Abstract
Heterogeneous nuclear ribonucleoprotein K (hnRNP K) is importantly involved in the regulation of development, DNA damage response, and several human diseases. The molecular mechanisms that control the expression of hnRNP K are largely unknown. In the present study, we investigated the detailed mechanism of the transcriptional regulation of human hnRNP K gene. Two activating and one repressive elements located in the proximal segment of the transcriptional initiation site were identified in hnRNP K gene. A 19 bp-region was responsible for the inhibitory activities of the repressor element. Twenty proteins were identified by DNA-affinity purification and mass spectrometry analyses as binding partners of the primary activating element in the hnRNP K promoter. Chromatin immunoprecipitation and EMSA analysis confirmed the binding of Sp1 with hnRNP K promoter. Sp1 enhanced the promoter activity, increased the expression of hnRNP K, and reduced the mRNA level of angiotensinogen, a gene known to be negatively regulated by hnRNP K. In summary, the current study characterized the promoter elements that regulate the transcription of human hnRNP K gene, identified 20 proteins that bind to the primary activating element of hnRNP K promoter, and demonstrated a functional effect of Sp1 on hnRNP K transcription.
Keywords: hnRNP K, Sp1, Promoter, Transcription, Gene expression
1. Introduction
Heterogeneous nuclear ribonucleoprotein K (hnRNP K), which is present in both cytoplasmic and nuclear compartments [1,2], has been demonstrated to be involved in a number of fundamental biological processes. HnRNP K can regulate the transcription of several genes [3–9]. It was found to bind to the single-stranded C-rich sequence within the poly (Pu/Py) tract [8] or CT-rich element in repeat 3 region [9] to promote the transcription of vascular endo-thelial growth factor (VEGF) or low density lipoprotein receptor (LDLR). It can also bind to the mRNAs of many genes such as glucose-6-phosphate dehydrogenase (G6PD) [10] and renin [11] to suppress splicing or enhance stabilization. Bomsztyk K et al. found hnRNP K was bound to elongation factor 1α (EF-1α) [12], and a series of studies showed a role of hnRNP K in protein translation. The tyrosine and Ser302 of hnRNP K were phosphorylated by angiotensin II through Ang II-AT1R-src-PKCδ pathway, which promoted the translation of VEGF mRNA [13,14]. Together with hnRNP E1and DEAD-box RNA helicase 6(DDX6), hnRNP K can also bind to the differentiation control element (DICE) in 3′ untranslated region (3′UTR) of 15-lipoxygenase mRNA to form a silencing complex, which could inhibit the translation of 15-lipoxygenase by control the assembly of 80s ribosomes [15,16]. Furthermore, hnRNP K contains more than seventy potential phosphorylation sites [17], suggesting it might be closely related to a variety of signal trans-duction pathways.
hnRNP K participates in the regulation of various physiological and pathological processes, including development [18–22], DNA damage response [23] and several human diseases [24–29]. It was reported recently that hnRNP K mRNA was gradually decreased in the rat central nervous system, but remained high in the peripheral nervous system throughout embryonic development [22]. Another study showed that the expression of hnRNP K was immediately induced by DNA damage [23]. Our previous study demonstrated that hnRNP K was up-regulated in the renal medulla of Consomic SS-Chr 13BN/Mcwi rat (SS–13BN) rats fed a 4% NaCl diet, but down-regulated in salt sensitive rat (SS) rats [30]. SS rats develop hypertension when fed high-salt diets. SS-13BN rats are Consomic rats in which chromosome 13 in SS rats has been replaced by the corresponding chromosome from Brown Norway (BN) rats. Blood pressure salt-sensitivity was significantly reduced in SS-13BN rats [30]. The down-regulation of hnRNP K in SS rats led to up-regulation of angiotensinogen (AGT) locally in the kidneys of SS rats [30,31], which might contribute to the development of salt-induced hypertension.
Given the important roles of hnRNP K in multiple physiological and pathological processes, it would be important to understand how the transcription of hnRNP K is regulated. However, little was known about the transcriptional mechanisms controlling the expression of hnRNP K. The purpose of current study is to identify and characterize the DNA sequence elements and transcriptional factors that regulate the transcription of hnRNP K.
2. Materials and methods
2.1. Cell culture
MCF-7, 293T and HK2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics (Invitrogen). Cells were maintained in a humidified 5% CO2 atmosphere at 37 °C, the cell media were changed every 2 days, and the cells were subjected to passage every 3–4 days.
2.2. Plasmids construction
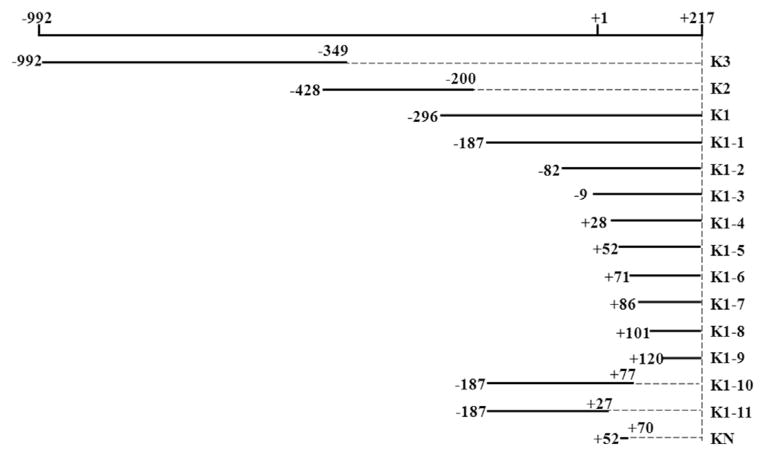
To construct luciferase reporter plasmids containing segments of hnRNP K proximal promoter (−992 to +217 bp, relative to the transcription start site +1), DNA fragments were amplified by PCR using Taq DNA polymerase (TaKaRa, Japan) with human genomic DNA as the template. The primer sequences are listed in Table 1. A Kpn I or an Xho I site was introduced into the sense and antisense primers respectively (underlined) and the length of each construct was shown in Fig. 1. The PCR conditions were as follows: 94 °C for 3 min; 30 cycles of 94 °C for 30sec, 60 °C for 30sec, and 72 °C for 90sec; and 72 °C for 10 min. PCR products were excised and purified from a 1.5% agarose gel, and cloned into the pMD 18T vector (TaKaRa, Japan). After identification by DNA sequencing, the target gene was recovered from the recombinant plasmid by digestion with Kpn I and Xho I and cloned into the pGL3-basic vector to construct the luciferase fusion plasmids. The fidelity of the promoter regions was further confirmed by the restriction enzyme digestion and DNA sequencing.
Table 1.
Oligonucleotides used for plasmid construction.a
| Oligo name | sequence(5′–3′) |
|---|---|
| K1-Forward | CGGGGTACCGCTTCGGGCACGAGTGTGGG |
| K1-Reverse | CCGCTCGAGGCCTTTCAGGGAGCCCCAACC |
| K2-Forward | CGGGGTACCGGGCGCCCCGGACCATTAC |
| K2-Reverse | CCGCTCGAGGGGACCGATGTTGCGCGAGG |
| K3-Forward | CGGGGTACCTGGGCGGAGGCTGGAAGGTT |
| K3-Reverse | CCGCTCGAGCGGTAAGTGCGGGCCGTCTG |
| K1–1-Forward | CGGGGTACCCTGTAAGGACAGGAACCGCCGC |
| K1–1-Reverse | CCGCTCGAGGCCTTTCAGGGAGCCCCAACC |
| K1–2-Forward | CGGGGTACCTCGGGGCTGGAAAACGCCC |
| K1–2-Reverse | CCGCTCGAGGCCTTTCAGGGAGCCCCAACC |
| K1–3-Forward | CGGGGTACCGGTTCGCCCCCTAGCCGCC |
| K1–3-Reverse | CCGCTCGAGGCCTTTCAGGGAGCCCCAACC |
| K1–4-Forward | CGGGGTACCGAGTGCGCGAACGAGAAAGGAGG |
| K1–4-Reverse | CCGCTCGAGGCCTTTCAGGGAGCCCCAACC |
| K1–5-Forward | CGGGGTACCGGGCGCTCCAGGCGACAGC |
| K1–5-Reverse | CCGCTCGAGGCCTTTCAGGGAGCCCCAACC |
| K1–6-Forward | CGGGGTACCACTGCAGACGCCATTATCCTCTGTTTC |
| K1–6-Reverse | CCGCTCGAGGCCTTTCAGGGAGCCCCAACC |
| K1–7-Forward | CGGGGTACCATCCTCTGTTTCTCTGCTGCACCG |
| K1–7-Reverse | CCGCTCGAGGCCTTTCAGGGAGCCCCAACC |
| K1–8-Forward | CGGGGTACCGCTGCACCGACCTCGACGTC |
| K1–8-Reverse | CCGCTCGAGGCCTTTCAGGGAGCCCCAACC |
| K1–9-Forward | CGGGGTACCCTTGCCTGTGTCCCACTTGTTCGC |
| K1–9-Reverse | CCGCTCGAGGCCTTTCAGGGAGCCCCAACC |
| K1–10-Forward | CGGGGTACCCTGTAAGGACAGGAACCGCCGC |
| K1–10-Reverse | CCGCTCGAGCTGCAGTGCTGTCGCCTGG |
| K1–11-Forward | CGGGGTACCCTGTAAGGACAGGAACCGCCGC |
| K1–11-Reverse | CCGCTCGAG ACTAGCTGGGGGGAGGGG |
| KN-sequence-F | CGGGCGCTCCAGGCGACAGCC |
| KN-sequence-R | TCGAGGCTGTCGCCTGGAGCGCCCGGTAC |
| K1–5spmut-F | GGG at GGGGAAGGGGCGT |
| K1–5spmut-R | CTCCCTGTGCGCGTGAACT |
| K2spmut-F | GGGCTGTGGTCTGTGGGCTG |
| K2spmut-R | GGG AT GGGGTAAGGGGCTTCG |
| AGT-forward | CTCACTATGCCTCTGACCTGGA |
| AGT-reverse | CATGGTCAGGTGGATGGTCC |
| 37 bp probe-F | GGAAAACGCCCCTTCCCCGCCCCCTCCCTGTGCGCGT |
| 37 bp probe-R | ACGCGCACAGGGAGGGGGCGGGGAAGGGGCGTTTTCC |
| Mutant probe-F | GGAAAACGCCCCTTCAAAAACCCCTCCCTGTGCGCGT |
| Mutant probe-R | ACGCGCACAGGGAGGGGTTTTTGAAGGGGCGTTTTCC |
A Kpn I or an Xho I site was introduced into the forward and reverse primers respectively(underlined).
Fig. 1.
All DNA segments analyzed in the current study. The numbers in the figure indicated the positions of start and end nucleotides, with +1/−1 indicating the transcription start site. The name of each construct was indicated on the right such as the K1–1 represents the sequence start from −187 bp to +217 bp.
2.3. Transient transfection
Sp1 overexpression plasmid was kindly provided by Dr. Gun-tram Suske at Philipps-University Marburg, Germany. The lucif-erase reporter plasmids were constructed as above. Cells were cultured at 24 or 6-well plates and transfected with the plasmids using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA). For co-transfection, two kinds of plasmids were equimolarly mixed before being combined with Lipofectamine™ 2000. All cells were harvested at 24 h after transfection.
2.4. Luciferase assay
Luciferase assay were carried out in 293T, MCF-7 and HK2 cells cultured in 24-well plates. The transfected cells were harvested and lysed. Luciferase assays were performed using a luciferase assay kit (Promega, USA). Renilla luciferase was used for normalization. Each experiment was repeated at least three times.
2.5. Western blot
Western blot was carried out in HK2 cells cultured in 6-well plates. Transfected cells were harvested and lysed by 8M urea. Western blot was performed as described previously28. HnRNP K antibody was from Santa Cruz Biotechnology and Sp1 antibody was from Cell Signal Technology.
2.6. Real time PCR
Real time PCR analysis using the SYBR chemistry (TaKaRa, Japan) was performed following the manufacturer’s protocol. Oligonu-cleotide sequences for human angiotensinogen are listed in Table 1.
2.7. In silico analysis
The conserved regions of hnRNP K gene are identified through a comparison of the 5000 bp sequence upstream and 220 bp downstream of the transcription initiation sites (including part of the untranslated regions) of the mouse, rat, and human hnRNP K genes using NCBI nucleotide database (Blast). Putative transcription factors were searched on Consite (http://consite.genereg.net/) using default settings. TFSEARCH (http://www.cbrc.jp./research/db/TFSEARCH.html) was also used to predict the transcription factor binding sites of the hnRNP K gene.
2.8. Nuclear extract preparation
Nuclear extracts were prepared from 293T cells using a nuclear extract kit (Sangon Biotech, China). Briefly, cells were grown to 90% confluence, harvested, and washed with phosphate-buffered saline supplemented 1 mM DTT, 0.05 mM PMSF and 1 mM phosphatase inhibitor. Cell pellets were then resuspended in hypotonic buffer with the same protease inhibitors for 10 min on ice, violently shaken for 10 s, and the lysate was centrifuged at 800 g for 5 min. The supernatant was discarded and the precipitated nuclei were washed again with hypotonic buffer and centrifuged at 2300 rpm for 5 min, and then resuspended in lysis buffer and incubated on ice for 20 min. The sample was centrifuged at 12,500 g for 10 min. Aliquots of the supernatant containing the nuclear extracts were stored at −80 °C.
2.9. Purification of DNA binding proteins
The 5′-end biotinylated and unlabeled probes (−187 bp to −9 bp) of hnRNP K promoter sequences were prepared by PCR using pfu DNA Polymerase (CW Biotech, China) and the fidelity of the sequences were confirmed by DNA sequencing. The following procedure is based on the interaction between biotin and strepta-vidin as described previously [5] with a few modifications. Briefly, 500 μl of 0.5 × SSC was added to 500 pM of probes. The streptavidin-paramagnetic particles (Promega) were washed three times with 0.5 × SSC and resuspended in 100 μl of 0.5 × SSC. Probes and particles were then combined and incubated for 15 min at room temperature. After three washes with 300 μl of buffer A (5 mM HEPES, 26% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM, PMSF, and 300 mM NaCl, 0.05% Triton X-100, pH 7.9), 1 mg of nuclear proteins was added to the affinity particles and incubated on an end to end rocking bed for 1 h at 4 °C. Then particles were consecutively washed with buffer A, buffer B (buffer A cantains 100 mM NaCl), and buffer C (buffer A without NaCl). Proteins bound to the particles were released by incubation with 20 μl of 1 × SDS for 10 min at 95 °C. In the control experiments, 5000 pM of unlabeled probes (as competitor) was used. The resulting protein solutions, with and without competitor, were separated by 15% SDS-PAGE and stained with Coomassie Blue R250. The differential protein bands contained potential transcription factors.
2.10. Proteomic identification and characterization of DNA binding proteins
The differential protein bands were cut out from SDS-PAGE gel. Following tryptic digestion, samples were reconstituted in 25 μl of the initial LC mobile phase (0.1% formic acid). Peptide separation was performed by Thermo Scientific Easy-nLC 1000 (Waltham, MA, USA). Digested peptides were separated on an HPLC column (C18, 2 μm, 100 Å, 75 μm × 15 cm, Waltham, MA, USA) with a 3%–90% linear gradient of solvent B (90% Acetonitrile and 0.1% Formic acid) for 90 min with a flow rate of 0.35 μl/min. For the study of the fragmentation pattern, a Thermo Scientific® (Waltham, MA, USA) Q Exactive mass spectrometer with an ESI ion source was used, using the following tune parameters: spray voltage, 2600 V; capillary temperature, 275 °C; and collision energy, 27% Higher Energy Collisional Dissociation (HCD). The instrument was calibrated in positive and negative ionization mode before any infusion experiment to ensure the mass accuracy of the results. The acquired mass data were analyzed by Proteome Discoverer 1.3 in Uniprot-human database.
2.11. Chromatin Immunoprecipitation
Chromatin Immunoprecipitation (ChIP) analysis was performed using the EZ ChIP™ kit from Upstate (Millipore). 293T cells were fixed by 1% formaldehyde and incubated with modest shaking for 30 min at room temperature. Cells were then washed twice with cold PBS. The pellet was resuspended and lysed, and nuclei were isolated and sonicated until the chromatin had an average length of 500 bp. After centrifugation, the supernatant was incubated with 5 μl of antibody against Sp1 (Cell Signal: 9389s) overnight at 4 °C for immunoprecipitation. Magnetic protein-G beads were then added and the mixture was further incubated at 4 °C for 1 h. After appropriate washing, the antibody-transcription factor-DNA complexes were eluted from the beads, formaldehyde cross-links were reversed, and proteins were digested with proteinase K at 65 °C overnight. DNA was purified and used for PCR with primers of K1 and K2.
2.12. Electrophoretic mobility shift assay (EMSA)
Nuclear proteins were extracted from HK2 cells. EMSA was performed with the Non-Radioactive EMSA Kit (Beyotime Biotech, Shanghai, China) according to the user manual. The Single-stranded oligonucleotides of 37 bp probe (−74 bp to −38 bp) which contained the putative Sp1-binding site (The sequence show in Table 1) was synthesized with and without a biotin label and then annealed to generate double-stranded probes. Five micrograms of cell nuclear extracts and various unlabeled competing probes were mixed and incubated at room temperature for 10 min followed by the addition of 1 μl of labeled probes for 20 min. For the Supershift assay, 5 μl of an anti-Sp1 polyclonal antibody (Cell Signal: 9389s) was added to the binding reaction.
3. Results
3.1. The key regulatory region of the hnRNP K promoter
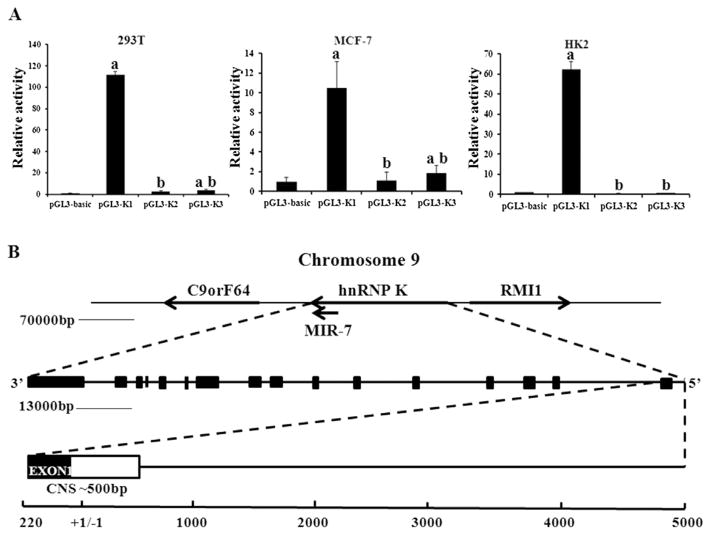
Three luciferase reporter plasmids: pGL3-K1 (−296 bp to +217 bp), pGL3-K2 (−428 bp to −200 bp) and pGL3-K3 (−349 bp to −992 bp) were constructed and tested by transient transfection and reporter assays in MCF-7 cells, 293T and HK2 cells. The pGL3-basic containing K1 (pGL3-K1) region of hnRNP K promoter significantly increased luciferase activity by approximately 30-fold, whereas plasmids containing the other two regions had less than 2-fold increases, compared to the empty vector control (Fig. 2A), indicating that the core promoter elements of hnRNP K gene were located within the K1 region.
Fig. 2.
Promoter analysis of the sequence upstream of the human hnRNP K gene. A. Promoter luciferase reporter analysis in 293T, MCF-7 and HK2 cells. The relative promoter activities are represented as the fold increase versus expression from the activities of renilla plasmid. Values represent the mean ± SD of three independent experiments. T-test was used for statistical analysis. (a) P < 0.05 versus pGL3-basic, (b) P < 0.01 versus pGL3-K1. B. Alignment analysis of the mouse/rat and human sequences identified one CNS in the 5′-upstream region of hnRNP K gene. The numbers at the bottom indicate the nucleotides, with position +1/−1 indicating the transcription start site.
Consistent with the result of the luciferase reporter assay, alignment analysis of the sequences from −5000 bp to +220 bp of the mouse, rat and human hnRNP K genes indicated that the conserved non-coding sequences (CNS) were located in the proximal region of the hnRNP K gene upstream of the transcriptional start site (Fig. 2B).
3.2. Multiple promoter elements within the K1 region
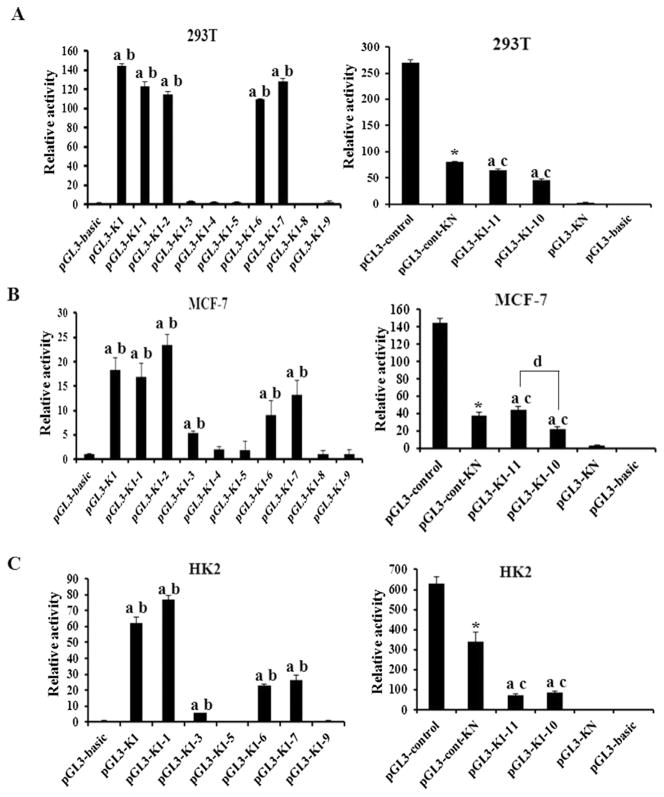
To narrow down the regulatory regions responsible for hnRNP K transcription, a series of chimeric constructs containing K1–1 to K1–9 regions (Fig. 1) were constructed to test the activity of each region in 293T cells by transient transfection assays. The activity of each chimeric constructs was showed in Fig. 3A. The results showed that pGL3-K1–1 and pGL3-K1–2 retained almost the full activity of the K1 region. However, the activity of pGL3-K1–3 decreased significantly compared with that of pGL3-K1–1 and pGL3-K1–2. pGL3-K1–4 and pGL3-K1–5 lost promoter activity completely. On the contrary, the activity of pGL3-K1–6 and pGL3-K1–7 increased significantly compared with pGL3-K1–4 and pGL3-K1–5 while the upstream sequences were further deleted. K1–8 and K1–9 did not have any significant promoter activity. The findings suggested that the sequence segments from −187 to −9 bp which present in pGL3-K1–1 and pGL3-K1–2 but not pGL3-K1–3, was an activating element that was important for the promoter activity. The sequence segments from +71 to +100 bp present in pGL3-K1–6 and pGL3-K1–7 but not pGL3-K1–8, was the second activating element, the promoter activity of which could be suppressed by the suppressive element that present in pGL3-K1–4 and pGL3-K1–5 but not pGL3-K1–6 (+52 to +70 bp). Collectively, there were three promoter elements in K1 sequence, two activating and one repressive.
Fig. 3.
Analysis of the elements within K1 sequence of hnRNP K promoter. Various truncated genomic regions were introduced into the promoter-less pGL3-basic or pGL3-control vector, and the reporter constructs were transiently transfected into 293T (A), MCF-7 (B) or HK2 (C) cells. The relative promoter activities are represented as the fold increase versus expression from the activities of renilla plasmid. Values represent the mean ± SD of three independent experiments. T-test was used for statistical analysis. (a) P < 0.05 versus pGL3-basic; and (b) P < 0.05 versus pGL3-K1–5; (c) P < 0.05 versus pGL3-KN; (d) P < 0.05 versus pGL3-K1–10 and *P < 0.05 versus pGL3-Control.
To identify the repressive element as well as the relationship between the negative regulatory sequence and the upstream element, the sequences of K1–6 and K1–4 were deleted from the K1–1 construct to produce pGL3-K1–10 (+77 bp to −187 bp) and pGL3-K1–11 (+27 bp to −187 bp) constructs, respectively. In addition, the sequence segment between K1–5 and K1–6 (+52 bp to +70 bp) was cloned into pGL3 or pGL3-SV40 (pGL3-control) to produce pGL3-KN or pGL3-cont-KN constructs (Fig. 1).
pGL3-KN did not have any significant promoter activity. The activity of pGL3-control containing KN sequence of hnRNP K promoter was significantly reduced compared with pGL3-control (p < 0.01) (Fig. 3A), indicating the negative regulatory effect of the KN segment. However, pGL3-K1–10 construct, which also contained the KN sequence, was still active. The activity of pGL3-K1–11 tended to be higher than pGL3-K1–10, but the difference did not reach statistical significance (p > 0.05). It suggested that the potentially repressive element in the KN sequence did not have substantial effects on the promoter activity of K1–1.
These results were further confirmed in MCF-7 and HK2 cells (Fig. 3B and C). From the results we noted that the activity of pGL3-K1–1 had different tendency in different cells compared with pGL3-K1when the upstream sequence was deleted. And it also shown that the activity of pGL3-K1–11 in MCF-7 cells was significantly higher than pGL3-K1–10 (p < 0.05). This hinted that the expression of hnRNP K was differently regulated in normal cells and cancer cells.
3.3. Transcription factors that bound to hnRNP K promoter
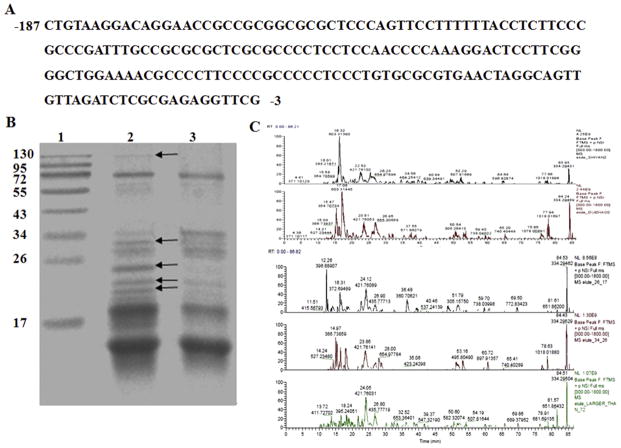
According to the above results, the primary promoter element of hnRNP K was the segments present in K1–1 and K1–2 but not K1–3 (Fig. 1). This 185 bp segment (−187 bp to −9 bp) (Fig. 4A) was amplified and used for DNA-affinity chromatography to identify transcriptional factors that could bind to it. Biotinylated DNA was tethered to streptavidin-coated paramagnetic particles, which were used as an affinity resin for binding proteins in nuclear extracts from 293T cell. Non-biotinylated DNA was used as control. The differential bands between sample and control lanes (Fig. 4B) were analyzed using Q-Exactive mass spectrometry (Fig. 4C) and bioinformatic analysis. 20 proteins were identified (Table 2).
Fig. 4.
Mass spectrometry analysis of DNA-binding proteins purified using an affinity column. A. The primary promoter element of hnRNP K. B. Purified samples were separated on 15% SDS-PAGE: Lane 1 Molecular mass marker; Lane 2 Sample; Lane 3 Control. The unique protein bands that were present only in the sample were marked by arrows and the bands numbered 1, 2, 3 and 4 (the last two bands). C. Q Exactive MS analysis of the unique bands on SDS-PAGE gel.
Table 2.
Analysis of the unique bands on SDS-PAGE gels by Q Exactive MS.a
| Sample number | Protein name | Accession number | Mol. masses (kDa) | Score | Unique peptides | Sequence coverage %. | Band number |
|---|---|---|---|---|---|---|---|
| 1 | TOP2A | P11388 | 174.3 | 0.00 | 1 | 0.72 | 1 |
| 2 | RFC1 | P35251-2 | 128.1 | 2.43 | 2 | 2.09 | 1 |
| 3 | DNTTIP2 | Q5QJE6 | 84.4 | 6.27 | 2 | 1.85 | 2 |
| 4 | TRIM28 | Q13263-2 | 79.4 | 4.63 | 2 | 25 | 1 |
| 5 | TIFβ/KAP1 | Q13263-2 | 74.9 | 4.63 | 2 | 2.79 | 1 |
| 6 | c-Jun | P05412 | 35.7 | 0.00 | 1 | 2.42 | 2 |
| 7 | PTRF | Q6NZI2-2 | 33.3 | 4.67 | 3 | 14.67 | 2 |
| 8 | HP | Q6NSB4 | 31.4 | 2.79 | 4 | 14.23 | 1 |
| 9 | Sp1 | C4PGM0 | 31.2 | 0.00 | 1 | 3.01 | 2 |
| 10 | ABT1 | Q9ULW3 | 31.1 | 0.00 | 1 | 4.04 | 2 |
| 11 | DDB1 | F5H581 | 29.2 | 0.00 | 1 | 3.37 | 2 |
| 12 | NPM1 | P06748-3 | 28.4 | 6.30 | 2 | 15.83 | 3 |
| 13 | PCBP C1/C2 | G3V576 | 25.2 | 4.15 | 2 | 9.52 | 3 |
| 14 | CREG1 | O75629 | 24.1 | 2.11 | 1 | 9.55 | 1 |
| 15 | PHB | C9JZ20 | 22.3 | 1.95 | 1 | 4.48 | 2 |
| 16 | TFIID | D6RIE8 | 21.8 | 6.45 | 3 | 15.66 | 3 |
| 17 | YBX-1 | A0JLU4 | 21.4 | 1.86 | 1 | 8.29 | 2 |
| 18 | Exportin-1 | C9JKM9 | 20.6 | 0.00 | 1 | 5.59 | 4 |
| 19 | SAP18 | O00422 | 17.5 | 2.06 | 1 | 7.84 | 4 |
| 20 | NF 1 X-type | K7ESG9 | 14.3 | 0.00 | 1 | 8.80 | 4 |
The abbreviations used in this table are: TOP2A, DNA topoisomerase 2-alpha; RFC1, Replication factor C subunit 1; DNTTIP2, Deoxynucleotidyltransferase terminal-interacting protein 2; TRIM28: Isoform 2 of Transcription intermediary factor 1-beta; TIFβ, Transcription intermediary factor 1-beta; KAP-1, KRAB-associated protein 1; c-Jun, Proto-oncogene c-Jun; PTRF, Polymerase I and transcript release factor; HP: HP protein; Sp1, Specificity protien 1; ABT1, Activator of basic transcription 1; DDB1: DNA damage-binding protein 1; NPM1, Nucleophosmin; PCBP C1/C2, Heterogeneous nuclear ribonucleoproteins C1/C2; CREG1, Cellular repressor of E1A-stimulated genes 1; PHB: Prohibitin; TFIID, Transcription initiation factor TFIID subunit 9; YBX-1, YBX1 protein; SAP18: Histone deacetylase complex subunit SAP18; NF 1 X-type, Nuclear factor 1 X-type.
3.4. Specific Sp1-binding site on the basal promoter is crucial for hnRNP K transcription
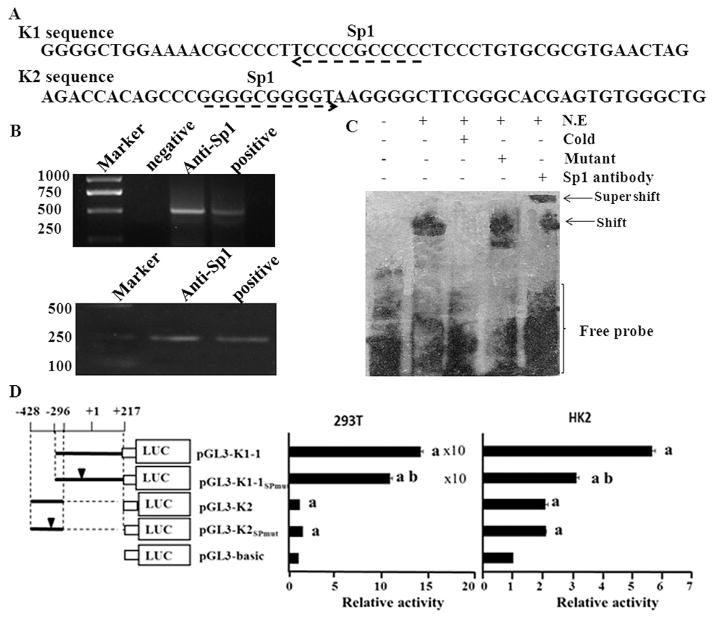
To confirm that the identified proteins bound to the promoter sequences, additional ChIPs were carried out. Sp1, which has predicted binding sites within K1 and K2 sequences (Fig. 5A), was chosen for ChIP analysis. Sp1, a ubiquitously expressed transcription factor, has important roles in the metabolic consequences of hyperinsulinemia and obesity associated metabolic syndrome [32]. It can be activated by stress such as osmolar [33] stress and oxidative stress [34,35] and regulate the transcription of downstream gene. And we are interested in the regulation of hnRNP K under oxidative stress. Sp1 was, therefore, chosen for additional analysis in the current study. Results showed that Sp1 could bind to the K1 and K2 sequences (Fig. 5B).
Fig. 5.
Analysis of the transcription factors that could bind to hnRNP K promoter. A. In silico analysis showed that at least two Sp1 binding sites in hnRNP K promoter. B. ChIP analysis verified the binding of Sp1 with hnRNP K promoter in the 293T cells. There were two controls, a non-DNA negative control and an input DNA sample. PCR was performed using primers recognizing the K1 and K2 sequences in the hnRNP K promoter and the amplified products were separated in agarose gel and visualized with EB. The sizes of the PCR products were 513 bp (K1) and 229 bp (K2). Similar results were observed in three independent experiments. C. EMSA confirmed the exact binding site of Sp1 in the 37 bp probe. N.E. denotes nuclear extract of 293T cells. Cold means cold competitor and mutant means mutant competitor. D. Promoter luciferase reporter analysis of the sequences with mutant Sp1 binding sites in 293T and HK2 cells. Values represent the mean ± SD of three independent experiments. T-test was used for statistical analysis. (a) P < 0.05 versus pGL3-basic; (b) P < 0.05 versus pGL3-K1–1.
EMSA was carried out for further confirming the exact binding site of Sp1 on K1 sequences using Sp1 antibody. The result of the EMSA was shown in Fig. 5C. The shift bands showed and the mutant probe confirmed that more than one nuclear protein extracted from 293T cells combined with the 37 bp probe. This binding activity could be blocked by 200 × unlabeled probes (cold probe). The Sp1 antibody blocked the mobility of the bands (Supershift bands), demonstrating that Sp1 was involved in the proteins binding to the probe.
Sp1 motif mutant constructs were generated in pGL3-K1–1 and pGL3-K2 and tested in 293T cells. Compared with wild-type construct, the promoter activity of mutant K1–1 was significantly reduced (p < 0.01). There was no significant difference between mutant and wild-type K2 sequence (p > 0.05). This suggested that Sp1 motif in the K1 sequence promoted the transcription of hnRNP K but the motif in the K2 sequence did not have significant regulatory function. These results were further confirmed in HK2 cells (Fig. 5D). On the contrary, the activity of mutant K1–1 was significantly increased (p < 0.01) in MCF-7 cells compared with wild-type construct (data not show), it suggested that Sp1 motif inhibited the transcription of hnRNP K, this might be due to the different binding factors in different cells.
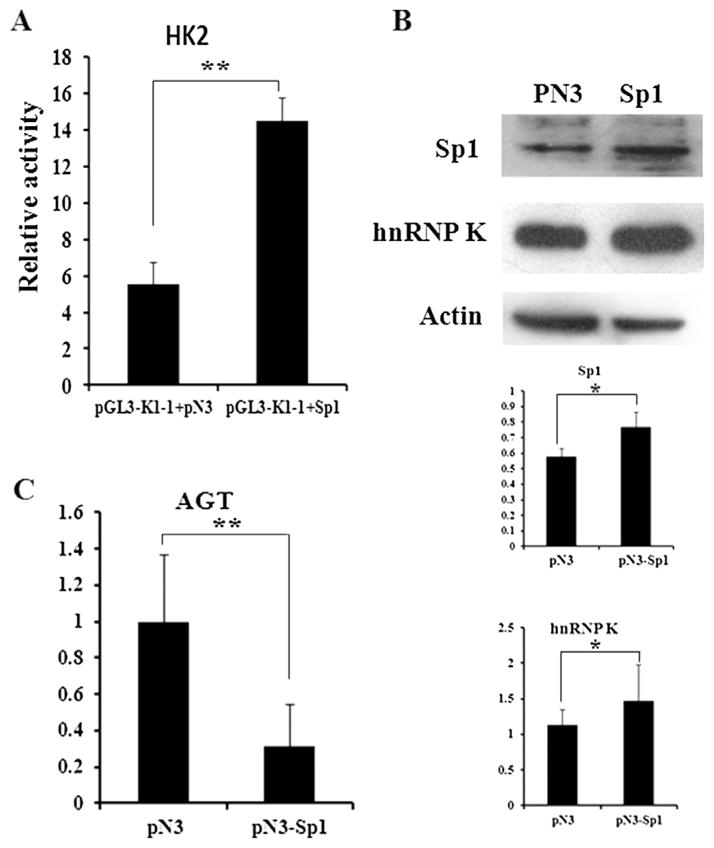
3.5. Sp1 up-regulated the expression of hnRNP K but down-regulated the mRNA level of AGT in kidney cell
The effect of Sp1 overexpression was examined to further test the regulatory function of Sp1. As shown in Fig. 6A, the promoter activity of K1–1 was significantly increased when Sp1 was over-expressed in HK2 cells (p < 0.01). Moreover, protein abundance of hnRNP K was up-regulated when Sp1 was overexpressed (p < 0.05) (Fig. 6B). These results suggested that Sp1 was capable of increasing hnRNP K transcription.
Fig. 6.
Effect of Sp1 overexpression on hnRNP K and AGT expression in HK2 cells. A. The promoter activity of hnRNP K was significantly enhanced by Sp1. B. Overexpression of Sp1 increased the protein level of hnRNP K. C. Overexpression of Sp1 significantly reduced the mRNA level of AGT. Values represent the mean ± SD of three independent experiments. T-test was used for statistical analysis.*P < 0.05; **P < 0.01.
HnRNP K has been reported to be a transcriptional factor that reduces the transcription of AGT [29]. As shown in Fig. 6C, in HK2 cells, the mRNA level of AGT was significantly reduced by Sp1 over-expression (p < 0.01), suggesting that Sp1 could reduce the mRNA level of AGT by up-regulating the expression of hnRNP K.
4. Discussion
The present study was the first to examine the transcriptional mechanisms regulating hnRNP K, a gene with important and diverse biological functions. We have identified the promoter elements and transcription factors that regulate the transcription of human hnRNP K gene. Our data showed that the promoter of hnRNP K gene existed in the proximal region of the transcriptional start site, and the expression of hnRNP K is controlled possibly by two activating and one repressive elements. Interestingly, the repressive element located at +52 to +70 bp abolished the activity of the activating element located at +71 to +100 bp, but did not have any significant effect on the activating element located at −187 to −9 bp in 293T and HK2 cells not MCF-7 cells. It suggests that the −187 to −9 bp element has strong promoter activities and might be critical for the transcription of hnRNP K in these cell lines. The different promoter regions in the K1 sequence may function in different conditions and provide the flexibility for combinatorial regulation of hnRNP K expression.
Our proteomic analysis identified 20 proteins that bound to the primary promoter element (Table 2), several of which warrant further investigation. For example, KRAB-associated protein 1(KAP1) may have an important role in DNA damage response [36–39]. Moumen et al. found that hnRNP K was rapidly induced by DNA damage in a manner that required DNA-damage signaling kinases ataxia telangiectasia mutated (ATM) or ATM and Rad3-related (ATR) [23]. Replication factor C subunit 1(RFC1) and KAP1 have been recognized as substrates of ATM and ATR [40]. It would be interesting to examine if KAP1 is involved in the hnRNP K expression in response to DNA damage.
For the responsiveness of Sp1 to oxidative stress, this factor was chosen for additional ChIP and EMSA analysis in the current study. Our data showed that Sp1 could bind to hnRNP K promoter and regulated the expression of hnRNP K and its downstream gene AGT in HK2 human kidney cells. Previous studies indicated that there was a higher level of reactive oxygen species (ROS) in the kidney of SS-rats compared with SS-13BN rats [41], and the protein level of hnRNP K was decreased in SS-rats but increased in SS-13BN rats in response to a high-salt diet [30]. As transcriptional factors, Sp1 also could be regulated by ROS [34,35,42]. The regulatory relationships among Sp1, ROS and hnRNP K remain to be further studied.
The binding factors of the other two elements in K1 sequence were not purified in current study, but these two elements may have important role in regulating the expression of hnRNP K during hemopoietic development. The putative transcription factor E2F which combined with the repressive element, a best-known cell cycle regulator, had an important role in angiogenesis [43]. Another binding factor, MZF-1 was also reported as a regulator during he-mopoietic development [44,45]. Interestingly, the possible binding factors of activating element present in +71 to +100 bp, GATA-1 and GATA-2, were also participated in hemopoietic development. All of the above data suggested that these two elements might be important in regulating the expression of hnRNP K in hemopoietic cells.
In summary, this study examined the detailed molecular mechanism of hnRNP K transcription for the first time. Three promoter elements regulated the transcription of hnRNP K and they had different function in HK2, 293T and MCF-7 cells. 20 transcriptional factors bind to the basal promoter element, in which Sp1 increased the expression of hnRNP K in HK2 and 293T cells but decreased the expression in MCF-7 cells.
Acknowledgments
We would like to thank Guntram Suske for pN3-Sp1 plasmid and Weihua Wang for human genomic DNA and Yujin Xu, Cun Zhang, Weina Li, Zhen Shu and Nan Mu for technical assistance.
This study was supported by the National Natural Science Foundation of China (NSFC) (Grant No. 31071029, 81270767, 81228002) and the US National Institutes of Health (Grant No. HL116264)
Abbreviations
- hnRNP K
heterogeneous nuclear ribonucleoprotein K
- VEGF
vascular endothelial growth factor
- LDLR
low density lipoprotein receptor
- G6PD
glucose-6-phosphate dehydrogenase
- AGT
angiotensinogen
- ANG II
angiotensin II
- AT1R
Angiotensin Type-1 Receptor
- DDX6
DEAD-box RNA helicase 6
- SS-rat
salt sensitive rat
- SS-13BN
Consomic SS-Chr 13BN/Mcwi rat
- EF-1α
elongation factor 1α
- SDS
sodium dodecyl sulfate
- AP-1
activator protein 1
- Sp1
specificity protein 1
- KAP-1
KRAB-associated protein 1
- ATM
ataxia telangiectasia mutated
- ATR
ATM and Rad3-related
- RFC1
Replication factor C subunit 1
- ROS
reactive oxygen species
- HCD
Higher Energy Collisional Dissociation
- CNS
conserved non-coding sequence
References
- 1.Dejgaard K, et al. Identification, molecular cloning, expression and chromosome mapping of a family of transformation upregulated hnRNP-K proteins derived by alternative splicing. J Mol Biol. 1994;236(1):33–48. doi: 10.1006/jmbi.1994.1116. [DOI] [PubMed] [Google Scholar]
- 2.Ostrowski J, et al. Purification, cloning, and expression of a murine phos-phoprotein that binds the kappa B motif in vitro identifies it as the homolog of the human heterogeneous nuclear ribonucleoprotein K protein. Description of a novel DNA-dependent phosphorylation process. J Biol Chem. 1994;269(26):17626–17634. [PubMed] [Google Scholar]
- 3.Gaillard C, Cabannes E, Strauss F. Identity of the RNA-binding protein K of hnRNP particles with protein H16, a sequence-specific single strand DNA-binding protein. Nucleic Acids Res. 1994;22(20):4183–4186. doi: 10.1093/nar/22.20.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch M, et al. hnRNP K binds a core polypyrimidine element in the eukaryotic translation initiation factor 4E (eIF4E) promoter, and its regulation of eIF4E contributes to neoplastic transformation. Mol Cell Biol. 2005;25(15):6436–6453. doi: 10.1128/MCB.25.15.6436-6453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi HS, et al. A proteomics approach for identification of single strand DNA-binding proteins involved in transcriptional regulation of mouse mu opioid receptor gene. Mol Cell Proteomics. 2008;7(8):1517–1529. doi: 10.1074/mcp.M800052-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SS, et al. Poly(C) binding protein family is a transcription factor in mu-opioid receptor gene expression. Mol Pharmacol. 2005;68(3):729–736. doi: 10.1124/mol.105.012245. [DOI] [PubMed] [Google Scholar]
- 7.Ko JL, Loh HH. Poly C binding protein, a single-stranded DNA binding protein, regulates mouse mu-opioid receptor gene expression. J Neurochem. 2005;93(3):749–761. doi: 10.1111/j.1471-4159.2005.03089.x. [DOI] [PubMed] [Google Scholar]
- 8.Uribe DJ, et al. Heterogeneous nuclear ribonucleoprotein K and nucleolin as transcriptional activators of the vascular endothelial growth factor promoter through interaction with secondary DNA structures. Biochemistry. 2011;50(18):3796–3806. doi: 10.1021/bi101633b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Liu J. Identification of heterogeneous nuclear ribonucleoprotein K as a transactivator for human low density lipoprotein receptor gene transcription. J Biol Chem. 2010;285(23):17789–17797. doi: 10.1074/jbc.M109.082057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffith BN, et al. Identification of hnRNPs K, L and A2/B1 as candidate proteins involved in the nutritional regulation of mRNA splicing. Biochim Biophys Acta. 2006;1759(11–12):552–561. doi: 10.1016/j.bbaexp.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skalweit A, et al. Posttranscriptional control of renin synthesis: identification of proteins interacting with renin mRNA 3′-untranslated region. Circ Res. 2003;92(4):419–427. doi: 10.1161/01.RES.0000059300.67152.4E. [DOI] [PubMed] [Google Scholar]
- 12.Bomsztyk K, et al. Diverse molecular interactions of the hnRNP K protein. FEBS Lett. 1997;403(2):113–115. doi: 10.1016/s0014-5793(97)00041-0. [DOI] [PubMed] [Google Scholar]
- 13.Feliers D, et al. Heterogeneous nuclear ribonucleoprotein K contributes to angiotensin II stimulation of vascular endothelial growth factor mRNA translation. Am J Physiol Ren Physiol. 2007;293(2):F607–F615. doi: 10.1152/ajprenal.00497.2006. [DOI] [PubMed] [Google Scholar]
- 14.Sataranatarajan K, et al. PKCdelta regulates the stimulation of vascular endothelial factor mRNA translation by angiotensin II through hnRNP K. Cell Signal. 2008;20(5):969–977. doi: 10.1016/j.cellsig.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostareck DH, et al. Lipoxygenase mRNA silencing in erythroid differentiation: the 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104(2):281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- 16.Naarmann IS, et al. DDX6 recruits translational silenced human reticulocyte 15-lipoxygenase mRNA to RNP granules. RNA. 2010;16(11):2189–2204. doi: 10.1261/rna.2211110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikula M, et al. Casein kinases phosphorylate multiple residues spanning the entire hnRNP K length. Biochim Biophys Acta. 2006;1764(2):299–306. doi: 10.1016/j.bbapap.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Wang N, et al. Hnrnpk, a protein differentially expressed in immature rat ovarian development is required for normal primordial follicle assembly and development. Endocrinology. 2011;152(3):1024–1035. doi: 10.1210/en.2010-0797. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Szaro BG. hnRNP K post-transcriptionally co-regulates multiple cyto-skeletal genes needed for axonogenesis. Development. 2011;138(14):3079–3090. doi: 10.1242/dev.066993. [DOI] [PubMed] [Google Scholar]
- 20.Thyagarajan A, Szaro BG. Dynamic endogenous association of neurofilament mRNAs with K-homology domain ribonucleoproteins in developing cerebral cortex. Brain Res. 2008;1189:33–42. doi: 10.1016/j.brainres.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Gervasi C, Szaro BG. A crucial role for hnRNP K in axon development in Xenopus laevis. Development. 2008;135(18):3125–3135. doi: 10.1242/dev.022236. [DOI] [PubMed] [Google Scholar]
- 22.Blanchette AR, Fuentes Medel YF, Gardner PD. Cell-type-specific and developmental regulation of heterogeneous nuclear ribonucleoprotein K mRNA in the rat nervous system. Gene Expr Patterns. 2006;6(6):596–606. doi: 10.1016/j.modgep.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Moumen A, et al. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123(6):1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 24.Wen F, et al. Higher expression of the heterogeneous nuclear ribonucleo-protein k in melanoma. Ann Surg Oncol. 2010;17(10):2619–2627. doi: 10.1245/s10434-010-1121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du Q, et al. The role of heterogeneous nuclear ribonucleoprotein K in the progression of chronic myeloid leukemia. Med Oncol. 2010;27(3):673–679. doi: 10.1007/s12032-009-9267-z. [DOI] [PubMed] [Google Scholar]
- 26.Zhang P, et al. HnRNP K and PDI marked response to chemotherapy to human colorectal cancer cells. Electrophoresis. 2010;31(10):1731–1738. doi: 10.1002/elps.200900495. [DOI] [PubMed] [Google Scholar]
- 27.Burnham AJ, Gong L, Hardy RW. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology. 2007;367(1):212–221. doi: 10.1016/j.virol.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Acevedo MJ, et al. Antibodies against heterogeneous nuclear ribonucleo-protein K in patients with cardiac allograft vasculopathy. J Heart Lung Transpl. 2011;30(9):1051–1059. doi: 10.1016/j.healun.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Mojtahedi Z, et al. Immunoproteomics of HER2-positive and HER2-negative breast cancer patients with positive lymph nodes. OMICS. 2011;15(6):409–418. doi: 10.1089/omi.2010.0131. [DOI] [PubMed] [Google Scholar]
- 30.Tian Z, et al. Renal regional proteomes in young Dahl salt-sensitive rats. Hypertension. 2008;51(4):899–904. doi: 10.1161/HYPERTENSIONAHA.107.109173. [DOI] [PubMed] [Google Scholar]
- 31.Kobori H, et al. Enhancement of intrarenal angiotensinogen in Dahl salt-sensitive rats on high salt diet. Hypertension. 2003;41(3):592–597. doi: 10.1161/01.HYP.0000056768.03657.B4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solomon SS, et al. A critical role of Sp1 transcription factor in regulating gene expression in response to insulin and other hormones. Life Sci. 2008;83(9–10):305–312. doi: 10.1016/j.lfs.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 33.Ramos A, et al. Hypo-osmolar stress induces p75NTR expression by activating Sp1-dependent transcription. J Neurosci. 2007;27(6):1498–1506. doi: 10.1523/JNEUROSCI.4806-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin M, et al. Regulation of cytochrome P450 2e1 expression by ethanol: role of oxidative stress-mediated pkc/jnk/sp1 pathway. Cell Death Dis. 2013;4:e554. doi: 10.1038/cddis.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sang J, et al. SUMO2 and SUMO3 transcription is differentially regulated by oxidative stress in an Sp1-dependent manner. Biochem J. 2011;435(2):489–498. doi: 10.1042/BJ20101474. [DOI] [PubMed] [Google Scholar]
- 36.Wang C, et al. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J. 2005;24(18):3279–3290. doi: 10.1038/sj.emboj.7600791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White DE, et al. KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res. 2006;66(24):11594–11599. doi: 10.1158/0008-5472.CAN-06-4138. [DOI] [PubMed] [Google Scholar]
- 38.Ziv Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8(8):870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 39.Gilmore-Hebert M, Ramabhadran R, Stern DF. Interactions of ErbB4 and Kap1 connect the growth factor and DNA damage response pathways. Mol Cancer Res. 2010;8(10):1388–1398. doi: 10.1158/1541-7786.MCR-10-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 41.Taylor NE, et al. NADPH oxidase in the renal medulla causes oxidative stress and contributes to salt-sensitive hypertension in Dahl S rats. Hypertension. 2006;47(4):692–698. doi: 10.1161/01.HYP.0000203161.02046.8d. [DOI] [PubMed] [Google Scholar]
- 42.Dhar A, Young MR, Colburn NH. The role of AP-1 NF-kappaB and ROS/NOS in skin carcinogenesis: the JB6 model is predictive. Mol Cell Biochem. 2002;234–235(1–2):185–193. [PubMed] [Google Scholar]
- 43.Weijts BG, et al. E2F7 and E2F8 promote angiogenesis through transcriptional activation of VEGFA in cooperation with HIF1. EMBO J. 2012;31(19):3871–3884. doi: 10.1038/emboj.2012.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong S, et al. Crucial roles of MZF1 and Sp1 in the transcriptional regulation of the peptidylarginine deiminase type I gene (PADI1) in human keratino-cytes. J Invest Dermatol. 2008;128(3):549–557. doi: 10.1038/sj.jid.5701048. [DOI] [PubMed] [Google Scholar]
- 45.Le Mee S, Fromigue O, Marie PJ. Sp1/Sp3 and the myeloid zinc finger gene MZF1 regulate the human N-cadherin promoter in osteoblasts. Exp Cell Res. 2005;302(1):129–142. doi: 10.1016/j.yexcr.2004.08.028. [DOI] [PubMed] [Google Scholar]