SUMMARY
The cytokinetic apparatus of bacteria is initially formed by the polymerization of the tubulin-like FtsZ protein into a ring structure at midcell. This so-called Z-ring facilitates the recruitment of many additional proteins to the division site to form the mature divisome machine. Although the assembly pathway leading to divisome formation has been well characterized, the mechanisms that trigger cell constriction remain unclear. In this report, we study a “forgotten” allele of ftsL from Escherichia coli, which encodes a conserved division gene of unknown function. We discovered that this allele promotes the premature initiation of cell division. Further analysis also revealed that the mutant bypasses the requirement for the essential division proteins ZipA, FtsK, and FtsN and partially bypasses the need for FtsA. These findings suggest that rather than serving simply as a protein scaffold within the divisome, FtsL may play a more active role in the activation of the machine. Our results support a model in which FtsL, along with its partners FtsB and FtsQ, function as part of a sensing mechanism that promotes the onset of cell wall remodeling processes needed for the initiation of cell constriction once assembly of the divisome complex is deemed complete.
Keywords: cell division, divisome, morphogenesis, cytokinesis, bacteriolysis
INTRODUCTION
Bacterial cell division or septation is an essential process in which new polar caps for the developing daughter cells are formed. In gram-negative organisms, this involves the coordinated constriction of both the inner and outer cell membranes as well as the synthesis and remodeling of the peptidoglycan (PG) cell wall layer located between them. This molecular construction project is carried out by a ring-shaped, multi-protein machine called the divisome or septal ring (Lutkenhaus et al., 2012). Over the years, dozens of proteins have been localized to this apparatus and interaction studies suggest that they are linked to one another via a complex web of connections that span the cell envelope (Lutkenhaus et al., 2012). Although many of these factors are known to be essential for cytokinesis, few of them have clearly defined functions.
Cell division can be broken down into four phases: divisome assembly, constriction initiation, active constriction, and septal closure/pole completion. The first step of the process is the most well understood (Lutkenhaus et al., 2012). In Escherichia coli, it begins with the assembly of polymers of the tubulin-like FtsZ protein along with its partners FtsA, ZipA, and other FtsZ-binding proteins into a cytoskeletal structure called the Z-ring (Bi and Lutkenhaus, 1991). Formation of the Z-ring at the inner face of the cytoplasmic membrane is then thought to promote the localization of the remaining septal ring components to the division site. Recruitment of the essential divisome components of E. coli to midcell has been shown to follow a mostly linear dependency pathway starting with FtsZ and ending with FtsN (FtsZ --> FtsA/ZipA --> FtsK --> FtsQ/FtsL/FtsB --> FtsW --> FtsI --> FtsN) (Wang et al., 1998; Hale and de Boer, 1999; Weiss et al., 1999; Chen and Beckwith, 2001; Mercer and Weiss, 2002; Buddelmeijer et al., 2002; Hale and de Boer, 2002; Schmidt et al., 2004; Goehring and Beckwith, 2005). In this pathway, the localization of each divisome component to the septal ring is dependent on the prior localization of all upstream proteins. It is important to note, however, that the simple hierarchical arrangement of the recruitment order belies a much more complicated interaction network that involves connections between components extending beyond nearest neighbors in the dependency scheme (Di Lallo et al., 2003; Corbin et al., 2004; Goehring et al., 2005; Karimova et al., 2005; Alexeeva et al., 2010; Busiek et al., 2012). Moreover, studies of the temporal sequence of protein recruitment to the divisome indicate that its assembly is likely to be a two-step process, with components of the Z-ring assembling early and persisting for about 20% of the cell cycle followed by the near simultaneous localization of the remaining “late” proteins at about the time the first signs of cell constriction become apparent (Aarsman et al., 2005).
Because missteps in cell envelope remodeling processes can have catastrophic consequences, the conversion of the divisome from a state of assembly to one of active constriction likely involves a number of regulatory inputs. Similarly, the active phase of constriction is likely to be closely monitored so that the development of potentially dangerous imbalances can be corrected. For example, cells must prevent the hydrolytic enzymes that remodel the cell wall from being activated out of sequence with the initiation of septal PG synthesis as constriction is triggered (Uehara et al., 2010; Peters et al., 2011; Desirée C Yang et al., 2011; Desirée C Yang et al., 2012). The division machinery also needs to prevent the two processes from becoming uncoupled as the new poles are formed. Otherwise, miscoordination of PG synthesis and hydrolysis is likely to induce the formation of substantial breaches in the PG matrix and cause cell lysis.
Surprisingly little is known about the mechanism of cell constriction by the divisome or the controls that govern its initiation, especially considering the extreme changes in cell envelope architecture that are ultimately elicited by the process. One of the major constraints limiting our understanding of divisome regulation has been the limited phenotypic range displayed by commonly studied division mutants. By and large, these mutants simply fail to constrict under non-permissive conditions and form smooth filaments (van de Putte et al., 1964; Hirota et al., 1970; Allen et al., 1974; Begg et al., 1980; Khattar et al., 1994; Begg et al., 1995). Such a phenotype indicates that the altered factor in question is important for division in general, but yields little information on what it is actually doing during the process. We have therefore been interested in studying mutants with phenotypes indicative of failures occurring after constriction is initiated. One such mutant is an allele of the ftsL gene encoding FtsL(E88K) isolated many years ago by Ishino and co-workers (Ishino et al., 1989; Ueki et al., 1992). This mutation was reported to result in cell lysis under non-permissive conditions, but outside of its initial identification, it was not studied further. We reasoned that revisiting the properties of this mutant allele, which we will refer to as ftsL*, would allow us to better understand the role of FtsL during cell division.
FtsL is a bitopic membrane protein of only 121 amino acids, with a short cytoplasmic N-terminus (residues 1-37), a transmembrane segment (residues 38-57), and a periplasmic domain (residues 58-121) that is predicted to consist mainly of a coiled-coil structure with a leucine zipper motif (Guzman et al., 1992). It is known to form a subcomplex with two other essential division proteins, FtsB and FtsQ, independently of their localization to the divisome (Buddelmeijer and Beckwith, 2004). The transmembrane segment and leucine zipper motif of FtsL have been shown to be required for optimal interaction with FtsB (Robichon et al., 2011; Khadria and Senes, 2013). Interestingly, homologs of ftsL, ftsB and ftsQ are well conserved among bacterial species (Gonzalez et al., 2010). Although they form a core component of the divisome, the physiological role of FtsL and its partners is still unknown. Bacterial two-hybrid experiments have detected interactions between FtsL and several other divisome components in E. coli (Karimova et al., 2005). Due to its involvement in many different protein-protein interactions, it has been suggested that the main role of FtsL may be as a structural or scaffolding protein important for divisome assembly and/or stability (Gonzalez et al., 2010).
Here, we show that the ftsL* mutation leads to a divisome malfunction that accelerates the division process, likely through the premature initiation of constriction. Mutant cells harboring this allele divide at a lower than normal cell volume, thus forming smaller daughter cells. This accelerated division phenotype is responsible for the growth defect and cell lysis observed when cells are shifted to non-permissive conditions. Consistent with this interpretation, mutations inactivating division proteins implicated in divisome function and/or stability were found to rescue the ftsL* growth defect. Furthermore, our suppressor analysis also revealed that the ftsL* allele can fully bypass the essential functions of the divisome proteins FtsK, FtsN, and ZipA, and partially bypass the need for FtsA activity. Given that FtsA and FtsN have both been previously implicated in the initiation of constriction (Geissler et al., 2003; Corbin et al., 2004; Goehring, Robichon, et al., 2007; Bernard et al., 2007; Gerding et al., 2009; Lutkenhaus, 2009; Pichoff et al., 2012; Busiek et al., 2012), our results support a model in which FtsL as part of the FtsQLB subcomplex also plays an important role in modulating divisome activation rather than merely serving as a scaffolding protein within the apparatus as has been proposed previously (Gonzalez et al., 2010). An accompanying study by Liu and co-workers in the de Boer laboratory also reports results supporting a role for the FtsQLB complex in the control of cell constriction stimulated by FtsN (Liu et al., 2014).
RESULTS
Phenotypic analysis of ftsL* mutants
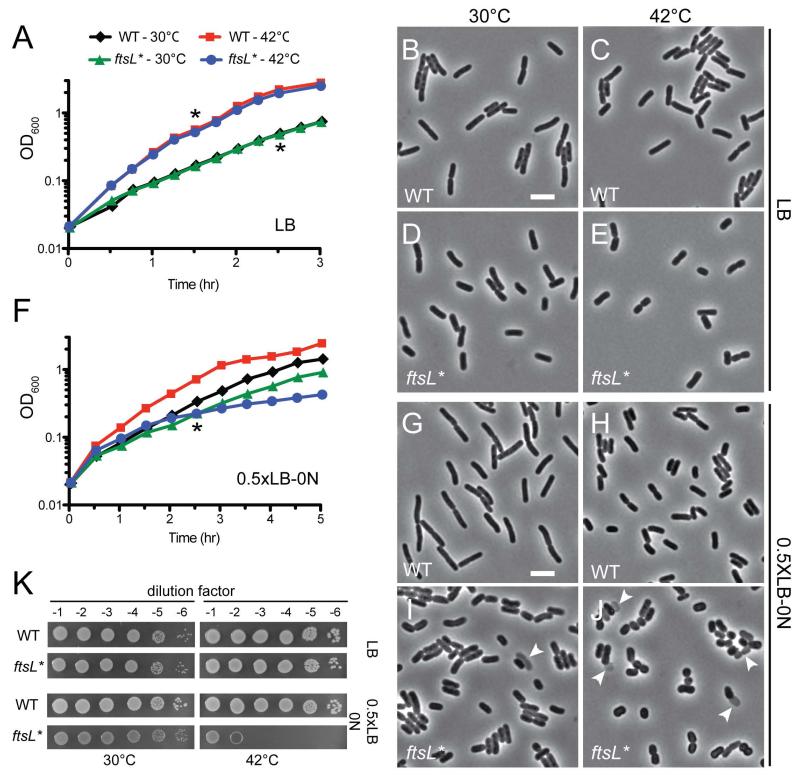
A mutant harboring the mutation ftsL(E88K), which we will refer to as ftsL*, was isolated several years ago by Ishino and co-workers (Ishino et al., 1989; Ueki et al., 1992). Rather than inducing a division block, this particular allele was reported to induce cell lysis at the non-permissive temperature of 42°C when cells were grown in LB medium with no added NaCl. Besides the initial description of its isolation, further characterization of the ftsL* mutant has not been reported. Given the unique phenotype induced by this allele, we thought a reinvestigation of its properties was warranted because it might yield new insight into the function of FtsL. Allelic replacement was therefore used to introduce ftsL* into a wild-type MG1655 strain background. When grown in standard LB medium (0.5% NaCl), the resulting strain MT10 [ftsL*] displayed a growth rate similar to wild-type at both 30°C and 42°C (Fig. 1A). However, the mutant cells were approximately 15-20% shorter and slightly wider than wild-type at both temperatures (Fig. 1B-E, Table 1). Mean cell volume was also significantly reduced in ftsL* cells relative to wild-type at 30°C (Table 1). In half-strength LB medium with no added NaCl (0.5×LB-0N), MT10 [ftsL*] grew at approximately the same rate as wild-type at 30°C, but displayed a severe reduction in growth rate at 42°C (Fig. 1F). Cell length and volume were reduced even further in ftsL* cells relative to wild-type in this medium (Fig. 1G-J, Table 1). The growth defect for strain MT10 [ftsL*] was accompanied by a higher frequency of lysed ghost cells in the culture and a striking change in morphology with the mutant cells becoming much shorter and approaching a spherical morphology (Fig. 1J). MT10 [ftsL*] plated with normal efficiency on standard LB agar at both 30°C and 42°C as well as on 0.5×LB-0N agar at 30°C, but displayed a severe plating defect on this medium at 42°C (Fig. 1K). Using RpoC-mCherry as a marker for the nucleoid, we investigated whether the lysis phenotype of the ftsL* mutant was due to improper chromosome segregation. However, obvious chromosome guillotining events were not observed, nor were anucleate cells, which would be indicative of chromosome segregation defects (data not shown). Our results thus confirm the original findings of Ishino and coworkers (Ishino et al., 1989) that the ftsL* allele results in a temperature-sensitive lethal phenotype distinct from typical cell filamentation. However, we did not observe dramatic lysis of liquid cultures upon shifting the mutant to the non-permissive temperature in 0.5×LB-0N medium. Instead, the primary effect of the mutation appeared to be a reduction in cell length that was observable at all temperatures, but was exacerbated at 42°C in 0.5×LB-0N where some cell lysis was also observed. We suspect that the lysis phenotype is much more severe on solid 0.5×LB-0N media at 42°C such that viability is significantly compromised.
Fig.1. Shape and growth defect of the ftsL* mutant.
(A) Overnight cultures of TB28 [WT] or MT10 [ftsL*] cells were diluted in fresh LB broth and grown at 30°C until mid-log. At time t = 0, each strain was diluted into fresh LB and grown at 30°C or shifted to 42°C. (B-E) Cells were removed from the TB28 (B, C) and MT10 (D, E) cultures at the time point indicated by the asterisks, fixed and examined by phase contrast microscopy. Bar = 4μm. (F) Growth of TB28 and MT10 was monitored as described in (A) except that growth medium was half-strength LB with no added NaCl (0.5×LB-0N). (G-J) Cells from the TB28 (G, H) or MT10 (I,J) cultures in (F) were removed at the time point indicated by the asterisks, fixed and examined by phase contrast microscopy. Bar = 4μm. Arrows point to lysed cells. (K) Cells of TB28 and MT10 were grown overnight in LB broth at 30°C. Following normalization for cell density (OD600 = 2), the resulting cultures were serially diluted (10−1 to 10−6), and 5 μl of each dilution was spotted on the indicated medium. Plates were incubated overnight at the indicated temperature and photographed.
Table 1. Mean cell length and mean cell volume of WT and ftsL* mutant cells under different conditions.
| LB (0.5% NaCI) | 0.5× LB (No NaCl) | |||||||
|---|---|---|---|---|---|---|---|---|
| genotype | 30°C | 42°C | 30°C | 42°C | ||||
|
| ||||||||
| Mean L (μm) [% WT length]a |
Mean Vol. (μm3) [% WT volume]a |
Mean L (μm) [% WT length] |
Mean Vol. (μm3) [% WT volume] |
Mean L (μm) [% WT length] |
Mean Vol. (μm3) [% WT volume] |
Mean L (μm) [% WT length] |
Mean Vol. (μm3) [% WT volume] |
|
| WT | 4.37 ± 0.06 [100%] |
3.93 ± 0.06 [100%] |
3.81 ± 0.09 [100%] |
3.65 ± 0.09 [100%] |
4.92 ± 0.04 [100%] |
3.98 ± 0.12 [100%] |
3.89 ± 0.03 [100%] |
3.54 ± 0.04 [100%] |
| ftsL* | 3.63 ± 0.01 [83.1%] ** |
3.51 ± 0.02 [89.3%] ** |
3.22 ± 0.05 [84.5%] ** |
3.70 ± 0.08 [101.4%] NS |
3.26 ± 0.06 [66.3%] *** |
3.26 ± 0.06 [81.9%] * |
2.76 ± 0.00 [71.0%] *** |
3.23 ± 0.07 [91.2%] * |
Overnight cultures of TB28 [WT] and MT10 [ftsL*] were diluted in the indicated liquid broth and grown to midlog at 30°C The cultures were then backdiluted to an OD600 ~ 0.02 in fresh medium and grown at 30°C or shifted to 42°C until an OD600 ~ 0.3 - 0.5. The cells were then imaged using phase contrast microscopy and analyzed using MicrobeTracker to determine cell length and cell volume. 600 cells were analyzed for each condition, n = 3. Shown are the average mean cell length or average mean cell volume ± standard error of the mean (SEM).
% = (Mean cell length (or cell volume) of ftsL* mutant or WT/ Mean cell length (or cell volume) of WT) * 100%
difference significant at p < 0.05
difference significant at p < 0.01
difference significant at p < 0.001
NS, difference not statistically significant
FtsL* promotes an accelerated division cycle
We reasoned that the dramatic effect of FtsL* on cell morphology could either be due to: (i) a change in the steady-state concentration of FtsL protein, (ii) a mis-localization of the variant protein that adversely affects cell elongation and rod-shape maintenance, or (iii) an alteration of divisome activity by FtsL*. In order to begin distinguishing between these possibilities, we determined the levels of the FtsL protein in both wild-type or ftsL* mutant cells. As shown by immunoblotting, the level of the FtsL* variant was only 2-fold higher than that of wild-type (Fig. S1A). In addition, expression of wild-type ftsL from a multicopy plasmid in a wild-type background does not induce the reduced cell length phenotype observed for the ftsL* mutant under non-permissive conditions (data not shown). Finally, the growth defect of the ftsL* mutant was rescued by the overproduction of wild-type FtsL, but not FtsL* (Fig. S1B). Thus, the FtsL* phenotype is unlikely to be caused by a change in FtsL concentration.
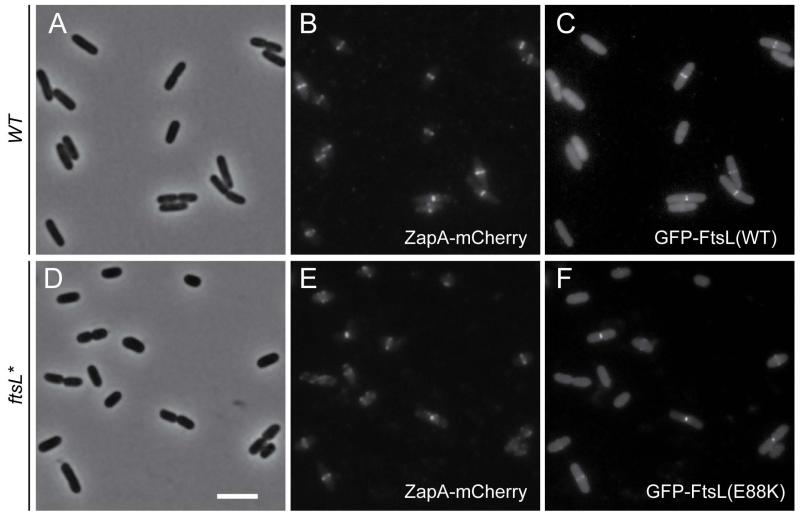
Next, we examined the subcellular localization of the mutant protein relative to wild-type FtsL and estimated the timing of its recruitment to the divisome by measuring the frequency with which the FtsL proteins co-localized with the early divisome recruit ZapA. To do so, we produced GFP-FtsL or GFP-FtsL* from a single-copy expression construct integrated at the attHK022 locus in either a wild-type or an ftsL* background respectively. The strains also encoded ZapA-mCherry expressed from the native zapA locus. When grown in minimal medium at 30°C, both FtsL variants displayed bright fluorescent bands at mid-cell (Fig. 2), indicating that the growth and morphological defects caused by the FtsL* variant are not likely to be due to the mis-localization of the protein. Consistent with this idea, proper mid-cell localization of the GFP-FtsL* variant was also observed under non-permissive conditions (42°C in 0.5×LB-0N) (Fig. S2). Strikingly, in minimal medium at 30°C, ZapA-mCherry rings were found to co-localize with a ring of GFP-FtsL* at a much higher percentage (76%) than with the wild-type GFP-FtsL fusion (57%) (Table 2). The fraction of cells with a ZapA-mCherry ring was found to be lower in ftsL* cells, suggesting that Z-rings form later in the cell cycle in the mutant relative to wild-type. This apparent delay in Z-ring formation likely reflects the fact that the mutant cells are born much shorter than normal cells and therefore take longer to achieve a cell length permissive for Z-ring assembly. Despite the delay in Z-ring formation, the increased co-localization of ZapA-mCherry and GFP-FtsL* suggests that the time between Z-ring assembly and the recruitment of the late component FtsL to the divisome is reduced in the ftsL* mutant.
Fig. 2. Localization of GFP-FtsL*.
Overnight cultures of TU211(attHKMT35) [WT zapA-mCherry (Plac::gfp-ftsL)] (A-C) and MT102(attHKMT36) [ftsL* zapA-mCherry (Plac::gfp-ftsL*)] (D-F) were diluted in minimal M9 medium supplemented with 0.2% maltose and 25 μM IPTG and grown at 30°C until an OD600 ~ 0.25 - 0.35. The cells were visualized on 2% agarose pads with phase contrast (A,D), mCherry (B,E) and GFP (C,F) optics. Bar = 4μm.
Table 2. Co-localization of FtsL or FtsL* with ZapA at the division site.
| genotype | % of total cells: |
||
|---|---|---|---|
| with stable ZapA-mCherry ring at the division site |
with GFP-FtsL ring at the division site |
with co-localized proteins |
|
|
WT zapA-mCherry
(Plac::gfp-ftsL) |
64.9 ± 1.1 | 38.1 ± 4.1 | 37.3 ± 4.4 |
|
ftsL* zapA-mCherry (Plac::gfp-ftsL*) |
54.3 ± 1.4 | 41.8 ± 2.7 | 41.3 ± 2.5 |
Overnight cultures of TU211 (attHKMT35) [WT zapA-mCherry (Plac::gfp-ftsL)] and MT102(attHKMT36) [ftsL* zapA-mCherry (Plac::gfp-ftsL*)] were diluted in minimal M9 medium supplemented with 0.2% maltose and 25uM IPTG and grown at 30°C until an OD600 ~ 0.25 - 0.35. The cells were imaged using phase contrast, mCherry and GFP optics and analyzed using the imaging software NIS-Elements (Nikon) to determine the number of cells with a ZapA-mCherry ring or a GFP-FtsL(wt or L*) ring or both at the division site. 1200 cells were analyzed for each condition, n = 3. Shown are the average values ± standard deviation.
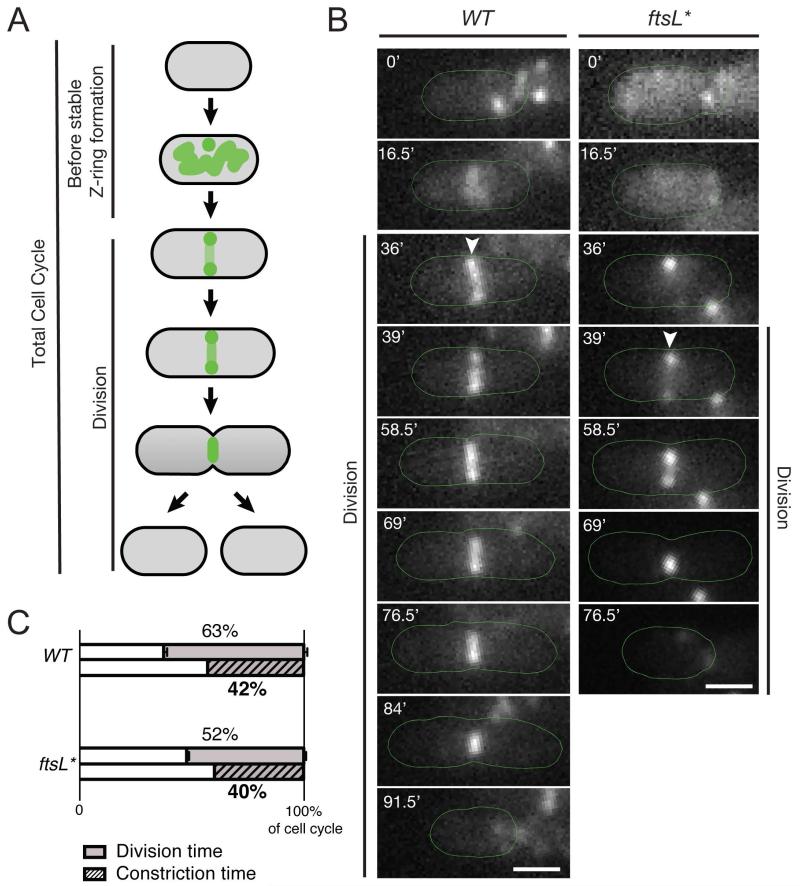
To examine the effect of FtsL* on the cell cycle further, we monitored the division process of wild-type and mutant cells (140 of each) using time-lapse microscopy. For this analysis, we used ZapA-GFP produced from the endogenous chromosomal locus as a proxy for cellular FtsZ dynamics. The division time was defined as the time between the formation of a stable Z-ring through the end of cell constriction and completion of daughter cell separation (Fig. 3A) (See Experimental Procedures for details). Using MicrobeTracker and SpotFinderZ (Sliusarenko et al., 2011) to quantify the relevant parameters, we determined that, when grown on minimal medium agarose pads at 30°C, the mean division time in the ftsL* mutant was significantly (ca. 25%) shorter than in wild-type (Fig. 3B-C, Table 3). In wild-type cells, the division process made up 63% of the total cell cycle time, while in the mutant, only 52% of the cell cycle was dedicated to division. Thus, cell division proceeded faster in the ftsL* mutant relative to wild-type cells. The accelerated division of ftsL* cells may either result from the premature initiation of constriction, an enhanced rate of constriction, or a combination of the two. In order to distinguish between these possibilities we compared the average cell constriction time for wild-type and the ftsL* mutant. Cells of each strain were grown to steady state in minimal medium at 30°C and the fraction of cells with a visible constriction was determined from DIC images (Table 4). Given steady-state growth, the constriction time as a percentage of the cell cycle was calculated as in a previous study (Aarsman et al., 2005). Under these conditions, both strains grew with similar doubling times but the mean cell length of ftsL* was less than that of wild-type as expected from the results described above. Interestingly, the constriction time of ftsL* was only slightly shorter than wild-type (Fig. 3C, Table 4), suggesting that the faster division of the ftsL* mutant is primarily due to the premature initiation of constriction. Because we also observed a shorter delay between the stabilization of the Z-ring and the recruitment of FtsL* to the divisome, we infer that the reduced cell length and cell volume of the ftsL* mutant likely result from the premature maturation of the divisome and the subsequent initiation of cell constriction at a lower than normal cell volume.
Fig. 3. Division time is shorter in the ftsL* mutant compared to wild-type.
(A) For the purposes of the analysis, total cell cycle time was broken down into the time prior to stable Z-ring formation and division time (the time from stable Z-ring formation to the completion of cell constriction). (B) Overnight cultures of HC261 [zapA-gfp] and MT90 [ftsL* zapA-gfp] were diluted in minimal M9 medium supplemented with 0.2% maltose and grown at 30°C until an OD600 ~ 0.15 - 0.3. The cells were spotted on separate sides of a 2% agarose pad that was cut in half and contained 1× M9 salts, 0.02% casamino acids and 0.2% maltose, the coverslip was sealed and a timelapse of both strains grown at 30°C was obtained using phase contrast and GFP optics, with frames taken every 1.5 min. Shown are representative time-lapse images for WT and ftsL* mutant cells. Bar = 1μm. Arrowheads mark the first time point where a stable FtsZ ring was formed. (C) Comparison of the division time and constriction time for WT and ftsL* mutant cells as a percentage of the total cell cycle. To determine division time, the timelapse movies described in (B) were analyzed using MicrobeTracker and SpotFinderZ (Sliusarenko et al., 2011) to determine the total cell cycle time, as well as the division time. 140 cells were analyzed for each strain, n = 3. Error bars indicate the standard error of the mean (SEM) (see data in Table 3). To determine constriction time, TB28 [WT] or MT10 [ftsL*] cells were grown to steady state in minimal M9 medium supplemented with 0.2% maltose at 30°C until an OD ~ 0.1. The cells were visualized on 2% agarose pads with DIC optics and the presence of a constriction was determined manually. The constriction time shown is the average of two independent replicates, with 800 cells total for each strain (see data in Table 4).
Table 3. Measured division times of WT and ftsL* cells.
| genotype | Mean cell cycle time (min) |
Mean division time (min) |
Mean division time as % of total cell cyclea |
|---|---|---|---|
| WT zapA-gfp | 87.5 ± 2.9 | 54.6 ± 0.8 | 62.5 ± 1.6 |
| ftsL* zapA-gfp | 79.0 ± 3.0 | 41.3 ± 1.3 | 52.3 ± 1.1 |
| NS | ** | ** |
Overnight cultures of HC261 [WT zapA-gfp] and MT90 [ftsL* zapA-gfp] were diluted in minimal M9 medium supplemented with 0.2% maltose and grown at 30°C until an OD600 ~ 0.15 - 0.3. The cells were spotted on two separate halves of a 2% agarose pad containing 1× M9 salts and 0.2% maltose, the coverslip was sealed and a timelapse of both strains grown at 30°C was obtained using phase contrast and gfp optics, with frames taken every 1.5 min. The timelapse was analyzed using MicrobeTracker and SpotFinderZ to determine the total cell cycle time, as well as the division time. Division time is defined as the time between the formation of a stable Z-ring (using ZapA-gfp as a proxy for FtsZ) and the end of constriction. 140 cells were analyzed for each strain, n = 3. Shown are the average mean cell cycle time, average mean division time (in min) and average mean division time (as % of total cell cycle time) ± standard error of the mean (SEM).
% = (mean division time / mean cell cycle time) * 100%
difference significant at p < 0.01
NS, difference not statistically significant
Table 4. Constriction time of WT and ftsL* cells.
| genotype | Mean cell length (μm)a |
Cell cycle analysisb | |||
|---|---|---|---|---|---|
|
| |||||
| Mass doubling time, Td (min) |
Fraction of cells with constriction, F(x) |
Constriction time, tc (min) |
Constriction as % of cell cyclec |
||
| WT | 2.93 ± 0.60 |
92 | 0.33 | 38 | 41.5 |
| ftsL* | 2.50 ± 0.48 |
94 | 0.32 | 37 | 39.5 |
TB28 [WT] and MT10 [ftsL*] were grown to steady-state in minimal M9 medium supplemented with 0.2% maltose at 30°C. The OD600 of the cultures was monitored at regular time intervals to determine the mass doubling time of each strain. At ODM600 ~ 0.1, the cells were visualized on 2% agarose pads with either phase contrast or DIC optics. The mean cell length was determined from the phase contrast images using MicrobeTracker. The presence of a constriction was determined manually from the DIC images using the imaging software NIS-Elements (Nikon). See Experimental Procedures for details.
Shown is the mean cell length ± standard deviation for a single experiment with 400 cells analyzed for each strain
Shown is the average values from two independent experiments, with 800 cells total analyzed for each strain
% = (tc / Td) * 100%
FtsL* causes a divisome malfunction
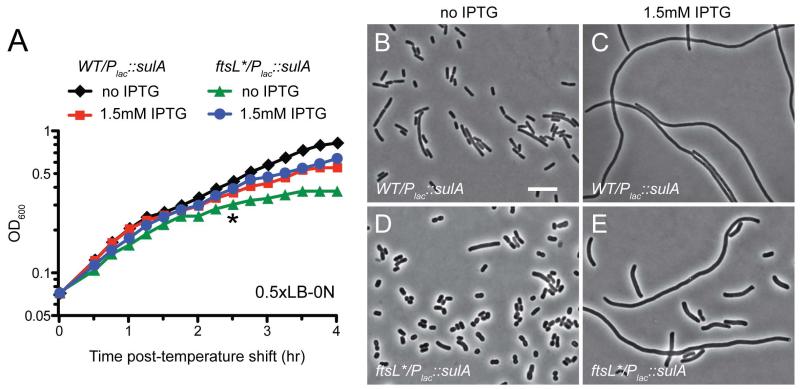
We suspected that the growth defect caused by the FtsL* variant under non-permissive conditions may stem from premature constriction initiation. We reasoned that if this were true, the defect should depend on an assembled divisome. Growth of ftsL* cells in 0.5×LB-0N at 42°C was therefore monitored with or without production of the FtsZ antagonist and division inhibitor SulA prior to shifting the cells to non-permissive conditions. After three hours of growth in the presence of IPTG to induce the sulA expression construct, wild-type cells displayed a mild growth defect relative to the uninduced control (Fig. 4A). As expected, these cells also formed long filaments indicative of a complete division block by SulA (Fig. 4B-C). Strikingly, induction of sulA had the opposite effect on the growth of the ftsL* mutant, with cells overexpressing sulA growing better than those lacking inducer (Fig. 4A). Notably, although the induction of sulA still impaired division in the ftsL* mutant, the division block appeared to be less robust than in wild-type cells. This observation suggests that the Z-ring may be more resistant to SulA in this background. Because blocking cell division relieved the negative effects of FtsL* on growth, we conclude that the variant protein is altering divisome function to induce the observed phenotypes.
Fig. 4. Overexpression of sulA suppresses the growth defect of the ftsL* mutant.
(A) Overnight cultures of TB28/pMT74 [WT/Plac::sulA] and MT10/pMT74 [ftsL*/Plac::sulA] were diluted in 0.5×LB-0N and grown to mid-log at 30°C. They were then diluted to a starting OD600 ~0.015 in fresh 0.5×LB-0N medium with or without 1.5 mM IPTG and grown at 30°C for 1h before shifting to 42°C at time t = 0. (B-E) Cells were removed from the TB28/pMT74 (B, C) and MT10/pMT74 (D, E) cultures at the time point indicated by the asterisk and examined by phase contrast microscopy. Bar = 10μm.
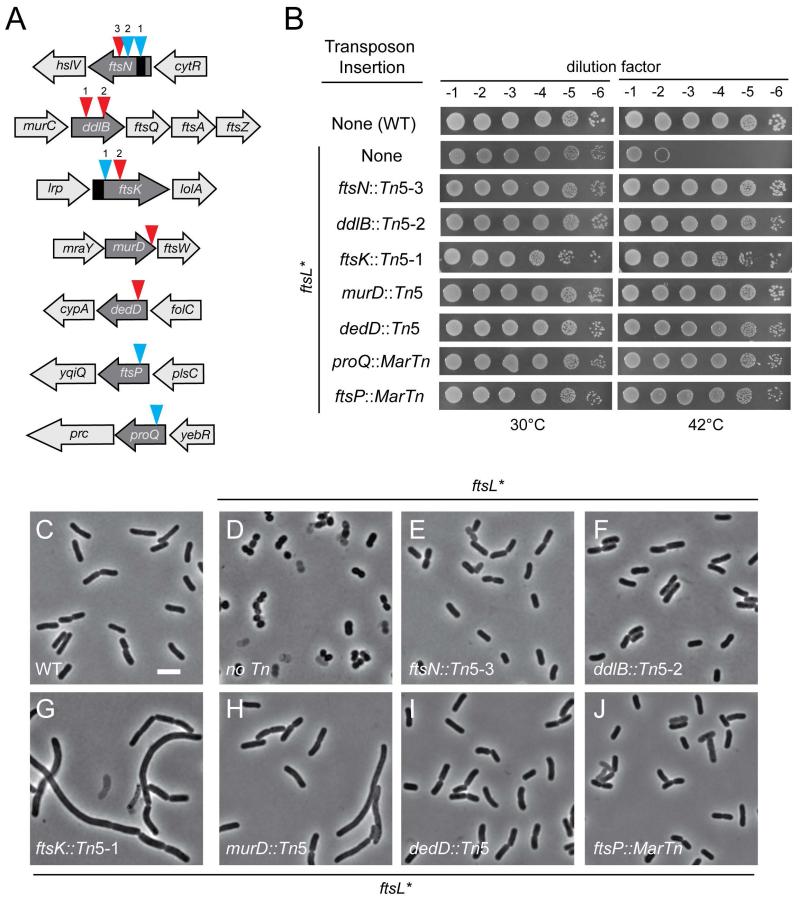
In order to learn more about the division defect induced by ftsL*, we selected for suppressors capable of growing on 0.5×LB-0N at 42°C following transposon mutagenesis of MT10 [ftsL*]. Surviving colonies were purified and the positions of the transposon insertions were mapped. The insertion alleles were also transduced to a fresh MT10 [ftsL*] background to confirm that suppression was indeed linked to the transposon. Several of the confirmed suppressors contained a transposon insertion in genes encoding divisome components implicated in the function and/or stability of the machine, such as ftsN, ftsK, dedD, and ftsP (Begg et al., 1995; Addinall et al., 1997; Samaluru et al., 2007; Gerding et al., 2009; Arends et al., 2010) (Fig. 5A-B). Three others had insertions in the complex dcw cluster of genes present at the two minute locus of the chromosome: two in ddlB and one at the 3′ end of murD (Fig. 5A-B). These insertions are likely to negatively impact division by affecting the expression of the many division genes located in this large operon. The final suppressor isolated was in proQ, which encodes an RNA-chaperone that post-transcriptionally regulates the osmoregulatory transporter ProP (G Chaulk et al., 2011) (Fig. 5A-B). This allele is likely to suppress the sensitivity of ftsL* cells to growth in medium with low osmolarity. Relative to the parental MT10 [ftsL*] strain, derivatives with transposon insertions in genes with division related functions appeared longer, approaching the length of wild-type cells or even longer when they were grown in 0.5×LB-0N at 42°C (Fig. 5C-J). Thus, as with sulA overexpression, transposon insertions that impair cell division suppress the growth phenotype of the ftsL* mutation. These findings along with the above microscopic analyses strongly suggest that the growth defect and morphological changes induced by the FtsL* protein are mediated by a malfunctioning of the divisome that promotes the premature initiation of cell constriction.
Fig. 5. Transposon insertions that suppress the shape and growth defect of the ftsL* mutant.
(A) Shown is a diagram of the gene context and approximate positions of the unique transposon insertion mutants isolated in the suppressor selection. The triangles indicate the position of the transposon. Red, the direction of transcription in the kanamycin resistance cassette of the transposon is in the same orientation as disrupted gene; blue, opposite direction. The black segments denote the essential domains of FtsN and FtsK. (B) Cells of TB28 [WT], MT10 [ftsL*(E88K)], and MT10 derivatives with the indicated transposon insertions were grown overnight in LB broth at 30°C. Following normalization for cell density (OD600 = 2), the resulting cultures were serially diluted (10−1 to 10−6), and 5 μl of each dilution was spotted on 0.5×LB-0N agar. The plates were incubated overnight at the indicated temperature and photographed. Tn5 and MarTn denote the two different transposons used for mutagenesis (see Experimental Procedures). (C-J) Overnight cultures of the strains from (B) were diluted in 0.5×LB-0N broth and grown to mid-log at 30°C. They were then diluted to a starting OD600 ~0.02 in fresh 0.5×LB-0N and grown at 42°C to an OD600 of 0.3 - 0.5 before they were visualized on 2% agarose pads by phase contrast microscopy. Bar = 4μm.
FtsL* bypasses the essential functions of FtsK and FtsN
FtsK and FtsN both contain multiple domains with only a portion of the protein being essential for cell division. FtsK has three identifiable domains: (i) a polytopic transmembrane domain at its N-termius, (ii) a long cytoplasmic linker domain, and (iii) a C-terminal AAA ATPase domain that functions as a DNA pump and also stimulate chromosome dimer resolution (Massey et al., 2006). Only the N-terminal domain of FtsK is essential for cell division, but its function remains unknown (Draper et al., 1998; Dubarry et al., 2010). FtsN is a bitopic membrane protein with a flexible linker in the periplasm followed by a C-terminal SPOR domain with PG-binding activity (Ji-Chun Yang et al., 2004; Ursinus et al., 2004). The essential domain of FtsN has been narrowed down to a small region of the periplasmic domain near the N-terminal transmembrane helix (Gerding et al., 2009).
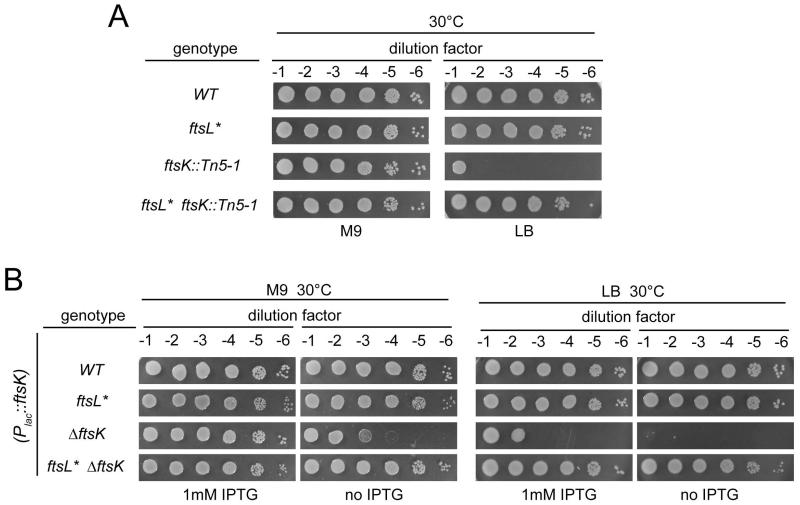
Of the ftsL* suppressors isolated, we were intrigued by the ftsK::Tn5-1 and ftsN::Tn5-1 alleles because the insertions mapped within or very near the essential domains of FtsK and FtsN (Fig. 5A). Further investigation showed that the ftsK::Tn5-1 insertion could only be transduced into a wild-type background on M9 minimal medium. On LB agar, the insertion resulted in a lethal division block (Fig. 6A). This growth defect was completely suppressed by the ftsL* mutation (Fig. 6A), suggesting that this allele was capable of promoting cell division in cells lacking the essential N-terminal domain of FtsK (Draper et al., 1998). To test this, we constructed an ftsK deletion strain possessing a second copy of the gene at an ectopic locus under control of the lac promoter (Plac). As expected, in an otherwise wild-type background, this strain was dependent on IPTG induction for growth on both rich and minimal medium (Fig. 6B). However, introduction of the ftsL* mutation into this background resulted in IPTG independent growth regardless of the medium. We were also able to delete ftsK in the presence of the ftsL* allele in a strain lacking the ftsK expression construct provided the strain was maintained on minimal medium (data not shown). We thus conclude that the ftsL* mutation can bypass the essential function of FtsK.
Fig. 6. The ftsL* allele suppresses the essentiality of ftsK.
(A) Cells of TB28 [WT], MT10 [ftsL*], MT38 [ftsK::Tn5-1] and MT30 [ftsL* ftsK::Tn5-1] were grown overnight in M9 maltose medium at 30°C. Following normalization for cell density (OD600 = 2), the resulting cultures were serially diluted (10−1 to 10−6), and 5 μl of each dilution was spotted on the indicated medium. The plates were incubated overnight at 30°C and photographed. (B) Cells of TB28, MT10, MT75 [ΔftsK] and MT76 [ftsL* ΔftsK] containing the integrated attHKMT117 plasmid (Plac::ftsK) were grown overnight in M9 maltose medium supplemented with 1mM IPTG, processed as in A, and spotted on the indicated medium with or without 1mM IPTG. The plates were incubated overnight at 30°C and photographed.
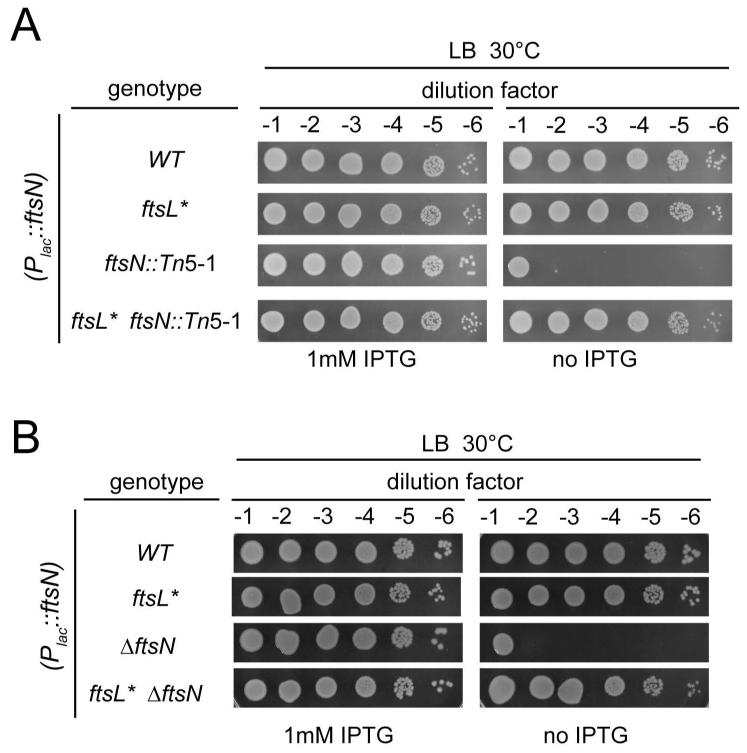
Similar to the results with ftsK::Tn5-1, we found that the ftsN::Tn5-1 allele, in which the insertion disrupts the essential domain of FtsN (Gerding et al., 2009), could only be transduced into a wild-type background if it possessed a second copy of ftsN under Plac control. Such transductants were completely dependent on IPTG for growth, but again this dependence could be suppressed upon introduction of the ftsL* mutation (Fig. 7A). To test whether ftsL* could suppress a more complete loss of FtsN function, we constructed an FtsN depletion strain that was deleted for the native ftsN gene and possessed the Plac::ftsN expression construct. In an otherwise wild-type background, the strain was dependent on IPTG for growth. Following introduction of the ftsL* allele, however, cell growth again became IPTG independent (Fig. 7B). Similar to the situation with ftsK, we could transduce an ftsN deletion into an ftsL* strain lacking the Plac::ftsN expression construct. However, the resulting strain grew very poorly, even on minimal medium (data not shown). We thus conclude that, as with FtsK, the ftsL* allele can bypass the loss of FtsN function.
Fig. 7. FtsL* bypasses the essential function of FtsN.
(A) Cells of TB28 [WT], MT10 [ftsL*], MT39 [ftsN::Tn5-1] and MT31 [ftsL* ftsN::Tn5-1] containing the integrated attHKNP102 plasmid (Plac::ftsN) were grown overnight in LB broth supplemented with 1 mM IPTG at 30°C. Following normalization for cell density (OD600 = 2), the resulting cultures were serially diluted (10−1 to 10−6), and 5 μl of each dilution was spotted on LB plates with or without 1 mM IPTG. The plates were incubated overnight at 30°C and photographed. (B) Cells of TB28, MT10, MT70 [ΔftsN] and MT71 [ftsL* ΔftsN] containing the integrated attHKNP102 plasmid (Plac::ftsN) were processed as in A.
FtsL* largely bypasses the essential function of FtsA
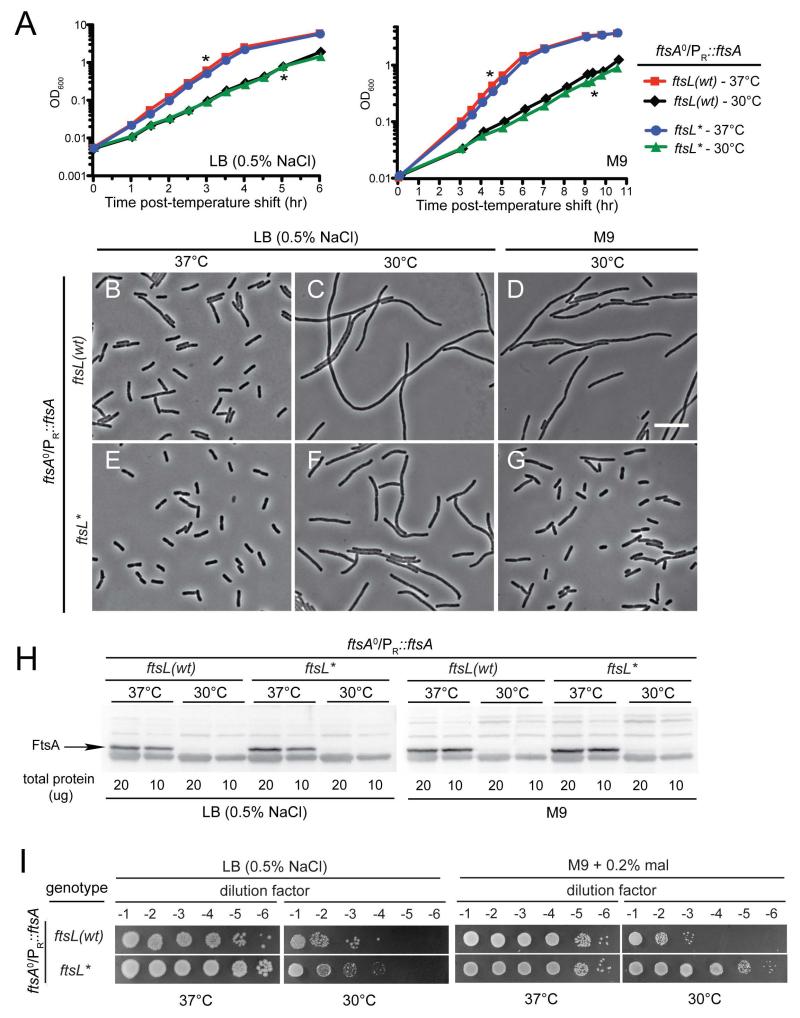
Our results with ftsL* were reminiscent of studies from Margolin and co-workers with an allele of ftsA called ftsA*, which encodes FtsA(R286W) (Geissler et al., 2003). Cells with the ftsA* mutation are shorter than normal and do not require the essential functions of several division proteins including ftsK, ftsN and zipA (Geissler et al., 2003; Geissler and Margolin, 2005; Geissler et al., 2007; Bernard et al., 2007). To further investigate the similarity between the phenotypes conferred by ftsA* and ftsL*, we assessed the ability of ftsL* to suppress the essentiality of zipA. Indeed, we found that a zipA deletion could be transduced into a strain containing either the ftsA* or ftsL* alleles but not wild-type (data not shown). Given the phenotypic similarities displayed by cells with the ftsL* and ftsA* alleles, FtsL and FtsA may lie within the same genetic pathway that modulates divisome activation and constriction initiation, possibly with FtsK, FtsN and ZipA acting upstream. If so, FtsL might either serve as an intermediary between FtsK/FtsN/ZipA and FtsA, or function downstream of FtsA to activate septal PG synthesis and cell constriction. To differentiate between these possibilities, we investigated the potential of ftsL* mutants to bypass the essential function of FtsA. For these experiments, we employed a previously characterized strain in which FtsA could be depleted by growth at low temperature (30°C) (Hale and de Boer, 1999). Cells of this strain harbor a frameshift mutation in the native ftsA gene, indicated as ftsA0, and possess a second copy of ftsA on a low copy plasmid under the control of the lambda PR promoter and a temperature-sensitive CI repressor (CI857). Thus, at 37°C, the repressor is inactivated, ftsA is expressed, and cells divide more or less normally. However, shifting the cells to 30°C restores repressor function and division is inhibited due to the depletion of FtsA. The observed phenotypes of the strain at 30°C: (i) division block, (ii) growth defect, and (iii) FtsA protein depletion as monitored by immunoblotting, were all observed regardless of whether cells were grown in minimal or rich medium, although the phenotypes were most severe in LB broth (Fig. 8). Strikingly, introduction of the ftsL* allele to the ftsA0/PR::ftsA strain dramatically improved its ability to divide at 30°C in both media types and completely restored plating efficiency at 30°C on M9 minimal agar without affecting the level of FtsA depletion achieved (Fig. 8). We were unable to transduce the ftsA0 allele into an ftsL* strain in the absence of the low-copy vector expressing ftsA (data not shown), suggesting that although the FtsA-requirement is dramatically reduced, a residual level of FtsA remains important for cell division in the presence of FtsL*. We thus conclude that the FtsL* protein largely bypasses the essential activity of FtsA. FtsL is therefore likely to be functioning downstream of FtsA in the proposed divisome activation pathway.
Fig. 8. The ftsL* mutation suppresses the growth defect resulting from FtsA depletion.
(A) Overnight cultures of MT78/pDB355 [ftsA0/PR::ftsA cITS] or MT79/pDB355 [ftsL* ftsA0/PR::ftsA cITS] cells were diluted in fresh LB or M9 maltose medium and grown at 37°C or shifted to 30°C. Growth at 30°C results in the repression of the PR promoter by CI and the concomitant depletion of wild-type FtsA. (B-G) Cells were removed from the MT78/pDB355 (B, C, D) and MT79/pDB355 (E, F, G) cultures at the time points indicated by the asterisks in (A) and examined by phase contrast microscopy. Bar = 10 μm. (H) At the time points indicated by the asterisks, cells were also harvested for whole-cell extract preparation. Proteins in the resulting extracts were separated by SDS-PAGE, transferred to PVDF, and FtsA was detected with anti-FtsA antisera. (I) Cells of MT78/pDB355 [ftsA0/PR::ftsA cITS] or MT79/pDB355 [ftsL* ftsA0/PR::ftsA cITS] were grown overnight in M9 maltose medium supplemented with spectinomycin at 37°C. Following normalization for cell density (OD600 = 2), the resulting cultures were serially diluted (10−1 to 10−6), and 5 μl of each dilution was spotted on LB or M9 maltose agar. The plates were incubated at 37°C or 30°C and photographed after 12-36 hours depending on the medium.
DISCUSSION
In E. coli, FtsL lies in the middle of the linear dependency pathway of divisome assembly and is a late recruit to the cytokinetic ring structure (Ghigo et al., 1999; Aarsman et al., 2005). It forms a complex with FtsB and FtsQ prior to localizing to the divisome (Buddelmeijer and Beckwith, 2004). Outside of the observation that these factors are essential for division, the function of FtsL and its partners in the FtsQLB complex remains unclear. Here, we studied the properties of a hypermorphic ftsL mutant we call ftsL* in an effort to better understand how FtsL promotes cell division. Unlike most mutant alleles of the essential fts genes that cause a division block and the formation of smooth filamentous cells at the non-permissive temperature, ftsL* [ftsL(E88K)] was reported to induce cell lysis (Ishino et al., 1989; Ueki et al., 1992). This observation suggested that ftsL* may encode a mutant protein that fails during the process of cell constriction rather than at a step prior to initiation. We therefore reconstructed an ftsL* strain and examined its growth and morphology under permissive [LB or M9 medium at all temperatures] or non-permissive [0.5×LB-0N at 42°C] growth conditions. In all media tested, the ftsL* mutant appeared significantly shorter and wider than the wild-type control strain, with the greatest differential between the two displayed in 0.5×LB-0N at 42°C. We did not observe a precipitous drop in culture optical density when the ftsL* mutant was shifted to non-permissive growth conditions, but lysed cell ghosts were apparent in the culture upon microscopic analysis. Although this lysis phenotype was relatively mild in liquid medium, we assume that it is likely to be exacerbated on solid 0.5×LB-0N agar at 42°C, resulting in the severe defect in plating efficiency observed under these conditions.
All of our microscopic and genetic results suggest that the size and growth phenotypes of the ftsL* mutant stem from an accelerated division process, most likely via the premature initiation of cell constriction. It is less clear why the mutant also appears to have a wider cell width and undergoes lysis in non-permissive conditions. However, given the importance of PG biogenesis for cell shape and integrity, these phenotypes probably result from defects in the assembly of the cell wall layer that are exacerbated upon growth in medium of low osmolarity at high temperatures. The problems with PG biogenesis in the mutant may be directly related to accelerated division, for example by the initiation of certain cell wall remodeling processes out of sequence or before the proper controls are in place. Alternatively, the shape and lysis phenotypes may result from an indirect and negative effect of a hyperactive divisome on the cell elongation machinery, possibly via a competition for cell wall precursors or protein components shared between the complexes. In either case, our findings point to a role for FtsL in the control of divisome function.
Previous studies in Bacillus subtilis have implicated FtsL as a possible point of regulation in the activity of the divisome (Goranov et al., 2005; Bramkamp et al., 2006). FtsL is highly unstable in B. subtilis and subject to degradation by the membrane protease YluC, a member of the site-2-protease family of proteases involved in regulated intramembrane proteolysis (Bramkamp et al., 2006). Importantly, inactivation of YluC or alterations of FtsL that prevent its turnover result in a short-cell phenotype, suggesting that FtsL levels are limiting for division (Bramkamp et al., 2006). This turnover of FtsL has also been implicated in blocking cell division in response to problems with DNA replication (Goranov et al., 2005). Given these results in B. subtilis, a simple explanation for the early division phenotype of the ftsL* mutant in E. coli is that the variant protein is stabilized and accumulates to higher than normal levels. However, although the steady-state level of FtsL* as assessed by immunoblot appears to be slightly higher than that of the wild-type protein, our genetic analysis indicates that the phenotypes are unlikely to be related to FtsL protein levels and/or stability. Overproduction of wild-type FtsL was not found to result in premature cell division. Furthermore, the expression of wild-type ftsL in the presence of ftsL* restored normal division and growth on 0.5×LB-0N at 42°C. Therefore, rather than affecting FtsL protein levels, the E88K substitution in the periplasmic coiled-coil domain of FtsL* is likely to result in an altered conformation of the protein that prematurely stimulates cell division. How the altered protein achieves this activation is not clear. One possibility is that it causes the FtsQLB complex to be recruited to the divisome early, thereby accelerating divisome maturation and the subsequent initiation of constriction. However, it is hard to envisage how merely speeding up the normal divisome assembly process could have such dramatic effects on cell shape and osmotic stability. We therefore favor an alternative explanation in which the FtsL* variant exerts its effects by short-circuiting the normal controls preventing the initiation of septal PG biogenesis and cell constriction prior to the proper completion of divisome assembly. The existence of a signaling process required for stimulating septal PG biogenesis following divisome assembly is consistent with recent work in Caulobacter cresentus showing that small membrane proteins produced in response to DNA damage can block division at the level of FtsI and FtsW activation (Modell et al., 2011; Modell et al., 2014). The FtsQLB complex is an attractive candidate for mediating this activation given its presence upstream of FtsW and FtsI in the recruitment dependency pathway (Goehring and Beckwith, 2005) and the potential interactions between these proteins suggested by two-hybrid analysis (Karimova et al., 2005). The properties of the FtsL* variant are consistent with a model in which the FtsQLB complex exists in one of two conformational states upon recruitment to the division site, “OFF” or “ON”, and that its conformation is somehow sensitive to the status of divisome assembly. Normally, the complex may only switch to the ON conformation to stimulate downstream events like septal PG synthesis once it “senses” that divisome assembly is complete. However, in the presence of FtsL* the complex may spontaneously convert to the ON state such that it stimulates constriction before the division machinery is capable of safely initiating the process, thus causing problems with septal PG biogenesis and remodeling and rendering cells sensitive to low osmolarity and extremes of temperature.
What signals might FtsQLB receive for its potential role in monitoring divisome assembly? Some clues were provided from our analysis of ftsL* suppressors as well as the recent literature. Because the essential functions of FtsN, FtsK and FtsA are completely or partially bypassed by the FtsL* variant, we propose that these proteins might communicate the status of divisome assembly to the FtsQLB complex. The bitopic membrane protein FtsN has long been thought to play a role in initiating constriction (Corbin et al., 2004) due to its status as the last essential protein to be recruited to the divisome in the dependency pathway (Addinall et al., 1997) and the ability of FtsN overexpression to stimulate division and suppress defects in numerous components of the divisome (Dai et al., 1993; Draper et al., 1998; Geissler and Margolin, 2005; Goehring, Petrovska, et al., 2007; Reddy, 2007). This idea was reinforced with the demonstration that FtsN is recruited to the divisome in a self-enhancing process involving its small, membrane-proximal, essential domain (EFtsN) and its C-terminal, PG-binding SPOR domain (SFtsN) (Gerding et al., 2009). Based on this observation, a model for a positive feedback loop that drives cell constriction was proposed in which EFtsN stimulates septal PG synthesis and remodeling to create the recruitment signal for SFtsN, which brings more EFtsN to the division site to stimulate more septal PG synthesis, and so on (Gerding et al., 2009). Thus, according to this model, it is the accumulation of EFtsN above a critical threshold at midcell that likely acts as one of the signals that divisome assembly is complete. The similarities between the phenotypes of FtsN overproduction and FtsL* in addition to the ability of FtsL* to bypass EFtsN function further suggests that the EFtsN accumulation signal works by either directly or indirectly converting the FtsQLB complex to an active complex that promotes septal PG synthesis and cell constriction.
Importantly, the isolation and analysis of ftsA mutants that bypass the normal requirement for other essential division proteins, including FtsN, indicates that the constriction initiation mechanism involves more than EFtsN recruitment (Geissler et al., 2003; Geissler and Margolin, 2005; Goehring, Petrovska, et al., 2007; Bernard et al., 2007). One of these mutants, ftsA*(R286W), is particularly relevant because it results in a short cell phenotype and has been shown to bypass the same set of divisome proteins as ftsL* (Geissler et al., 2003; Geissler and Margolin, 2005; Geissler et al., 2007). Thus, similar to our proposal for the FtsQLB complex, FtsA may also exist in one of two possible conformations, OFF and ON, that may participate in the control of constriction initiation. Recent results from Pichoff and Lutkenhaus suggest that this conformational change may be related to the polymeric status of FtsA with the polymeric form being inhibitory and the monomeric or reduced polymeric form being stimulatory for division (Pichoff et al., 2012). In addition, evidence from the Margolin group has been accumulating indicating that the cytoplasmic N-terminus of FtsN interacts directly with FtsA in such a way that it would compete with the formation of FtsA polymers (Corbin et al., 2004; Busiek et al., 2012; Szwedziak et al., 2012; Busiek and Margolin, 2014). Taken together, these results have led to the suggestion that the N-terminus of FtsN promotes a conformation of FtsA that can stimulate constriction (Busiek et al., 2012; Busiek and Margolin, 2014). The observation that FtsL* can largely bypass the need for FtsA in addition to FtsN indicates that the FtsQLB complex likely to be downstream of FtsA in this putative signaling pathway (Fig. 9). The complex may therefore be sensing FtsA conformation in addition to EFtsN recruitment as a means of assessing the status of divisome assembly prior to stimulating constriction. Additional support for this working model for the control of constriction initiation can be found in the accompanying report from Liu and coworkers (Liu et al., 2014). What is missing from these models is a definitive role for FtsK. Defects in FtsK are suppressed by increased expression of FtsN, FtsB, FtsQ as well as the ftsA* and ftsL* alleles (Geissler and Margolin, 2005), suggesting its essential N-terminal domain is providing an input upstream of the FtsQLB complex in the putative signaling pathway. However, whether it is working through an effect on FtsA, an interaction with the FtsQLB complex (Dubarry et al., 2010), or both is unclear.
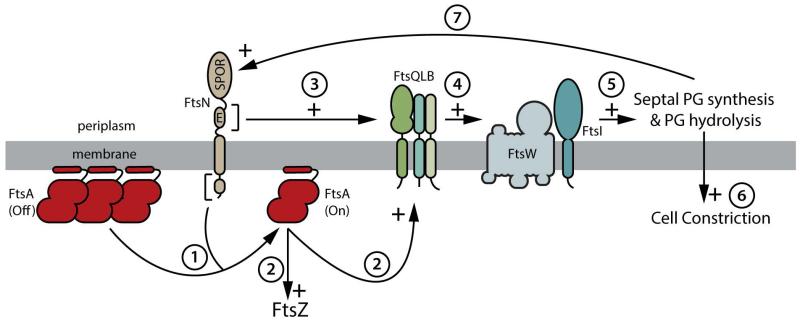
Figure 9. A potential signaling system involved in controlling constriction initiation.
Shown is an illustration highlighting the major features of a model for a signaling pathway involved in controlling the initiation of cell constriction by the divisome. The transition of FtsA from an OFF to an ON conformation appears to be one of the major signals and may involve a change in polymerization state (step 1). The ON conformation of FtsA may then promote an altered, more stable, form of the Z-ring as well as directly or indirectly signaling its altered status to the FtsQLB complex (step 2). Because reduced FtsA self interaction bypasses the essential functions of FtsN, FtsN may normally facilitate the disruption of FtsA polymers to stimulate constriction (step 1). The essential domain of FtsN (E) is also likely to either communicate the status of divisome assembly directly to the FtsQLB complex to stimulate constriction (step 3) or stimulate the synthesis of septal PG by activating the septal PG biogenesis machinery (FtsW and FtsI) directly in a manner that is redundant with FtsQLB activity (not shown). Similarly, given that FtsA and FtsL variants can bypass FtsK function, FtsK may also play a role in modulating FtsA conformation and or the communication of divisome assembly status directly to FtsQLB (not shown). The input from upstream signals received by FtsQLB is likely to cause a conformational change in the complex that is, in turn, communicated to FtsW, FtsI, and the rest of the PG synthetic apparatus to stimulate cell wall remodeling and cell constriction (steps 4, 5, and 6). The entire process is likely to be reinforced by positive feedback loops involving FtsN recruitment (step 7) and possibly other as yet to be identified feedbacks. See text for details, and also the accompanying paper from Liu and co-workers for additional support for this general scheme (Liu et al., 2014).
It is important to note that, based on the results with FtsN from the de Boer lab (Gerding et al., 2009), the entire constriction initiation system proposed in Figure 9 is likely to be self-enhancing and intimately connected with the recruitment of the relevant components to the divisome. Thus, it may only be a few molecules of each factor that are recruited to midcell to initiate the cycle, but their activity is expected to be rapidly amplified to attract additional components and stimulate a sustained and visible constriction of the cell envelope. Such a possibility may explain the observed two-step divisome assembly process and the near simultaneous arrival of the late proteins coincident with the onset of constriction (Aarsman et al., 2005).
In summary, studies of division mutants with phenotypes other than filamentation such as ftsA* (Geissler et al., 2003) and ftsL* are revealing what appears to be a complex signaling pathway that promotes the initiation of cell constriction by the divisome. The isolation and characterization of additional alleles in this class as well as other types of mutants that fail post-initiation is likely to shed additional light on the function of essential divisome components and the mechanism of cell constriction.
EXPERIMENTAL PROCEDURES
Media, bacterial strains and plasmids
Cells were grown in LB [1% tryptone, 0.5% yeast extract, 0.5% NaCl], 0.5×LB-0N [0.5% tryptone, 0.25% yeast extract] or minimal M9 medium (Miller, 1972) supplemented with 0.2% casamino acids and 0.2% maltose. Unless otherwise indicated, antibiotics were used at 25 (chloramphenicol; Cm), 25 (kanamycin; Kan), 10 (tetracycline; Tet) or 50 (spectinomycin, Spec) μg/ml. For M9 medium, 50μg/ml Kan and 12.5μg/ml Tet were used.
The bacterial strains used in this study are listed in Table S1. All E. coli strains used in the reported experiments are derivatives of MG1655 (Guyer et al., 1981). Plasmids used in this study are listed in Table S2. PCR was performed using KOD polymerase (Novagen) for cloning purposes and Taq DNA polymerase (NEB) for diagnostic purposes, both according to the manufacturer’s instructions. Unless otherwise indicated, MG1655 chromosomal DNA was used as the template. Plasmid DNA and PCR fragments were purified using the Zyppy plasmid miniprep kit (Zymo Research) or the Qiaquick PCR purification kit (Qiagen), respectively.
Allelic replacement
A strain harboring the chromosomal ftsL* mutation was constructed by allelic exchange using a modification of a procedure that was previously described (Philippe et al., 2004). The pir-dependent suicide plasmid pMT17 [sacB CmR] contained the ftsL* gene with large regions of flanking homology. It was introduced into the recipient strain TB28(attHKMT15) [WT(Plac::ftsL TetR)] by conjugative transfer from the donor strain SM10(λpir). Briefly, 100 μl each of overnight cultures of the donor and recipient strains were mixed with fresh LB broth. After approximately 6 hours of incubation at 37°C, cells were diluted in fresh LB broth and plated on minimal M9 maltose plates supplemented with chloramphenicol, tetracycline and 1mM IPTG to select for TB28(attHKMT15) exconjugants with pMT17 integrated into the chromosome via a single cross-over. After incubation at 30°C for approximately 36 hours, one colony from this plate was picked, diluted in 10 mM MgSO4 and serial dilutions were plated on minimal M9 maltose plates with 1 mM IPTG and 6% sucrose. After incubation at 30°C for several days, 30 isolates were purified on minimal M9 maltose plates supplemented with 1mM IPTG and either chloramphenicol or 6% sucrose. The SucR CmS isolates were tested for a temperature-sensitive growth defect on 0.5×LB-0N indicative of the ftsL* allele replacing the wild-type copy of ftsL at the native chromosomal locus. Presence of the allele was then confirmed by PCR and sequencing. Strain MT10 was obtained by P1-mediated transduction of the ftsL* allele from the primary isolate into the chromosome of CH43 [leu::Tn10] by selecting for Leu+ transductants and screening for temperature-sensitive growth on 0.5×LB-0N medium.
Recombineering
The ΔftsK::KanR allele was constructed by replacing the region between the 2nd codon and the 7th codon from the stop codon of ftsK with the KanR cassette as described previously (Yu et al., 2000; Baba et al., 2006). The KanR cassette was amplified from pKD13 (Datsenko and Wanner, 2000) using the primers 5′-ATCGGGCAGGAAAAGCCTGTAACCTGGAGAGCCTTTCTTGATTCCGGGGATCCGTCGAC C-3′ and 5′-CCGGCATACGATGCATTAGTTAGTCAAACGGCGGTGGGGCTGTAGGCTGGAGCTGCTTC G-3′. The resulting PCR product was purified and electroporated into strain TB28(attHKMT117)/pKD46 as described previously (Bernhardt and de Boer, 2004) and the recombinants were selected at 30°C on a minimal M9 plate containing 50 μg/ml kanamycin and 1 mM IPTG to generate the chromosomal deletion.
Suppressor selection
To select for suppressors of the lytic temperature sensitive phenotype of MT10 [ftsL*], the strain was mutagenized either with the EzTn-Kan2 transposome (Epicentre) or with a Mariner-based transposon as previous described (Chiang and Rubin, 2002; Bernhardt and de Boer, 2004). Mutants were selected for kanamycin resistance at 30°C, yielding a library of ~ 70,000 or ~ 300,000 independent transposon insertions, respectively. The mutant libraries were plated on 0.5×LB-0N agar and incubated at 42°C to identify mutants capable of growing under these non-permissive conditions. The frequency at which survivors arose was approximately 10−5 for spontaneous suppressors and 10−4 for the transposon libraries. The sites of the transposon insertions in the suppressors were identified by arbitrarily primed PCR followed by sequencing (Bernhardt and de Boer, 2004), and the transposons were transduced into a fresh MT10 background to confirm that the suppression phenotype was linked to the insertion in question.
Microscopy and image analyses
Both light and fluorescence microscopy were performed as described previously (Uehara et al., 2009). See figure legends for specific growth conditions employed for each experiment. For the determination of constriction time shown in Figure 3 and Table 4, overnight cultures of TB28 [WT] and MT10 [ftsL*] were diluted to a starting OD600 ~ 0.03 in minimal M9 medium supplemented with 0.2% maltose and grown at 30°C until an OD600 ~ 0.15 was reached. These cultures were then back-diluted to OD600 ~ 0.005 in fresh M9 + 0.2% maltose and growth was continued at 30°C. The OD600 of the cultures was monitored at regular time intervals to determine the mass doubling time of each strain. At specific time points, cells were imaged on 2% agarose pads using phase contrast optics and cell length was measured using MicrobeTracker. Cultures were considered to be at steady state if the cell length distribution remained constant over time. At OD600 ~ 0.1, the cells were visualized on 2% agarose pads with DIC optics. The presence of a constriction was determined manually using the imaging software NIS-Elements (Nikon). Given that the cultures were in steady-state growth, the period of visible constriction was determined as described (Aarsman et al., 2005). Briefly, the constriction time (tc) was calculated using the formula [tc = (Td * ln [1 + F(x)]) / ln2], where Td is the mass doubling time and F(x) is the fraction of cells with a visible constriction.
Time-Lapse Analysis
Overnight cultures of HC261 [WT zapA-gfp] and MT90 [ftsL* zapA-gfp] were diluted in minimal M9 medium supplemented with 0.2% maltose and grown at 30°C until an OD600 ~ 0.15 - 0.3. The cells were spotted on two separate halves of a 2% agarose pad containing 1× M9 salts, 0.02% casamino acids and 0.2% maltose, the coverslip was sealed and a timelapse of both strains grown at 30°C (using a heated objective) was obtained using phase contrast and gfp optics, with frames taken every 1.5 min. The timelapse was analyzed using MicrobeTracker and SpotFinderZ to determine the total cell cycle time, as well as the division time. SpotFinderZ was used to identify diffraction-limited ZapA-GFP spots within each cell. Division time is defined as the time between the formation of a stable Z-ring (using ZapA-GFP as a proxy for FtsZ) and the end of constriction. The criteria for a stable Z-ring is two ZapA-GFP spots that are aligned perpendicular to the long axis of the cell and that remain for at least three consecutive frames. During the segmentation, the end of constriction was manually determined as the moment when no ZapA-GFP spot was visible at the division site.
Immunoblotting
Strains were grown as described in the figure legends. At the designated times, cells were harvested and whole-cell extracts were prepared as described previously (Hale and de Boer, 1999). The protein concentration of each extract was determined using the non-interfering protein assay (Genotech) according to the manufacturer’s instructions. Protein concentrations were normalized between extracts and the indicated amount of total protein from each extract was separated on a 12% or 15% SDS-PAGE gel. Proteins were transferred to a PVDF membrane (Whatman) and the membrane was blocked with Rapid-Block (Amresco) for 5 minutes. The membrane was incubated with primary antibodies diluted in Rapid-Block (either 1:10,000 dilution for anti-FtsA antibodies or 1:2,500 dilution for anti-FtsL antibodies) overnight at 4°C. In the case of the anti-FtsA antibodies, the same primary antibody solution was re-used for incubation with multiple blots. The next day, the primary antibody solution was removed and the membrane was quickly rinsed with TBST (10mM Tris-HCl pH 7.5, 100mM NaCl, 0.1% Tween-20) and then thoroughly washed three times with 25ml TBST for 10 minutes each wash. Following the final wash, the membrane was incubated with the secondary goat anti-rabbit antibodies conjugated to horseradish peroxidase (Rockland) diluted 1:40,000 in Rapid-Block for 1 hour with gentle agitation at room temperature. After this incubation period, the secondary antibody solution was discarded and the membrane was again quickly rinsed with TBST and then thoroughly washed an additional four times with 25ml TBST for 10 minutes each. The blot was developed using the Super Signal West Pico system (Pierce) according to the manufacturer’s protocol. Chemiluminescence was detected using a BioRad Chemidoc system.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank all members of the Bernhardt and Rudner laboratories for helpful comments and suggestions. We would also like to thank Piet de Boer for strains, the anti-FtsA antiserum, helpful comments, and for communicating results prior to publication. Thanks also to Jon Beckwith and the Beckwith lab for providing the anti-FtsL antiserum. This work was supported by the National Institute of Allergy and Infections Diseases of the National Institutes of Health (R01 AI083365).
REFERENCES
- Aarsman MEG, Piette A, Fraipont C, Vinkenvleugel TMF, Nguyen-Distèche M, Blaauwen, den T. Maturation of the Escherichia coli divisome occurs in two steps. Molecular Microbiology. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- Addinall SG, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Molecular Microbiology. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- Alexeeva S, Gadella TWJ, Jr, Verheul J, Verhoeven GS, Blaauwen, den T. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Molecular Microbiology. 2010;77:384–398. doi: 10.1111/j.1365-2958.2010.07211.x. [DOI] [PubMed] [Google Scholar]
- Allen JS, Filip CC, Gustafson RA, Allen RG, Walker JR. Regulation of bacterial cell division: genetic and phenotypic analysis of temperature-sensitive, multinucleate, filament-forming mutants of Escherichia. J Bacteriol. 1974;117:978–986. doi: 10.1128/jb.117.3.978-986.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends SJR, Williams K, Scott RJ, Rolong S, Popham DL, Weiss DS. Discovery and characterization of three new Escherichia coli septal ring proteins that contain a SPOR domain: DamX, DedD, and RlpA. J Bacteriol. 2010;192:242–255. doi: 10.1128/JB.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg KJ, Dewar SJ, Donachie WD. A new Escherichia coli cell division gene, ftsK. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg KJ, Hatfull GF, Donachie WD. Identification of new genes in a cell envelope-cell division gene cluster of Escherichia coli: cell division gene ftsQ. J Bacteriol. 1980;144:435–437. doi: 10.1128/jb.144.1.435-437.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard CS, Sadasivam M, Shiomi D, Margolin W. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Molecular Microbiology. 2007;64:1289–1305. doi: 10.1111/j.1365-2958.2007.05738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PAJ. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Molecular Microbiology. 2004;52:1255–1269. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bramkamp M, Weston L, Daniel RA, Errington J. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Molecular Microbiology. 2006;62:580–591. doi: 10.1111/j.1365-2958.2006.05402.x. [DOI] [PubMed] [Google Scholar]
- Buddelmeijer N, Beckwith J. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Molecular Microbiology. 2004;52:1315–1327. doi: 10.1111/j.1365-2958.2004.04044.x. [DOI] [PubMed] [Google Scholar]
- Buddelmeijer N, Judson N, Boyd D, Mekalanos JJ, Beckwith J. YgbQ, a cell division protein in Escherichia coli and Vibrio cholerae, localizes in codependent fashion with FtsL to the division site. Proc Natl Acad Sci USA. 2002;99:6316–6321. doi: 10.1073/pnas.092128499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busiek KK, Margolin W. A role for FtsA in SPOR-independent localization of the essential Escherichia colicell division protein FtsN. Molecular Microbiology. 2014;92:1212–1226. doi: 10.1111/mmi.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busiek KK, Eraso JM, Wang Y, Margolin W. The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J Bacteriol. 2012;194:1989–2000. doi: 10.1128/JB.06683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Beckwith J. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Molecular Microbiology. 2001;42:395–413. doi: 10.1046/j.1365-2958.2001.02640.x. [DOI] [PubMed] [Google Scholar]
- Chiang SL, Rubin EJ. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene. 2002;296:179–185. doi: 10.1016/s0378-1119(02)00856-9. [DOI] [PubMed] [Google Scholar]
- Corbin BD, Geissler B, Sadasivam M, Margolin W. Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J Bacteriol. 2004;186:7736–7744. doi: 10.1128/JB.186.22.7736-7744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Xu Y, Lutkenhaus J. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts) J Bacteriol. 1993;175:3790–3797. doi: 10.1128/jb.175.12.3790-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lallo G, Fagioli M, Barionovi D, Ghelardini P, Paolozzi L. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology. 2003;149:3353–3359. doi: 10.1099/mic.0.26580-0. [DOI] [PubMed] [Google Scholar]
- Draper GC, McLennan N, Begg K, Masters M, Donachie WD. Only the N-terminal domain of FtsK functions in cell division. J Bacteriol. 1998;180:4621–4627. doi: 10.1128/jb.180.17.4621-4627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubarry N, Possoz C, Barre F-X. Multiple regions along the Escherichia coli FtsK protein are implicated in cell division. Molecular Microbiology. 2010;78:1088–1100. doi: 10.1111/j.1365-2958.2010.07412.x. [DOI] [PubMed] [Google Scholar]
- G Chaulk S, Smith Frieday MN, Arthur DC, Culham DE, Edwards RA, Soo P, et al. ProQ Is an RNA Chaperone that Controls ProP Levels in Escherichia coli. Biochemistry. 2011;50:3095–3106. doi: 10.1021/bi101683a. [DOI] [PubMed] [Google Scholar]
- Geissler B, Margolin W. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Molecular Microbiology. 2005;58:596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Elraheb D, Margolin W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci USA. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Shiomi D, Margolin W. The ftsA* gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology (Reading, Engl) 2007;153:814–825. doi: 10.1099/mic.0.2006/001834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding MA, Liu B, Bendezú FO, Hale CA, Bernhardt TG, de Boer PAJ. Self-enhanced accumulation of FtsN at Division Sites and Roles for Other Proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol. 2009;191:7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo JM, Weiss DS, Chen JC, Yarrow JC, Beckwith J. Localization of FtsL to the Escherichia coli septal ring. Molecular Microbiology. 1999;31:725–737. doi: 10.1046/j.1365-2958.1999.01213.x. [DOI] [PubMed] [Google Scholar]
- Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15:R514–26. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Goehring NW, Gueiros-Filho F, Beckwith J. Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli. Genes Dev. 2005;19:127–137. doi: 10.1101/gad.1253805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Petrovska I, Boyd D, Beckwith J. Mutants, suppressors, and wrinkled colonies: mutant alleles of the cell division gene ftsQ point to functional domains in FtsQ and a role for domain 1C of FtsA in divisome assembly. J Bacteriol. 2007;189:633–645. doi: 10.1128/JB.00991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Robichon C, Beckwith J. Role for the nonessential N terminus of FtsN in divisome assembly. J Bacteriol. 2007;189:646–649. doi: 10.1128/JB.00992-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez MD, Akbay EA, Boyd D, Beckwith J. Multiple Interaction Domains in FtsL, a Protein Component of the Widely Conserved Bacterial FtsLBQ Cell Division Complex. J Bacteriol. 2010;192:2757–2768. doi: 10.1128/JB.01609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goranov AI, Katz L, Breier AM, Burge CB, Grossman AD. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc Natl Acad Sci USA. 2005;102:12932–12937. doi: 10.1073/pnas.0506174102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer MS, Reed RR, Steitz JA, Low KB. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Barondess JJ, Beckwith J. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PA. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CA, de Boer PAJ. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J Bacteriol. 2002;184:2552–2556. doi: 10.1128/JB.184.9.2552-2556.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota Y, Mordoh J, Jacob F. On the process of cellular division in Escherichia coli. 3. Thermosensitive mutants of Escherichia coli altered in the process of DNA initiation. J Mol Biol. 1970;53:369–387. doi: 10.1016/0022-2836(70)90072-0. [DOI] [PubMed] [Google Scholar]
- Ishino F, Jung HK, Ikeda M, Doi M, Wachi M, Matsuhashi M. New mutations fts-36, lts-33, and ftsW clustered in the mra region of the Escherichia coli chromosome induce thermosensitive cell growth and division. J Bacteriol. 1989;171:5523–5530. doi: 10.1128/jb.171.10.5523-5530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol. 2005;187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadria AS, Senes A. The Transmembrane Domains of the Bacterial Cell Division Proteins FtsB and FtsL Form a Stable High-Order Oligomer. Biochemistry. 2013;52:7542–7550. doi: 10.1021/bi4009837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khattar MM, Begg KJ, Donachie WD. Identification of FtsW and characterization of a new ftsW division mutant of Escherichia coli. J Bacteriol. 1994;176:7140–7147. doi: 10.1128/jb.176.23.7140-7147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Persons L, Lee L, de Boer PAJ. Roles for both FtsA and the FtsQLB subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Molecular Microbiology. 2014 doi: 10.1111/mmi.12906. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. FtsN--trigger for septation. J Bacteriol. 2009;191:7381–7382. doi: 10.1128/JB.01100-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton. 2012;69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Löwe J. Double-Stranded DNA Translocation: Structure and Mechanism of Hexameric FtsK. Mol Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Mercer KLN, Weiss DS. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J Bacteriol. 2002;184:904–912. doi: 10.1128/jb.184.4.904-912.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; New York: 1972. [Google Scholar]
- Modell JW, Hopkins AC, Laub MT. A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 2011;25:1328–1343. doi: 10.1101/gad.2038911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell JW, Kambara TK, Perchuk BS, Laub MT. A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol. 2014;12:e1001977. doi: 10.1371/journal.pbio.1001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NT, Dinh T, Bernhardt TG. A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. J Bacteriol. 2011;193:4973–4983. doi: 10.1128/JB.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe N, Alcaraz J-P, Coursange E, Geiselmann J, Schneider D. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid. 2004;51:246–255. doi: 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Shen B, Sullivan B, Lutkenhaus J. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA’s self-interaction competes with its ability to recruit downstream division proteins. Molecular Microbiology. 2012;83:151–167. doi: 10.1111/j.1365-2958.2011.07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy M. Role of FtsEX in cell division of Escherichia coli: viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. J Bacteriol. 2007;189:98–108. doi: 10.1128/JB.01347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichon C, Karimova G, Beckwith J, Ladant D. Role of Leucine Zipper Motifs in Association of the Escherichia coli Cell Division Proteins FtsL and FtsB. J Bacteriol. 2011;193:4988–4992. doi: 10.1128/JB.00324-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaluru H, SaiSree L, Reddy M. Role of SufI (FtsP) in cell division of Escherichia coli: evidence for its involvement in stabilizing the assembly of the divisome. J Bacteriol. 2007;189:8044–8052. doi: 10.1128/JB.00773-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt KL, Peterson ND, Kustusch RJ, Wissel MC, Graham B, Phillips GJ, Weiss DS. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J Bacteriol. 2004;186:785–793. doi: 10.1128/JB.186.3.785-793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Molecular Microbiology. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Freund SM, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Parzych KR, Dinh T, Bernhardt TG. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 2010;29:1412–1422. doi: 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki M, Wachi M, Jung HK, Ishino F, Matsuhashi M. Escherichia coli mraR gene involved in cell growth and division. J Bacteriol. 1992;174:7841–7843. doi: 10.1128/jb.174.23.7841-7843.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursinus A, van den Ent F, Brechtel S, de Pedro M, Höltje J-V, Löwe J, Vollmer W. Murein (peptidoglycan) binding property of the essential cell division protein FtsN from Escherichia coli. J Bacteriol. 2004;186:6728–6737. doi: 10.1128/JB.186.20.6728-6737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de PUTTE P, van DILLEWIJN, ROERSCH A. THE SELECTION OF MUTANTS OF ESCHERICHIA COLI WITH IMPAIRED CELL DIVISION AT ELEVATED TEMPERATURE. Mutat Res. 1964;106:121–128. doi: 10.1016/0027-5107(64)90014-4. [DOI] [PubMed] [Google Scholar]
- Wang L, Khattar MK, Donachie WD, Lutkenhaus J. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol. 1998;180:2810–2816. doi: 10.1128/jb.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M, Bernhardt TG. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acad Sci USA. 2011;108:E1052–E1060. doi: 10.1073/pnas.1107780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Tan K, Joachimiak A, Bernhardt TG. A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division. Molecular Microbiology. 2012;85:768–781. doi: 10.1111/j.1365-2958.2012.08138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J-C, van den Ent F, Neuhaus D, Brevier J, Löwe J. Solution structure and domain architecture of the divisome protein FtsN. Molecular Microbiology. 2004;52:651–660. doi: 10.1111/j.1365-2958.2004.03991.x. [DOI] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.