findings suggest that U1A and U1A-derived proteins have inherent selectivity for a particular RNA hairpin over the analogous DNA and tRNAs. By virtue of this unique feature, U1A overcomes challenges faced by some previously reported RNA-binding small and medium-sized molecules. The best TAR RNA-binding synthetic RRMs described in this work likely represent very useful starting points for protein evolution experiments aimed to evolve RRMs with dramatically improved TAR RNA affinity and selectivity. Efforts toward this end are currently underway and will be reported in due course.
Experimental Procedures
General Information: 5′-fluorescein tagged nucleic acids were purchased from Integrated DNA Technologies (ITD). LB Miller's broth, bacterial agar, nickel resin and black 384-well polystyrene plates were purchased from Fisher. Chemically competent BL21(DE3) E.coli, and Q5 DNA polymerase were purchased from New England Biolabs. Carbenicillin and IPTG were purchased from Gold Biotechnologies. 15% and 7% TBE Ready Gel precast gels were purchased from Bio-Rad. Pfu Turbo DNA polymerase was purchased from Agilent technologies. Gel imaging was performed on a Gel Doc XR+ system (Bio-Rad). All protein mutants were expressed in E. coli and purified by nickel-EDTA column chromatography. Nucleic acids were purchased from IDT as RNAse free HPLC purified pellets. Nucleic acids were purchased equipped with a 5′-fluorescein and 6 carbon linker. Total cellular tRNAs from E.coli were purchased from Sigma-Aldrich. The sequences of nucleic acids used in this work are as follows: TAR RNA: 5′-GGCAGAUCUGAGCCUGGGAGCUCUCUGCC-3′; TAR DNA: 5′-GGCAGATCTGAGCCTGGGAGCTCTCTGCC-3′; U1hpII RNA: 5′-AGCTTATCCATTGCACCGGATAAGCT-3′. Fluorescence Polarization measurements: Were performed and analyzed as previously described.(46)
Supplementary Material
Figure 4.
Binding affinities for the three best synthetic RNA Recognition Motifs (U1A E19S, U1A E19F, and U1A E19S:Y13Q) for TAR RNA (black), TAR RNA and 10 molar equivalents of total tRNA from E. coli (red), TAR DNA (green), U1hpII RNA (blue). Binding affinities are shown as reciprocal dissociation constant (1/KD).
Acknowledgments
This work was supported in part by institutional funds provided by Colorado State University and the Colorado Center for Drug Discovery. Plasmids encoding U1A or U1A ΔK50ΔM51 were generously provided by Professor Ite Laird-Offringa (University of Southern California). We Acknowledge Professor Olve Peerson for use of fluorescence polarization instrumentation.
Footnotes
Supporting Information. Detailed experimental procedures, fluorescence polarization data, PAGE characterization of TAR RNA-binding synthetic RNA Recognition Motifs (RRMs). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osborne RJ, Thornton CA. RNA-dominant diseases. Human Molecular Genetics. 2006;15:R162–169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M. Non-coding RNAs in human disease. Nature Rev Gen. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 4.Gallego J, Varani G. Targeting RNA with small-molecule drugs: therapeutic promise and chemical challenges. Acc Chem Res. 2001;34:836–843. doi: 10.1021/ar000118k. [DOI] [PubMed] [Google Scholar]
- 5.Aboul-ela F. Strategies for the design of RNA-binding small molecules. Future Med Chem. 2010;2:93–119. doi: 10.4155/fmc.09.149. [DOI] [PubMed] [Google Scholar]
- 6.Hermann T, Tor Y. RNA as a target for small-molecule therapeutics. Expert Opin Ther Pat. 2005;15:49–62. [Google Scholar]
- 7.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 8.Pushechnikov A, Lee MM, Childs-Disney JL, Sobczak K, French JM, Thornton CA, Disney MD. Rational Design of Ligands Targeting Triplet Repeating Transcripts That Cause RNA Dominant Disease: Application to Myotonic Muscular Dystrophy Type 1 and Spinocerebellar Ataxia Type 3. J Am Chem Soc. 2009;131:9767–9779. doi: 10.1021/ja9020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee MM, Pushechnikov A, Disney MD. Rational and Modular Design of Potent Ligands Targeting the RNA That Causes Myotonic Dystrophy 2. ACS Chem Biol. 2009;4:345–355. doi: 10.1021/cb900025w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Childs-Disney JL, Wu ML, Pushechnikov A, Aminova O, Disney MD. A small molecule microarray platform to select RNA internal loop-ligand interactions. ACS Chem Biol. 2007;2:745–754. doi: 10.1021/cb700174r. [DOI] [PubMed] [Google Scholar]
- 11.Gareiss PC, Sobczak K, McNaughton BR, Palde PB, Thornton CA, Miller BL. Dynamic Combinatorial Selection of Molecules Capable of Inhibiting the (CUG) Repeat RNA-MBNL1 Interaction In Vitro: Discovery of Lead Compounds Targeting Myotonic Dystrophy (DM1) J Am Chem Soc. 2008;130:16254–16261. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNaughton BR, Gareiss PC, Miller BL. Identification of a selective small-molecule ligand for HIV-1 frameshift-inducing stem-loop RNA from an 11,325 member resin bound dynamic combinatorial library. J Am Chem Soc. 2007;129:11306–+. doi: 10.1021/ja072114h. [DOI] [PubMed] [Google Scholar]
- 13.Palde PB, Ofori LO, Gareiss PC, Lerea J, Miller BL. Strategies for Recognition of Stem-Loop RNA Structures by Synthetic Ligands: Application to the HIV-1 Frameshift Stimulatory Sequence. J Med Chem. 2010;53:6018–6027. doi: 10.1021/jm100231t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer ST, Hergenrother PJ. Small Molecule Ligands for Bulged RNA Secondary Structures. Org Lett. 2009;11:4052–4055. doi: 10.1021/ol901478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas JR, Liu XJ, Hergenrother PJ. Size-specific ligands for RNA hairpin loops. J Am Chem Soc. 2005;127:12434–12435. doi: 10.1021/ja051685b. [DOI] [PubMed] [Google Scholar]
- 16.Liu XJ, Thomas JR, Hergenrother PJ. Deoxystreptamine dimers bind to RNA hairpin loops. J Am Chem Soc. 2004;126:9196–9197. doi: 10.1021/ja048936l. [DOI] [PubMed] [Google Scholar]
- 17.McCoy LS, Roberts KD, Nation RL, Thompson PE, Velkov T, Li J, Tor Y. Polymyxins and Analogues Bind to Ribosomal RNA and Interfere with Eukaryotic Translation in Vitro. ChemBioChem. 2013;14:2083–2086. doi: 10.1002/cbic.201300496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaur M, Rupasinghe CN, Klosi E, Spaller MR, Chow CS. Selection of heptapeptides that bind helix 69 of bacterial 23S ribosomal RNA. Bioorgan Med Chem. 2013;21:1240–1247. doi: 10.1016/j.bmc.2012.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, Duc ACE, Klosi E, Pattabiraman S, Spaller MR, Chow CS. Selection of Peptides That Target the Aminoacyl-tRNA Site of Bacterial 16S Ribosomal RNA. Biochemistry. 2009;48:8299–8311. doi: 10.1021/bi900982t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong CH, Fu Y, Ramisetty SR, Baranger AM, Zimmerman SC. Selective inhibition of MBNL1-CCUG interaction by small molecules toward potential therapeutic agents for myotonic dystrophy type 2 (DM2) Nucleic Acids Res. 2011;39:8881–8890. doi: 10.1093/nar/gkr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryson DI, Zhang WY, McLendon PM, Reineke TM, Santos WL. Toward Targeting RNA Structure: Branched Peptides as Cell-Permeable Ligands to TAR RNA. ACS Chem Biol. 2012;7:210–217. doi: 10.1021/cb200181v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maris C, Dominguez C, Allain FHT. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 23.Clery A, Blatter M, Allain FHT. RNA recognition motifs: boring? Not quite. Curr Opin Struc Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Varani G. RNA-protein intermolecular recognition. Acc Chem Res. 1997;30:189–195. [Google Scholar]
- 25.Chen Y, Varani G. Engineering RNA-binding proteins for biology. FEBS J. 2013;280:3734–3754. doi: 10.1111/febs.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Wang ZF, Hall TMT. Engineered proteins with Pumilio/fem-3 mRNA binding factor scaffold to manipulate RNA metabolism. FEBS J. 2013;280:3755–3767. doi: 10.1111/febs.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Varani G. Finding the Missing Code of RNA Recognition by PUF Proteins. Chem Biol. 2011;18:821–823. doi: 10.1016/j.chembiol.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Kortemme T, Robertson T, Baker D, Varani G. A new hydrogen-bonding potential for the design of protein-RNA interactions predicts specific contacts and discriminates decoys. Nucleic Acids Res. 2004;32:5147–5162. doi: 10.1093/nar/gkh785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ban T, Ke J, Chen R, Gu X, Tan MH, Zhou XE, Kang Y, Melcher K, Zhu JK, Xu HE. Structure of a PLS-class pentatricopeptide repeat protein provides insights into mechanism of RNA recognition. J Biol Chem. 2013;288:31540–31548. doi: 10.1074/jbc.M113.496828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin P, Li Q, Yan C, Liu Y, Liu J, Yu F, Wang Z, Long J, He J, Wang HW, Wang J, Zhu JK, Shi Y, Yan N. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013;504:168–171. doi: 10.1038/nature12651. [DOI] [PubMed] [Google Scholar]
- 31.Allain FHT, Howe PWA, Neuhaus D, Varani G. Structural basis of the RNA-binding specificity of human U1A protein. EMBO J. 1997;16:5764–5774. doi: 10.1093/emboj/16.18.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal-Structure at 1.92 Angstrom Resolution of the Rna-Binding Domain of the U1a Spliceosomal Protein Complexed with an Rna Hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 33.Hall KB. Interaction of Rna Hairpins with the Human U1a N-Terminal Rna-Binding Domain. Biochemistry. 1994;33:10076–10088. doi: 10.1021/bi00199a035. [DOI] [PubMed] [Google Scholar]
- 34.Kranz JK, Hall KB. RNA recognition by the human U1A protein is mediated by a network of local cooperative interactions that create the optimal binding surface. J Mol Biol. 1999;285:215–231. doi: 10.1006/jmbi.1998.2296. [DOI] [PubMed] [Google Scholar]
- 35.Hall KB, Stump WT. Interaction of N-Terminal Domain of U1a Protein with an Rna Stem Loop. Nucleic Acids Res. 1992;20:4283–4290. doi: 10.1093/nar/20.16.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kormos BL, Benitex Y, Baranger AM, Beveridge DL. Affinity and specificity of protein U1A-RNA complex formation based on an additive component free energy model. J Mol Biol. 2007;371:1405–1419. doi: 10.1016/j.jmb.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kormos BL, Baranger AM, Beveridge DL. Protein-RNA recognition: Insight into U1A-RNA from molecular dynamics simulations. J Biomol Struct Dyn. 2007;24:694–694. [Google Scholar]
- 38.Law MJ, Lee DS, Lee CS, Anglim PP, Haworth IS, Laird-Offringa IA. The role of the C-terminal helix of U1A protein in the interaction with U1hpII RNA. Nucleic Acids Res. 2013;41:7092–7100. doi: 10.1093/nar/gkt326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law MJ, Rice AJ, Lin P, Laird-Offringa IA. The role of RNA structure in the interaction of U1A protein with U1 hairpin II RNA. RNA. 2006;12:1168–1178. doi: 10.1261/rna.75206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katsamba PS, Myszka DG, Laird-Offringa IA. Two functionally distinct steps mediate high affinity binding of U1A protein to U1 hairpin II RNA. J Biol Chem. 2001;276:21476–21481. doi: 10.1074/jbc.M101624200. [DOI] [PubMed] [Google Scholar]
- 41.Law MJ, Linde ME, Chambers EJ, Oubridge C, Katsamba PS, Nilsson L, Haworth IS, Laird-Offringa IA. The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Res. 2006;34:275–285. doi: 10.1093/nar/gkj436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Law MJ, Chambers EJ, Katsamba PS, Haworth IS, Laird-Offringa IA. Kinetic analysis of the role of the tyrosine 13, phenylalanine 56 and glutamine 54 network in the U1A/U1 hairpin II interaction. Nucleic Acids Res. 2005;33:2917–2928. doi: 10.1093/nar/gki602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan Y. Ph D. University of Illinois; 2009. Exploring Protein-RNA Interactions with Site-Directed Mutagenesis and Phage Display. [Google Scholar]
- 44.Williams DJ, Hall KB. RNA hairpins with non-nucleotide spacers bind efficiently to the human U1A protein. J Mol Biol. 1996;257:265–275. doi: 10.1006/jmbi.1996.0161. [DOI] [PubMed] [Google Scholar]
- 45.Katsamba PS, Bayramyan M, Haworth IS, Myszka DG, Laird-Offringa IA. Complex role of the beta(2)-beta(3) loop in the interaction of U1A with U1 hairpin II RNA. J Biol Chem. 2002;277:33267–33274. doi: 10.1074/jbc.M200304200. [DOI] [PubMed] [Google Scholar]
- 46.Blakeley BD, Shattuck J, Coates MB, Tran E, Laird-Offringa IA, McNaughton BR. Analysis of Protein-RNA Complexes Involving a RNA Recognition Motif Engineered To Bind Hairpins with Seven- and Eight-Nucleotide Loops. Biochemistry. 2013;52:4745–4747. doi: 10.1021/bi400801q. [DOI] [PubMed] [Google Scholar]
- 47.Laird-Offringa IA, Belasco JG. Analysis of RNA-binding proteins by in vitro genetic selection: identification of an amino acid residue important for locking U1A onto its RNA target. Proc Natl Acad Sci, USA. 1995;92:11859–11863. doi: 10.1073/pnas.92.25.11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laird-Offringa IA. In vitro genetic analysis of RNA-binding proteins using phage display. Methods in Molecular Biology. 1999;118:189–216. doi: 10.1385/1-59259-676-2:189. [DOI] [PubMed] [Google Scholar]
- 49.Aboulela G, Karn J, Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge (vol 24, pg 3974, 1996) Nucleic Acids Res. 1996;24:4598–4598. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-Termination of Transcription within the Long Terminal Repeat of HIV-1 by Tat Gene-Product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 51.Bonnard V, Pascale L, Azoulay S, Di Giorgio A, Rogez-Kreuz C, Storck K, Clayette P, Patino N. Polyamide Amino Acids trimers as TAR RNA ligands and anti-HIV agents. Bioorgan Med Chem. 2010;18:7432–7438. doi: 10.1016/j.bmc.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 52.Blount KF, Tor Y. A tale of two targets: Differential RNA selectivity of nucleobase-aminoglycoside conjugates. ChemBioChem. 2006;7:1612–1621. doi: 10.1002/cbic.200600109. [DOI] [PubMed] [Google Scholar]
- 53.Velagapudi SP, Pushechnikov A, Labuda LP, French JM, Disney MD. Probing a 2-Aminobenzimidazole Library for Binding to RNA Internal Loops via Two-Dimensional Combinatorial Screening. ACS Chem Biol. 2012;7:1902–1909. doi: 10.1021/cb300213g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mammen M, Choi SK, Whitesides GM. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Edit. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 55.Arambula JF, Ramisetty SR, Baranger AM, Zimmerman SC. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc Natl Acad Sci, USA. 2009;106:16068–16073. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson WD, Ratmeyer L, Zhao M, Strekowski L, Boykin D. The Search for Structure-Specific Nucleic-Acid Interactive Drugs - Effects of Compound Structure on Rna Versus DNA Interaction Strength. Biochemistry. 1993;32:4098–4104. doi: 10.1021/bi00066a035. [DOI] [PubMed] [Google Scholar]
- 57.Overington JP, Al-Lazikani B, Hopkins AL. Opinion - How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 58.Dervan PB. Molecular recognition of DNA by small molecules. Bioorgan Med Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.