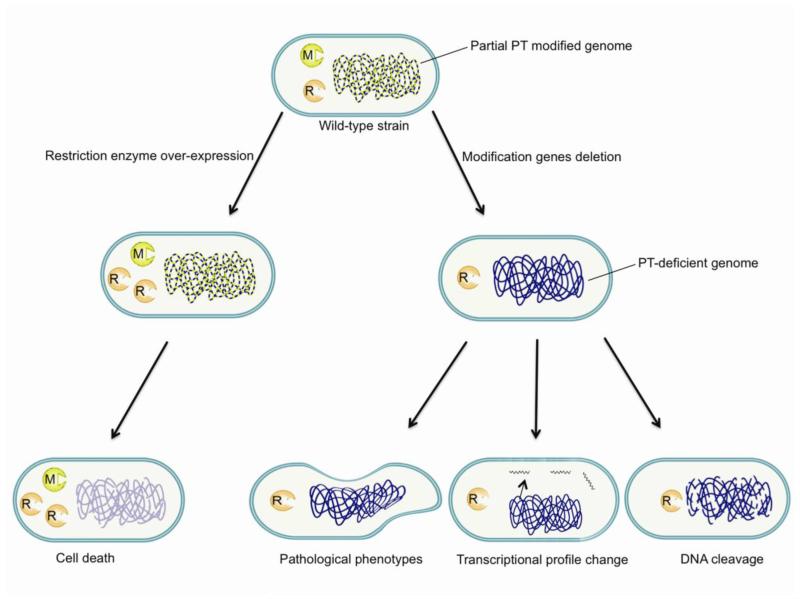
Summary
Prokaryotes protect their genomes from foreign DNA with a diversity of defense mechanisms, including a widespread restriction-modification (R-M) system involving phosphorothioate (PT) modification of the DNA backbone. Unlike classical R-M systems, highly partial PT-modification of consensus motifs in bacterial genomes suggests an unusual mechanism of PT-dependent restriction. In Salmonella enterica, PT modification is mediated by four genes dptB-E, while restriction involves additional three genes dptF-H. Here, we performed a series of studies to characterize the PT-dependent restriction, and found that it presented several features distinct with traditional R-M systems. The presence of restriction genes in a PT-deficient mutant was not lethal, but instead resulted in several pathological phenotypes. Subsequent transcriptional profiling revealed the expression of >600 genes was affected by restriction enzymes in cells lacking PT, including induction of bacteriophage, SOS response and DNA repair-related genes. These transcriptional responses are consistent with the observation that restriction enzymes caused extensive DNA cleavage in the absence of PT modifications in vivo. However, over-expression of restriction genes was lethal to the host in spite of the presence PT modifications. These results point to an unusual mechanism of PT-dependent DNA cleavage by restriction enzymes in the face of partial PT modification.
Introduction
Prokaryotes have evolved numerous systems to defend against foreign DNA, such as toxin-antitoxin (TA) pairs (Gerdes et al., 2005), abortive infection (Abi) (Chopin et al., 2005), clustered regularly interspaced short palindromic repeats-CRISPR-associated proteins (CRISPR-Cas) (Datsenko et al., 2012), and silencing by the histone-like nucleoid structuring (H-NS) proteins (Lucchini et al., 2006; Navarre et al., 2006). The best characterized of these defense systems are the methylation-based restriction-modification (R-M) systems, in which a DNA methyltransferase modifies the host DNA in a specific pattern while a restriction enzyme recognizes and cleaves unmodified foreign DNA (Wilson and Murray, 1991). Among the tremendous diversity of Type I-III R-M systems, all known DNA modifications were thought to occur on the nucleobases until our recent discovery of phosphorothioate (PT) modifications of the DNA backbone, in which a non-bridging phosphate oxygen is replaced with sulfur (Wang et al., 2007).
Modification of DNA with sequence- and stereo-specific PT involves a complex biosynthetic pathway requiring four horizontally-transferred genes termed dndA, dndC, dndD and dndE in Streptomyces lividans 66, with gene nomenclature based on the DNA degradation (dnd) phenotype observed during electrophoresis of genomic DNA from bacteria possessing the dnd gene cluster (Zhou et al., 2005). Both the PT modifications and the dnd genes are distributed in a wide range of bacterial and archaeal genomes (Ou et al., 2009; Wang et al., 2011). In terms of function, the protein product of dndB is predicted to be a transcriptional regulator (Zhou et al., 2005) and its absence leads to increased levels of PT modifications (Wang et al., 2007; Wang et al., 2011). DndA acts as a cysteine desulfurase and assembles DndC a 4Fe-4S cluster protein (You et al., 2007), with some bacteria replacing DndA with a homologous cysteine desulfurase activity, such as IscS in Escherichia coli (An et al., 2012).DndC possesses ATP pyrophosphatase activity (Zhou et al., 2005), while a DndD homologue in Pseudomonas fluorescens Pf0-1, SpfD, has ATPase activity that is possibly related to DNA structure alteration or nicking during PT incorporation (Yao et al., 2009). DNA nicking activity may also be associated with E. coli DndE (Hu et al., 2012). Many bacterial strains have been found to possess the dnd homologs, including wide-ranging genera from diverse habitats, such as soil-dwelling and marine species, aerobic and anaerobic microbes, and human saprophytes and pathogens (Ou et al., 2009).
Recent evidence points to host defense as a major function for PT modifications. We recently demonstrated that PT modifications protect Salmonella enterica Serovar Cerro 87 against unmodified plasmid DNA during transformation, albeit with a lower efficiency compared to the traditional R-M system (Xu et al., 2010). In S. enterica, a gene cluster homologous to dndB-E, namely dptB-E, is required for PT modification, with an endogenous cysteine desulfurase homologous to E. coli iscS replacing dndA (An et al., 2012). The PT-dependent restriction phenotype in S. enterica is conferred by an additional 3-gene cluster, named as dptF-H (Xu et al., 2010). The gene clusters of dptB-E and dptF-H are present in a wide variety of bacteria with a similar genomic organization (Xu et al., 2010), which suggests widespread use of PT modifications-dependent R-M system in prokaryotes.
A highly unusual feature of the PT-based R-M system is the fact that the only apparent consensus sequence for the bistranded modification sites in Escherichia coli B7A, which shares dnd/dpt genes that are highly homologous to S. enterica, involves GPTAAC/GPTTTC and only 12% of these sites contain a PT modification (Cao et al., 2014). This partial modification phenomenon led us to pursue a series of experiments to characterize the dptF-H PT-dependent restriction system, with the discovery of several unusual features that differ from classical R-M systems.
Results
Phenotypic changes caused by loss of dptB-E in the face of active dptF-H
To understand the PT-dependent restriction mechanism, we undertook studies to define the effects of expression of the dptF-H restriction genes in S. enterica mutants lacking PT modifications (ΔdptB-E). Interestingly, unlike many restriction systems, the presence of dptF-H in the PT-deficient mutant was not lethal, but resulted in a range of abnormal phenotypes. All of the pathological phenotypes were resolved by either additional deletion of dptF-H or by complementation of the mutant strain with a dptB-E PT modification gene cluster-containing plasmid. A summary of the colony and cell morphology and growth phenotypes altered by dptF-H in the absence of dptB-E is shown in Figure 1.
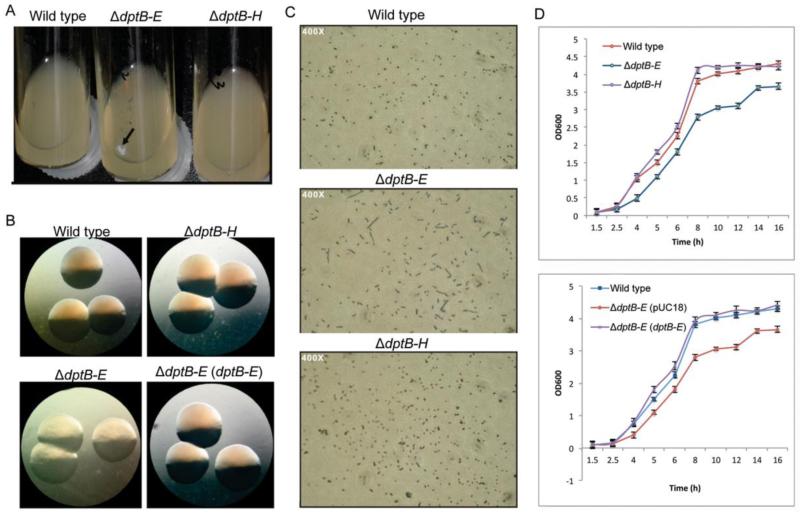
Figure 1.
Phenotypic alterations in the ΔdptB-E mutant. (A) White mucoid material was observed in ΔdptB-E mutant grown in liquid culture, but not ΔdptB-H. (B) Altered colony morphology on the solid media, with recovery provided by the dptB-E-containing plasmid (pJTU1238). (C) Microscopic analysis of cell mobility and division. (D) Growth rate curves in liquid culture.
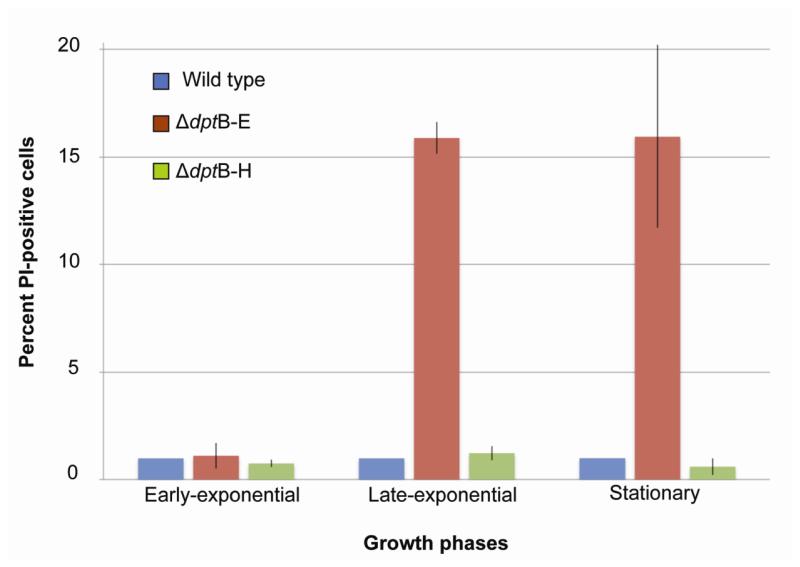
One feature of the growth of the ΔdptB-E mutant was the accumulation of a white mucoid material during late-exponential phase in liquid culture (Fig. 1A). In terms of macroscopic phenotypes, the ΔdptB-E mutant displayed altered colony morphology on solid medium, with a faint yellow color and a rougher surface, suggesting defective growth (Fig. 1B). When grown in liquid culture, microscopic analysis revealed an elongated morphology for the mutant cells, which is consistent with failed cytokinesis (Fig. 1C). Meanwhile, ΔdptB-E grew at a slower rate than the wild-type strain and ΔdptB-H mutant (Fig. 1D). Based on these changes and on a proteomic analysis of mucoid material apparent late-exponential phase in liquid culture (Fig. 1A, 1B), which showed enrichment in elongation factors, RecA, ribosomal proteins, and outer membrane proteins (Table S1), we assessed the membrane integrity of ΔdptB-E mutant by propidium iodide (PI) fluorescence flow cytometry (Gregori et al., 2001; Su et al., 2009). This approach is based on the fact that DNA-binding PI is excluded from cells with intact membranes, while cells with damaged membranes allow entry and DNA-binding by PI, resulting in an increased cellular fluorescence that can be quantified by flow cytometry. Both the wild-type strain and the ΔdptB-H mutant showed identical levels PI fluorescence and no changes as a function of cell state, as shown in Figure 2 (representative flow cytometry histograms are shown in Fig. S1). In contrast, ΔdptB-E showed a shift to higher levels of PI fluorescence (~16%) during late-exponential and stationary phase (Fig. 2).
Figure 2.
Analysis of membrane defects in the ΔdptB-E mutant at different growth phases. Membrane permeability was quantified by PI staining and flow cytometry, as described in Materials and Methods. Data represent mean ±SD for three biological replicates.
Effects of unrestrained PT-dependent restriction activity on gene expression
To further explore the observations of abnormal phenotypes of the PT-deficient mutant, we performed comprehensive transcriptional profiling to identify changes in gene expression caused by loss of dptB-E in S. enterica. In this study, we used 8797 probes that covered all genes of published S. enterica genomes in Genbank, including the seven dpt genes, and three independent biological replicates with cell cultures in early-exponential, late-exponential and stationary phases. The resulting microarray data reflected 5413 genes, which is consistent with other Salmonella strains possessing ~4000-6000 genes (Chiu et al., 2005; Deng et al., 2003; McClelland et al., 2001; Parkhill et al., 2001). With the criterion of at least a ±2-fold change with a P value of ≤0.05 to define significantly up- or down-regulated genes, the filtered data from the ΔdptB-E and ΔdptB-H mutants at each growth phase can be found at the NCBI Gene Expression Omnibus website (Geo Accession GSE59225; http://www.ncbi.nml.nih.gov/geo/).
Based on these statistical parameters, the ΔdptB-H mutant displayed a transcriptional profile similar to the wild-type strain, with 23 and 17 differentially expressed genes at the early- and late-exponential phase, respectively. However, relative to wild-type cells, the ΔdptB-E strain showed 116 genes altered at their early-exponential phase, and 235 and 683 at late-exponential and stationary phase, respectively (Fig. S2). Analysis of these changes in gene expression by hierarchical clustering is shown in Figure 3, from which it is apparent that most of the up-regulated genes in the early-exponential phase were continuously expressed at higher levels throughout the course of growth. Meanwhile, up-regulation of other genes began at the late-exponential phase. Notably, down-regulated genes were significantly outnumbered by up-regulated genes. The microarray data were corroborated by real-time reverse transcription-polymerase chain reaction analysis of six representative genes, including the PT restriction gene dptF, flagellar biosynthesis gene fliH and the four DNA repair genes recA, recN, ruvA and nerE (Fig. S3).
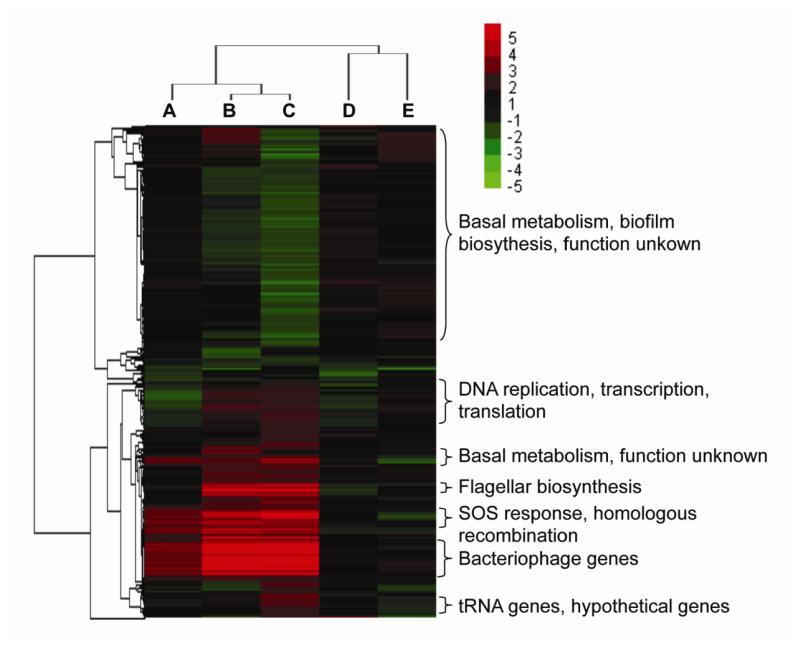
Figure 3.
Hierarchical clustering analysis of microarray data of ΔdptB-E and ΔdptB-H mutants in different growth phases. Clustering was performed on fold-change data for each gene (rows), with green indicating down-regulation and red indicating up-regulation relative to the wild-type strain; the inset shows the color code for the fold-change values. Columns A, B, C represent the ΔdptB-E strain grown at early-exponential, late-exponential and stationary phase, respectively. Columns D and E represent the ΔdptB-H strain at early- and late-exponential phases, respectively.
To gain insight into the functional significance of the dptF-H-dependent changes in gene expression, we then annotated and categorized the significantly changed genes according to the NCBI Clusters of Orthologous Groups (COG) database. Strikingly, one of the most prominent groups of up-regulated genes represented DNA repair pathways, including base-excision repair, homologous recombination, and the SOS response genes (Fig. 3). These genes were induced at the early-exponential phase and increased to a significantly high lever at stationary phase, which is consistent with DNA damage arising from DptF-H in the ΔdptB-E mutant. Furthermore, 31 prophage genes were also induced at early-exponential phase and accumulated to an expression level 100-fold higher than that in the wild-type strain (Fig. 3). The ΔdptB-E mutant also showed up-regulation of genes involved in cell mobility (23 genes), including flagellar biosynthesis; DNA replication, transcription and translation (41 genes); energy production and conversion (39 genes), especially anaerobic metabolism; membrane proteins and colanic acid capsullar biosynthesis (8 genes); as well as 101 genes with unknown function. The down-regulated genes corresponded to basal metabolism (166 genes), cell division (2 genes), biofilm formation (3 genes), and 97 genes of unknown function. These transcriptional changes are consistent with the protein content of the mucoid debri (Table S1), with up-regulation of 30 ribosomal protein genes, an elongation factor, RecA, and 17 outer membrane proteins. For the slowed cell division phenotype, we found that the sulA gene for an SOS-dependent cell division inhibitor was up-regulated by 21-fold, while two cell division genes (ftsA, ftsQ) were down-regulated. Reduction of biofilm formation regulatory gene bssR (yliH) and activation of the lytic bacteriophage are consistent with the rough, less shiny colony morphology (Fig. 1) (Levine, 1957; Park et al., 2007), while genes responsible for cell division suppression, lytic bacteriophage activation and SOS response could cause the slower growth phenotype (Fig. 1).
dptF-H expression in the absence of PT is associated with DNA cleavage in vivo
Given the phenotypic and gene expression evidence for a DNA damage response in the ΔdptB-E mutant strain, we used a derivative of the TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick and labeling) assay to quantify DNA strand breaks in wild-type and mutant S. enterica strains. As a routinely used approach to monitor DNA strand breaks in vivo in both eukaryotic and prokaryotic cells, the TUNEL assay is based on TdT-mediated labeling of the 3′-OH of DNA breaks with fluorescently-labeled dUTP (Loo, 2011; Rohwer and Azam, 2000), here using FITC-dUTP. Single-cell fluorescence analysis was then accomplished by flow cytometry, using PI staining to gate out cell-sized particulate lacking DNA. Three independent samples of the wild-type strain and the ΔdptB-E and ΔdptB-H mutants were collected at three different growth phases for quantification of DNA breaks using the TUNEL assay, with flow cytometrixc analysis of a total of 30,000 events for each sample.
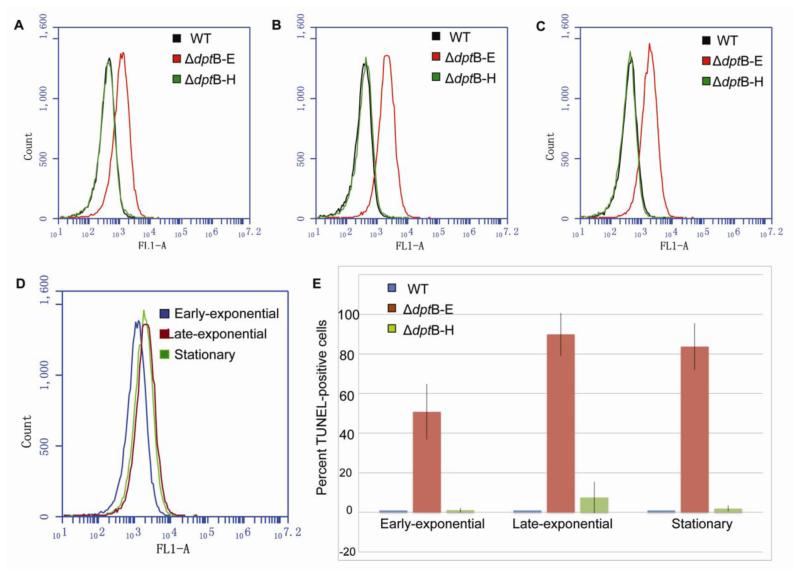
As shown in Figure 4, ΔdptB-E cells exhibited a significantly higher level of TUNEL signal than ΔdptB-H and wild-type strain in all three growth phases, and the level of TUNEL signal kept increasing along the course of its growth. In order to quantitatively analyze the cells containing DNA strand breaks, we defined them as TUNEL-positive cells that showed FITC fluorescent signal exceeding that of 99% of the wide-type cells. With this definition, we found that ~50% of ΔdptB-E cells were observed as TUNEL-positive at early-exponential phase, while the percentage increased to more than 80% when grown to late-exponential and stationary phases. These results indicate that DptF-H induces DNA breaks in the absence of PT modifications, suggesting a PT-dependent, DNA cleavage-associated R-M system.
Figure 4.
TUNEL assay analysis of DptF-H-induced DNA stand-breaks in the dptB-E mutant at early-exponential (A), late-exponential (B) and stationary phase (C). (D) Comparison of TUNEL-positive cells of dptB-E mutant in different growth phases. (E) Percentage of TUNEL-positive cells in dptB-E mutant at different growth phases.
Over-expression of dptF-H is lethal to the host in the presence of PT modifications
Given that dptF, G, and H reside within an operon in Salmonella, these three enzymes are likely to assemble and perform their function as a complex rather than as individual proteins. We then attempted to heterologously express the dptF, G and H operon in E. coli BL21 (DE3), a strain that does not possess dnd/dpt genes. Not surprisingly, the dptFGH-containing plasmid could not be transformed into the host E. coli lacking PT modifications, suggesting a lethal effect to the host by the restriction enzymes in the absence of PT modification. However, over-expression of the restriction gene operon also resulted in a lethal effect (i.e., failure of transformation) in host strains possessing PT modifications, such as wild-type Salmonella or an E. coli strain harbouring a dptB-E-containing plasmid. Considering the partial or incomplete state of PT modifications in bacterial genomes (Cao et al., 2014) and the fact that, in most R-M systems, the presence of the DNA modification prevented DNA cleavage by restriction enzymes, the lethal effects of over-expression of DndF-H restriction enzymes in the presence of a normal complement of PT modifications suggests that the PT modification-based R-M system involves controlled and balanced restriction activity.
Discussion
Of the diverse defense mechanisms in bacteria, R-M systems are the only ones that discriminate foreign DNA directly by modification of self DNA. In most R-M systems, restriction and modification enzymes occur in parallel since the restriction enzyme usually causes lethal effects in the modification-deficient strains (Boyer, 1964; Suri and Bickle, 1985). All known R-M systems are based on DNA nucleobase modifications until the recent discovery of the PT-dependent restriction system in bacteria (Xu et al., 2010). However, unlike traditional R-M systems, each member of the set of all possible PT modification sites in a bacterial cell is not always modified, such that any cell will contain unmodified consensus sites that should be susceptible to restriction enzyme cleavage (Cao et al., 2014). Considering the fact that, in most R-M systems, all possible sites are modified to prevent DNA cleavage by restriction enzymes, the existence of partial modification with PT points to a highly unusual restriction mechanism in the PT R-M system and some kind of regulation of the restriction enzymes in the wild-type strain. This is entirely consistent with our observations that PT-dependent restriction enzymes presented several unusual features in the PT-deficient mutant, as summarized in Figure 5.
Figure 5.
Summary of the effects of PT-dependent restriction enzymes on the host strain.
Both the transcriptional profiling data and evidence for DNA cleavage establish that the PT-based restriction system involves DNA cleavage. It is thus not surprising that expression of restriction enzymes in a PT-deficient S. enterica mutant led to a pathological phenotype, which is best explained as unrestrained DNA cleavage by restriction enzymes in the absence of DNA modifications. This is consistent with similar observations in other R-M systems (Makovets et al., 1999).
The presence of DNA-cleaving restriction activity in cells containing unmodified consensus sites points to some kind of mechanism for controlling the activity of the PT-based restriction system. This is illustrated by the observation that over-expression of the restriction genes is lethal even in a host strain possessing normal levels of PT modifications, which is consistent with lethal DNA cleavage when restriction enzymes are expressed at levels that exceed the capacity of the cell to control their activity. In most R-M systems, all possible consensus sites are modified to prevent cleavage by restriction enzymes. So the toxic consequences of DptF-H over-expression in the presence of PT modifications and the phenomenon of partial modification with PT in wild-type cells suggest some kind of mechanism for regulating restriction activity in the PT R-M system. For example, Clp protease has been shown to control restriction endonuclease activity in a Type I R-M system in E. coli and to prevent lethal DNA cleavage in the absence of DNA modifications (Makovets et al., 1999).
In summary, we have presented evidence that, while traditional and PT-dependent R-M systems share a common function of restriction of unmodified foreign DNA, the PT-dependent restriction enzymes present several unusual features. Deletion of PT modification genes was not lethal to the host strain and caused phenotypic alterations, transcriptional profiling change and DNA damage, with over-expression of restriction enzymes causing lethality even in the presence of the PT modifications. These results provide new insights into an unusual R-M system in bacteria.
Experimental Procedures
Bacterial strains and culture conditions
S. enterica serovar Cerro 87 wild-type strain harbored both the dptB-E gene cluster (modification genes) and the dptF-H (restriction genes); the ΔdptB-E mutant strain was named XTG102 in our previous study (Xu et al., 2010); ΔdptB-E mutants harboring a complementary dptB-E in pUC18 (pJTU1238; Xu et al., 2010), namely ΔdptB-E (dptB-E), and the empty pUC18, namely ΔdptB-E (pUC18), were used for phenotypic description. The ΔdptB-H mutant was constructed with the same strategy used for generation of XTG102, which was based on double-crossover homologous recombination (Xu et al., 2010). Briefly, the left and right arms for homologous recombination to generate ΔdptB-H mutant were amplified by PCR using genomic DNA as the template. PCR primers dptBLL/dptBLR (dptLL: 5′-GACCTCGAGTTGGTTTTCAATA-3′, XhoI underlined; dptLR: 5′-AAGCAACCGTGTCAAGGTAATTGGCGTTGCTGCGTGGTTA-3′) were used for the 752 bp left arm and dptHRR/dptHRL (dptHRR 5′-CCGAGGCTCCAGCATTCACAGTGGTCGCACTAACACTGGA-3′; dptHRL: 5′-CAAGGATCCCAAAGAAATGTTCGCAAC-3′, BamHI) were for right arm. The two 40 bp overlapped arms were ligated by PCR and then cloned into the thermo-sensitive plasmid pKOV-Kan through the designed endonuclease sites. The plasmid construct was then transformed into wild-type S. enterica to generate the ΔdptB-H mutant, which grew on LA plates with15% sucrose at 43 °C (Xu et al., 2010). Cells were grown at 37 °C on solid or liquid Luria-Bertani (LB) medium supplemented with appropriate antibiotics unless otherwise stated. To obtain cells from different growth phases, the overnight bacterial cultures were diluted into 5 ml of LB medium at a concentration of 5000 cells/ml and the cell growth was monitored by the optical density 600 nm (OD600). The cells reached OD600 of 0.8, 2.0 and 3.5 were considered to be at their early-, late-exponential and stationary phase, respectively.
Phenotypic characterization of bacterial strains
For microscopic analysis of the colony morphology on solid media, strains were diluted to an appropriate concentration and grown overnight on LA plates. The colonies forming on the plates during overnight growth were then monitored directly by microscopy. To study cell mobility and division, the strains were grown in liquid LB medium to exponential phase and the morphology was then observed by microscopy following methylene blue staining (400× magnification). To study growth rate, 1,000 cells (based on OD600) of each strain were introduced into 10 ml fresh LB medium and growth was monitored by OD600 every 1-2 hr. After 16 hr, growth curves were plotted based on three independent replicates for each strain.
Proteomic analysis of mucoid material accumulated by ΔdptB-E mutant
To gain insight into the mucoid material produced by ΔdptB-E mutant during late-exponential phase in liquid culture, we performed a proteomic analysis of trypsin-derived peptides using chromatography (HPLC)-coupled tandem mass spectrometry. In this assay, the digestion mixture containing trypsin-digested mucoid material was resolved on an Agilent Zorbax 300SB-C8 column (2.1 × 150 mm, 3.5 μm) with a gradient 5%–95% acetonitrile in 0.1% formic acid over 60 min at a flow of 0.2 ml/min, and then monitored the peptides by Agilent 6410 QTOF MS (4 GHz high resolution, 50–1700 m/z mass range mode, positive ion mode) and MS-MS analysis (30 eV for fragmentation). Agilent MassHunter software was used to analyze the peptide mass values. All the data from MS were submitted to the online peptide database (http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=PMF) to identify the peptides from the digested mucoid material.
RNA isolation and purification
Total RNA from bacteria in different growth phases was extracted using TRIZOL Reagent (Life technologies, Carlsbad, CA, US) following the manufacturer’s instructions and RNA integrity was determined using an Agilent Bioanalyzer 2100 and the resulting RIN number (Agilent technologies, Santa Clara, CA, US). Quantified total RNA was further purified using an RNeasy micro kit (QIAGEN, GmBH, Germany) and RNase-Free DNase Set (QIAGEN, GmBH, Germany).
cDNA synthesis, labeling, and microarray hybridization
Purified total RNA was amplified and labeled using the Low RNA Input Linear Amplification kit (Agilent technologies, Santa Clara, CA, US), 5-(3-aminoallyl)-UTP (Ambion, Austin, TX, US), Cy3 NHS ester (GE healthcare Biosciences, Pittsburgh, PA, US), following the manufacturer’s instructions. Labeled cDNA were purified by RNeasy mini kit (QIAGEN, GmBH, Germany). A designed 8*15K DNA Microarray (Design ID 030218, Agilent), with 8797 probes that covered all the genes of 13 published Salmonella genome, was applied to microarray hybridization. During the hybridization, each slide was hybridized with 1.65 μg of Cy3-labeled cRNA using Gene Expression Hybridization Kit (Agilent technologies, Santa Clara, CA, US) in Hybridization Oven (Agilent technologies, Santa Clara, CA, US), according to the manufacturer’s instructions. After 17 h hybridization, slides were washed in staining dishes (Thermo Shandon, Waltham, MA, US) with Gene Expression Wash Buffer Kit (Agilent technologies, Santa Clara, CA, US), Stabilization and Drying Solution (Agilent technologies, Santa Clara, CA, US), following the manufacturer’s instructions.
Data acquisition and analysis of the microarray
After hybridization, microarray slides were scanned using an Agilent Microarray Scanner (Agilent technologies, Santa Clara, CA, US) and Feature Extraction software 10.7 (Agilent technologies, Santa Clara, CA, US) with default settings: scan resolution, 5 μm; PMT, 100%/10%. For data analysis, signal intensities from each array were normalized using the Quantile algorithm in Gene Spring Software 11.0 (Agilent technologies, Santa Clara, CA, US). Changes in gene expression were expressed as signal log ratios (base 2), and analysis of variance (ANOVA) was used to compare gene expression values in each mutant strain, using wild-type cells as the baseline for comparison. Experiments for array analysis were performed based on biological replicates with three independent experiments for each strain. Transcripts were filtered based on a P value, obtained from t-test by R-software (http://www.r-project.org/), threshold of 0.05 as well as a fold-change threshold of 2. Differentially regulated genes were annotated and categorized according to general function by the NCBI Clusters of Orthologous Groups (COG) database. In addition, Gene Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/) and Java TreeView (http://jtreeview.sourceforge.net/) tools were used to generate the heat map.
Quantitative real-time reverse-transcription PCR (qRT-PCR)
For qRT-PCR, RNA isolation and purification were performed as described above. To synthesize cDNA, 3 μg of purified total RNA was amplified in a 20 μl reaction volume using RevertAid™ H Minus Reverse Transcriptase and Random Hexamer Primer (Fermentas, Inc.). cDNA (50 ng) was used as template for each qPCR experiment using the primers listed in Table S2. The 23S rRNA gene and the 16S rRNA gene were used as multiple internal references. The qPCR reactions were performed according to the protocol of the Maxima® SYBR Green/ROX qPCR Master Mix, and the program was carried out in the Applied Biosystems 7500 Fast Real-Time PCR System. The relative transcript copy number for each target gene was shown as the ratio between the samples in different growth phases and the wild-type controls. It was calculated using the comparative Ct method (Pfaffl et al., 2002). For each gene, the transcript copy number of wild-type strain in 8 h growth phase was assigned a value of 1.0. Accordingly, in the other sample and growth phases, values higher than 2 or lower than 0.5 suggested that the transcription of the target gene was significantly changed. All the quantitative real-time PCR assays were carried out in triplicate for each culture and then repeated three times with RNA isolated from independent cultures.
Membrane integrity test assay
The fluorescent reporter dye propidium iodide (PI) was used to monitor the membrane permeability of the bacterial strains at different growth phase. Three independent biological replicates of each strain were grown as described above and ~107 cells were harvested at each different growth phases. After washing twice with PBS, the cells were incubated in the dark at ambient temperature for 20 min and then analyzed by flow cytometry. The data were collected with the FSC threshold set at 40,000 on a Becton Dickinson Accuri C6 flow cytometer. 30,000 qualified events were collected for each sample with their FL-2 (488 -> 550 nm) values recorded. FlowJo software was then used to analyze fluorescence values. Data analysis of the fluorescence values of PI-stained cells was averaged from three independent biological replicates.
Detection of DNA strand breaks
Three independent biological replicates for each strain were grown as described above, with ~107 cells at different growth phases collected and washed twice with PBS. The cells were then re-suspended in 1 ml of 4% paraformaldehyde and incubated for 30 min on ice. The cells was subsequently collected by centrifugation, washed twice with PBS, and re-suspended in 200 ml of ice-cold PBS. Then 1 ml ice-cold 70% ethanol (v/v, in H2O) was added to each sample and the samples were stored at −20°C overnight for permeabilization. The next day, the cells were collected by centrifuge at 8000 g for 2 min and washed once with PBS. For the TUNEL assay, the Apo-Direct Kit (BD Bioscience) was used to detect the DNA strand-breaks in each strain, according to the manufacturer’s instructions. After staining, the samples were analysed using a Becton Dickinson Accuri C6 flow cytometer. The data were collected using a dual threshold of FSC 40,000 and FL-2 500, with both FL-1 channel (488 nm→510 nm) and FL-2 channel (488 nm→550 nm) recorded, and then analyzed with the FlowJo software. A TUNEL-positive cell was defined as one whose FITC fluorescence intensity exceeded that detected in 99% of the wild-type cells at each growth phase.
Over-expression of the dptF-H operon
The dptF-H operon was derived from the plasmid pJTU3818, which contained both dptB-E and dptF-H gene clusters (Xu et al., 2010). Two strategies were used to clone the dptF-H operon: NsiI digested dptF-H-containing fragment from pJTU3818 was cloned to the PstI (NsiI isocandamer) treated pBluScript SK+ (Life Technologies, NY); or the dptB-E gene cluster in pJTU3818 was removed by ScaI and PvuII partial digestion, with dptF-H left alone in the plasmid. Both plasmid constructs were then transformed into E.coli DH10b, E. coli BL21 or wild-type Salmonella enterica by electroporation (Micropulser, Bio-Rad).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation of China (31170085 and 31070058); the Ministry of Science and Technology (973 and 863 Programs); Shanghai Pujiang Program from the Shanghai Municipal Council of Science and Technology (12PJD021); China Scholarship Council; and the National Science Foundation of the United States (CHE-1019990).
Footnotes
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1111/mmi.12692
References
- An X, Xiong W, Yang Y, Li F, Zhou X, Wang Z, Deng Z, Liang J. A novel target of IscS in Escherichia coli: participating in DNA phosphorothioation. PLoS One. 2012;7:e51265. doi: 10.1371/journal.pone.0051265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. Genetic Control of Restriction and Modification in Escherichia Coli. J Bacteriol. 1964;88:1652–1660. doi: 10.1128/jb.88.6.1652-1660.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Lin K, Zhang Z, Chen L, Shi X, Cao C, Wang Z, Liang J, Deng Z, Wu G. Purification, crystallization and preliminary X-ray analysis of the DndE protein from Salmonella enterica serovar Cerro 87, which is involved in DNA phosphorothioation. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67:1440–1442. doi: 10.1107/S1744309111036694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CH, Tang P, Chu C, Hu S, Bao Q, Yu J, Chou YY, Wang HS, Lee YS. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 2005;33:1690–1698. doi: 10.1093/nar/gki297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin MC, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr Opin Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- Deng W, Liou SR, Plunkett G, 3rd, Mayhew GF, Rose DJ, Burland V, Kodoyianni V, Schwartz DC, Blattner FR. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J Bacteriol. 2003;185:2330–2337. doi: 10.1128/JB.185.7.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Gregori G, Citterio S, Ghiani A, Labra M, Sgorbati S, Brown S, Denis M. Resolution of viable and membrane-compromised bacteria in freshwater and marine waters based on analytical flow cytometry and nucleic acid double staining. Appl Environ Microbiol. 2001;67:4662–4670. doi: 10.1128/AEM.67.10.4662-4670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Wang C, Liang J, Zhang T, Hu Z, Wang Z, Lan W, Li F, Wu H, Ding J, Wu G, Deng Z, Cao C. Structural insights into DndE from Escherichia coli B7A involved in DNA phosphorothioation modification. Cell Res. 22:1203–1206. doi: 10.1038/cr.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Mutations in the temperate phage P22 and lysogeny in Salmonella. Virology. 1957;3:22–41. doi: 10.1016/0042-6822(57)90021-1. [DOI] [PubMed] [Google Scholar]
- Loo DT. In situ detection of apoptosis by the TUNEL assay: an overview of techniques. Methods Mol Biol. 2011;682:3–13. doi: 10.1007/978-1-60327-409-8_1. [DOI] [PubMed] [Google Scholar]
- Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovets S, Doronina VA, Murray NE. Regulation of endonuclease activity by proteolysis prevents breakage of unmodified bacterial chromosomes by type I restriction enzymes. Proc Natl Acad Sci U S A. 1999;96:9757–9762. doi: 10.1073/pnas.96.17.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- Ou HY, He X, Shao Y, Tai C, Rajakumar K, Deng Z. dndDB: a database focused on phosphorothioation of the DNA backbone. PLoS One. 2009;4:e5132. doi: 10.1371/journal.pone.0005132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KH, Song M, Choy HE. Initial characterization of yliH in Salmonella typhimurium. J Microbiol. 2007;45:558–565. [PubMed] [Google Scholar]
- Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O’Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F, Azam F. Detection of DNA damage in prokaryotes by terminal deoxyribonucleotide transferase-mediated dUTP nick end labeling. Appl Environ Microbiol. 2000;66:1001–1006. doi: 10.1128/aem.66.3.1001-1006.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su HL, Chou CC, Hung DJ, Lin SH, Pao IC, Lin JH, Huang FL, Dong RX, Lin JJ. The disruption of bacterial membrane integrity through ROS generation induced by nanohybrids of silver and clay. Biomaterials. 2009;30:5979–5987. doi: 10.1016/j.biomaterials.2009.07.030. [DOI] [PubMed] [Google Scholar]
- Suri B, Bickle TA. EcoA: the first member of a new family of type I restriction modification systems. Gene organization and enzymatic activities. J Mol Biol. 1985;186:77–85. doi: 10.1016/0022-2836(85)90258-x. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen S, Xu T, Taghizadeh K, Wishnok JS, Zhou X, You D, Deng Z, Dedon PC. Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol. 2007;3:709–710. doi: 10.1038/nchembio.2007.39. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen S, Vergin KL, Giovannoni SJ, Chan SW, DeMott MS, Taghizadeh K, Cordero OX, Cutler M, Timberlake S, Alm EJ, Polz MF, Pinhassi J, Deng Z, Dedon PC. DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc Natl Acad Sci U S A. 2011;108:2963–2968. doi: 10.1073/pnas.1017261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GG, Murray NE. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- Xu T, Yao F, Zhou X, Deng Z, You D. A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res. 2010;38:7133–7141. doi: 10.1093/nar/gkq610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F, Xu T, Zhou X, Deng Z, You D. Functional analysis of spfD gene involved in DNA phosphorothioation in Pseudomonas fluorescens Pf0-1. FEBS Lett. 2009;583:729–733. doi: 10.1016/j.febslet.2009.01.029. [DOI] [PubMed] [Google Scholar]
- You D, Wang L, Yao F, Zhou X, Deng Z. A novel DNA modification by sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in Streptomyces lividans. Biochemistry. 2007;46:6126–6133. doi: 10.1021/bi602615k. [DOI] [PubMed] [Google Scholar]
- Zhou X, He X, Liang J, Li A, Xu T, Kieser T, Helmann JD, Deng Z. A novel DNA modification by sulphur. Mol Microbiol. 2005;57:1428–1438. doi: 10.1111/j.1365-2958.2005.04764.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.