Abstract
Human genetic factors predispose to tuberculosis (TB). We studied 7.6 million genetic variants in 5,530 pulmonary TB patients and 5,607 healthy controls. In the combined analysis of these subjects and the follow-up cohort (15,087 TB patients and controls altogether), we found association between TB and variants located in introns of the ASAP1 gene on chromosome 8q24 (P = 2.6 × 10−11 for rs4733781; P = 1.0 × 10−10 for rs10956514). Dendritic cells (DCs) showed high level of ASAP1 expression, which was reduced after M. tuberculosis infection, and rs10956514 was associated with the level of reduction of ASAP1 expression. The ASAP1 protein is involved in actin and membrane remodeling and has been associated with podosomes. The ASAP1-depleted DCs showed impaired matrix degradation and migration. Therefore, genetically determined excessive reduction of ASAP1 expression in M. tuberculosis-infected DCs may lead to their impaired migration, suggesting a potential novel mechanism that predisposes to TB.
Introduction
Tuberculosis (TB) is a major public health problem in the developing world and an increasing threat in developed countries1. While 2 billion people are estimated to be infected with the TB pathogen, Mycobacterium tuberculosis, only ~10% of infected individuals eventually develop active TB1. A majority can contain the pathogen; the infection remains latent and clinical disease never occurs. Case observations, twin studies and mouse models indicate that host genetic factors are important in determining susceptibility to M. tuberculosis2,3. Recent genome wide association studies (GWAS) reported two loci on chromosomes 11p13 and 18q11 that contain single nucleotide polymorphisms (SNPs) associated with TB4,5. However, the pathophysiological mechanisms underpinning these genetic associations remain unknown.
Results
Genome-wide association study
To identify novel genes associated with susceptibility to TB, we initially studied 5,914 HIV-negative patients with active pulmonary TB confirmed by culture of M. tuberculosis from sputum, as well as 6,022 healthy adult controls from Russia (see Methods). We genotyped their DNA samples using Affymetrix Genome-Wide Human SNP Array 6.0. After rigorous quality control procedures (Methods, Supplementary Figs. 1 and 2) 5,530 TB patients and 5,607 controls with high quality genotypes at 707,452 SNPs remained (Set 1). We then imputed genotypes at 7,614,862 SNPs into the Set 1 subjects using data from the 1000 Genomes project. We tested these SNPs for association with TB using logistic regression corrected for 4 principal components, and did not observe significant inflation of test statistics (λGC = 1.10), indicating that our cases and controls are well matched (Supplementary Fig. 3).
We first examined our association results in the context of previously published TB GWAS in two African populations4,5. We did not detect any association at the previously reported 18q11 locus. However, we did see association at the 11p13 locus (rs2057178, P = 0.00068, Supplementary Table 1), as we reported previously for the overlapping Russian dataset5. This is also consistent with another recently published TB GWAS in the South African population6 and suggests that the 11p13 locus is associated with TB risk in various populations, whereas the 18q11 locus may be population-specific. We next studied candidate genes previously reported to be associated with TB and found weak signals in several genes, including the HLA-DQA1 region (Supplementary Table 2). We then looked for the overlap with published genetic associations in another human mycobacterial infectious disease, leprosy7,8, and in inflammatory bowel disease (IBD) that is known to be enriched with genes involved in defense against mycobacteria9. We found that, except for the MHC region, none of those loci was significantly associated with TB after Bonferroni correction (Supplementary Tables 3 and 4).
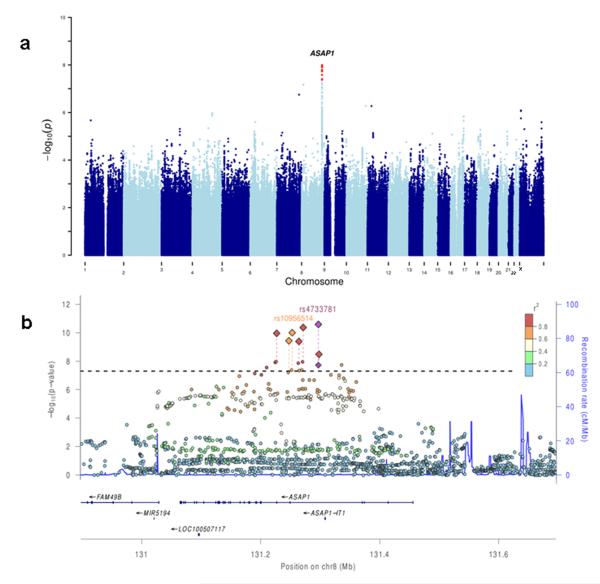
Our only genome-wide significant TB-associated locus was on chromosome 8q24, where 11 SNPs located in introns of the ASAP1 gene had P < 5 × 10−8 (Fig. 1); the strongest association was at rs2033059 (P = 1.1 × 10−8). To rule out possible technical artefacts, and to attempt to replicate our discovery, we designed individual Taqman genotyping assays for seven out of the most significantly associated ASAP1 SNPs. We genotyped these SNPs in Set 1 and, additionally, in 1,085 pulmonary TB patients and 2,865 controls from Russia (Set 2). In the combined analysis of 15,087 subjects in Sets 1 and 2 all seven SNPs showed convincing evidence of association with TB (Table 1); the most significant association was at rs4733781 (P = 2.6 × 10−11). In order to try to identify which of these seven SNPs is likely the functional polymorphism affecting risk to pulmonary TB, we employed a Bayesian fine-mapping approach (Methods). Three SNPs had less than 5% posterior probability of being causal, leaving four likely candidates (Table 1), with rs4733781 having the highest posterior (43.6%). This narrowing to four candidates likely represents the limit of genetic resolution in our Russian dataset.
Figure 1. Genome-wide association study of susceptibility to TB.
(a) Manhattan plot showing TB association of genotyped and imputed SNPs across the genome. SNPs with P < 5 × 10−8 are shown in red.
(b) Regional plot showing SNPs in the ASAP1 gene region. Circles show P-values obtained in GWAS in Set 1; diamonds show P-values obtained in the follow-up study of 7 SNPs in Sets 1 and 2 combined. Horizontal line corresponds to P < 5 × 10−8. Colors indicate linkage disequilibrium (r2) between the most-associated SNP rs4733781 and other SNPs.
Table 1. Association between the ASAP1 gene polymorphisms and tuberculosis in the Russian datasets.
| rs1017281 | rs10956514 | rs1469288 | rs17285138 | rs2033059 | rs4733781 | rs12680942 | |
|---|---|---|---|---|---|---|---|
| Alleles (min./maj.) | A/G | G/A | A/G | A/T | T/C | C/A | G/A |
| MAF (cont., cases) | 35.7, 32.2 | 35.8, 32.3 | 31.5, 28.0 | 31.5, 28.3 | 31.3, 27.7 | 31.4, 27.8 | 31.2, 27.8 |
| Set 1 (GWAS set re-typed) | |||||||
| n cont. | 5,526 | 5,503 | 5,346 | 5,494 | 5,266 | 5,533 | 5,369 |
| n cases | 5,466 | 5,425 | 5,272 | 5,420 | 5,416 | 5,457 | 5,314 |
| P-value | 3.7 × 10−08 | 1.9 × 10−08 | 1.4 × 10−08 | 5.6 × 10−08 | 1.6 × 10−09 | 3.6 × 10−09 | 7.6 × 10−09 |
| OR a | 0.85 | 0.85 | 0.84 | 0.85 | 0.83 | 0.84 | 0.84 |
| 95% CI | 0.81-0.90 | 0.80-0.90 | 0.79-0.89 | 0.80-0.90 | 0.79-0.88 | 0.79-0.89 | 0.79-0.89 |
|
| |||||||
| Set 2 (follow-up) | |||||||
| n cont. | 2,538 | 2,535 | 2,390 | 2,717 | 2,692 | 2,549 | 2,522 |
| n cases | 970 | 971 | 897 | 1,048 | 1,038 | 975 | 1,030 |
| P-value | 6.6 × 10−03 | 3.5 × 10−03 | 2.3 × 10−02 | 2.2 × 10−02 | 1.7 × 10−02 | 7.8 × 10−03 | 3.5 × 10−02 |
| OR a | 0.85 | 0.84 | 0.87 | 0.88 | 0.87 | 0.85 | 0.88 |
| 95% CI | 0.76-0.96 | 0.75-0.95 | 0.77-0.98 | 0.78-0.98 | 0.78-0.98 | 0.76-0.96 | 0.79-0.99 |
|
| |||||||
| Sets 1 and 2 combined | |||||||
| n cont. | 8,064 | 8,038 | 7,736 | 8,211 | 7,958 | 8,082 | 7,891 |
| n cases | 6,436 | 6,396 | 6,169 | 6,468 | 6,454 | 6,432 | 6,344 |
| P-value | 3.7 × 10−10 | 1.0 × 10−10 | 1.1 × 10−10 | 3.3 × 10−09 | 4.3 × 10−11 | 2.6 × 10−11 | 4.1 × 10−10 |
| OR a | 0.85 | 0.85 | 0.84 | 0.86 | 0.84 | 0.84 | 0.85 |
| 95% CI | 0.81-0.90 | 0.81-0.89 | 0.80-0.89 | 0.82-0.90 | 0.80-0.89 | 0.80-0.88 | 0.81-0.89 |
|
| |||||||
| PPP | 0.030 | 0.108 | 0.118 | 0.004 | 0.271 | 0.436 | 0.033 |
ORs for minor alleles are shown
cont. – controls; min. – minor; maj. – major; MAF – minor allele frequency; OR – odds ratio; 95% CI – 95% confidence interval; PPP - proportion of posterior probability, SNPs with PPP > 0.05 are shown in bold.
We then analyzed the seven ASAP1 SNPs in the Ghanaian (971 TB cases and 988 controls) and Gambian (1,306 TB cases and 1,372 controls) GWAS datasets that were published previously4,5 (Table 2). Despite that these SNPs were less frequent in the African populations than in Russians, resulting in lower statistical power to detect association, this analysis provided further support for TB association of the ASAP1 SNPs, e.g. for rs4733781 effects in the Ghanaian and Gambian datasets were similar to those in the Russian dataset (ORs = 0.89, 0.89 and 0.84, respectively; PGhana+Gambia = 0.052 and the overall PRussia+Ghana+Gambia = 3.6 × 10−12).
Table 2. Association between the ASAP1 gene polymorphisms and tuberculosis in the Ghanaian and Gambian datasets.
| minor/major | Ghana |
Gambia |
Ghana + Gambia |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | allele | MAF | ORa (95% CI) | P | MAF | ORa (95% CI) | P | MAF | ORa (95% CI) | Pb | χ 2 |
| rs4733781 | C/A | 11.6 | 0.89 (0.73-1.09) | 0.26 | 9.2 | 0.89 (0.74-1.08) | 0.23 | 10.2 | 0.89 (0.78-1.02) | 0.052 | 2.636 |
| rs12680942 | G/A | 11.9 | 0.89 (0.73-1.09) | 0.26 | 9.3 | 0.89 (0.74-1.08) | 0.24 | 10.4 | 0.89 (0.78-1.03) | 0.055 | 2.547 |
| rs17285138 | A/T | 11.8 | 0.90 (0.73-1.09) | 0.28 | 9.3 | 0.89 (0.73-1.07) | 0.21 | 10.3 | 0.89 (0.78-1.02) | 0.053 | 2.612 |
| rs2033059 | T/C | 7.3 | 0.91 (0.71-1.16) | 0.47 | 8.6 | 0.88 (0.73-1.07) | 0.21 | 8.1 | 0.89 (0.76-1.04) | 0.074 | 2.085 |
| rs10956514 | G/A | 24.5 | 0.95 (0.82-1.10) | 0.48 | 21.2 | 1.02 (0.90-1.16) | 0.75 | 22.6 | 0.99 (0.9-1.09) | 0.845 | 0.038 |
| rs1017281 | A/G | 24.2 | 0.94 (0.82-1.09) | 0.45 | 21.1 | 1.02 (0.90-1.17) | 0.72 | 22.4 | 0.99 (0.9-1.09) | 0.832 | 0.045 |
| rs1469288 | A/G | 19.9 | 0.97 (0.83-1.14) | 0.74 | 17.0 | 0.96 (0.83-1.11) | 0.58 | 18.2 | 0.97 (0.87-1.08) | 0.272 | 0.367 |
SNP – single nucleotide polymorphism; MAF – minor allele frequency; OR – odds ratio; 95% CI – 95% confidence interval
ORs for minor alleles are shown
When ORs in the Russian, Ghanaian and Gambian populations were in the same direction, we used one-tailed χ2 test to calculate P-values. Otherwise, we used two-tailed χ2 test (for SNPs rs10956514 and rs1017281).
Analyses of ASAP1 expression and function
The ASAP1 gene (also known as AMAP1 or DDEF1) encodes an Arf GTPase-activating protein (Arf GAP), a multifunctional scaffold protein that induces hydrolysis of GTP bound to the ADP ribosylation factor family GTP-binding (Arf) proteins10. The Arf and Arf GAP proteins are known to regulate coordinated actin and membrane remodelling11. In NIH 3T3 fibroblasts localization of the ASAP1 protein was associated with focal adhesions, circular dorsal ruffles, podosomes and invadopodia, i.e. large macromolecular complexes that connect intracellular actin cytoskeleton to the extracellular matrix and are important for cell adhesion and migration, as well as invasion of cancer cells11. The transition of a cell from a quiescent to a migratory phenotype is characterized by the dissolution of focal adhesions and the formation of podosomes or invadopodia at the sites of contact with extracellular matrix12. In vitro knockdown and reduction of ASAP1 expression in the transformed fibroblast and prostate cancer cell lines has been shown to prevent formation of invadopodia and suppress cell migration and invasiveness13,14. Overexpression of ASAP1 has been associated with metastasis in several cancers14-17. While invadopodia are found in transformed cells, podosomes are similar structures that are present in several types of normal cells, including macrophages and dendritic cells (DCs)12. These immune cells play critical roles in TB pathogenesis. Thus, the infection is initiated when inhaled M. tuberculosis bacilli are phagocytosed by alveolar macrophages, which provide a cellular niche for the expansion of mycobacteria. The onset of adaptive immune response leads to killing of mycobacteria and sterilization of regional lesions18. DCs are crucial for the initiation of the adaptive immunity: infected DCs migrate to the lung-draining lymph nodes and activate T cells19. M. tuberculosis is known to inhibit migration as well as other functions of DCs, and the adaptive immunity in TB is delayed compared with other microbial infections20-22.
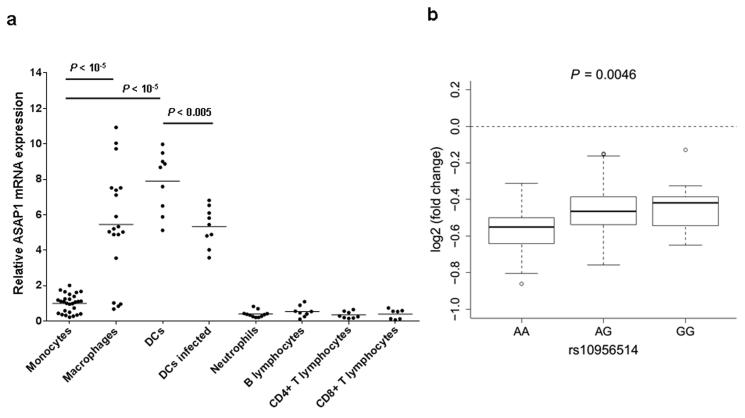
The role of ASAP1 in macrophages, DCs and other immune cells has not been studied previously. To investigate it, we initially characterized the ASAP1 mRNA expression by quantitative reverse transcriptase-PCR (qRT-PCR) in primary leukocytes of healthy donors and found that monocytes, B lymphocytes, CD4+ and CD8+ T lymphocytes and neutrophils express relatively low levels of ASAP1. However, when monocytes were differentiated into macrophages or DCs, the expression of ASAP1 increased (Fig. 2a). We then infected DCs with Mycobacterium bovis BCG and found that it led to the reduction of ASAP1 expression (Fig. 2a). We also analyzed ASAP1 expression in DCs from 65 subjects that were previously studied using microarrays23. Similar to our results, in this dataset the ASAP1 level in the M. tuberculosis-infected DCs was significantly lower than in non-infected DCs (log2 fold change = −0.49, Pcorrected = 2.9 × 10−18, ref 23). To study if any of the TB-associated SNPs in the ASAP1 gene affects its expression, we first analyzed rs10956514 and rs4733781 that were genotyped in this dataset. The SNP rs10956514 showed evidence of association with ASAP1 expression in non-infected samples (P = 4.6 × 10−3) and with the relative level of reduction in ASAP1 expression after M. tuberculosis infection (P = 4.6 × 10−3). Homozygotes of allele A, which was associated with higher TB risk in our GWAS, showed stronger reduction of ASAP1 expression after infection than homozygotes of allele G, which was associated with lower TB risk (Fig. 2b). We imputed genotypes of other TB-associated ASAP1 SNPs and found that they had weaker association with the level of reduction of ASAP1 expression after M. tuberculosis infection than rs10956514 (Supplementary Table 5). The lack of association between ASAP1 expression and rs4733781 reflects different linkage disequilibrium patterns in this region (e.g. r2 between rs4733781 and rs10956514 was 0.8 in the Russian dataset, but only 0.62 in 65 non-Russian Caucasians23). These results indicate that larger studies in different populations will be needed to pinpoint the causative variant affecting ASAP1 expression, and thence the risk of pulmonary TB. Nevertheless, our results suggest that either rs10956514 or a polymorphism in strong linkage disequilibrium with it may be responsible for the control of ASAP1 expression in DCs. Since rs10956514 is also one of the candidates associated with the risk of active pulmonary TB (Table 1), genetically regulated levels of ASAP1 expression in DCs are likely to be important in TB pathogenesis.
Figure 2. ASAP1 expression in peripheral blood leukocytes and monocyte-derived macrophages and dendritic cells (DCs).
(a) Levels of the ASAP1 mRNA expression in primary leukocytes of healthy volunteers each measured in triplicate by quantitative reverse transcriptase PCR and shown relative to the level of expression in monocytes. CD14+ monocytes were isolated from peripheral blood of healthy volunteers and cultured either with M-CSF for 5 days to derive macrophages or with GM-CSF and IL-4 for 6 days to derive DCs. DCs were infected with live M. bovis BCG at multiplicity of infection 3:1 for 24 hours. Horizontal bars show mean levels. P-values were calculated using two-tailed Student T test. Unadjusted P-values are shown.
(b) Box plot showing the level of reduction of ASAP1 mRNA expression in DCs after M. tuberculosis infection for 65 subjects with different rs10956514 genotypes, 23 (AA), 30 (AG) and 12 (GG). The bold line in the boxplot is the median; the whiskers are 1.5 IQR away from the first and third quartiles. Data are from ref 23 after adjustment for 5 and 8 principle components in non-infected and infected DCs, respectively (see Methods).
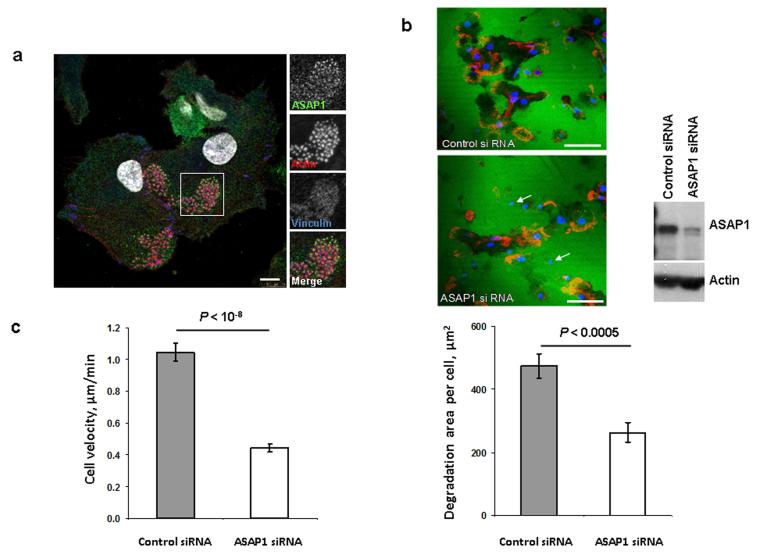
To investigate the role of the ASAP1 protein in DCs, we first studied its cellular localization. Confocal microscopy showed that the ASAP1 protein was localized in the cytoplasm and partly was associated with podosomes (Fig. 3a). Therefore, we hypothesized that reduced ASAP1 expression may affect matrix degradation by DCs and their migration. To test this, we isolated primary monocytes from healthy volunteers, derived DCs in vitro and transfected them with siRNA to reduce ASAP1 expression. We found that degradation of a gelatine matrix by the ASAP1-depleted DCs was impaired (Fig. 3b) and such cells migrated significantly slower than control cells (Fig. 3c).
Figure 3. In DCs, the ASAP1 protein is partly associated with podosomes and its depletion leads to impaired matrix degradation and migration.
(a) Confocal microscopy showing ASAP1 (green), actin (red), vinculin (blue) and nuclei (white) in DCs derived from monocytes of a healthy volunteer. Podosomes are structures characterized by the actin core (visible as red dots) surrounded by other proteins. Boxed area is shown in grayscale enlarged on the right. Scale bar represents 7.5μm.
(b) Representative confocal microscopy images (left panel) showing monocyte-derived DCs treated with either ASAP1 or control siRNA and seeded on FITC-conjugated gelatine (green) for 4 hours. The cells were stained to visualize actin (phalloidin-red) and nuclei (DAPI-blue). White arrows show examples of the DAPI-stained nuclei of cells that do not degrade matrix. Scale bar represents 50μm.
In the same experiment cell lysates were collected for western blotting and probed with anti-ASAP1 and anti-actin antibodies (right panel) to assess knockdown efficiency.
Area of degradation of the FITC-conjugated gelatine matrix by DCs (lower panel). In total 394 and 317 DCs treated with either ASAP1 or control siRNA, respectively, were counted in 4 independent experiments.
(c) Migration velocity of DCs in a Dunn chamber. In total 162 and 186 DCs treated with either ASAP1 or control siRNA, respectively, were tracked in 5 independent experiments.
Graphs show mean values ± SEM. P-values were calculated using two-tailed Student T test. Unadjusted P-values are shown.
Discussion
Taken together, our results suggest that impaired migration of M. tuberculosis-infected DCs, caused by the genetically determined excessive reduction of ASAP1 expression, may contribute to TB pathogenesis. It may be one of the mechanisms that lead to the slow migration of DCs to lymph nodes and delay the adaptive immune response during early stages of TB infection. Furthermore, given that latent TB is a state of dynamic equilibrium, it is likely that trafficking of immune cells in granulomas has to be constantly maintained19. Therefore, impaired migration of DCs and/or macrophages due to the reduced ASAP1 expression may contribute also at late stages of infection leading to reactivation of latent TB.
To date, GWAS of susceptibility to TB and other infectious diseases identified fewer associated loci than studies of other complex diseases. This indicates that effects of common polymorphisms predisposing to infection are usually small, and current GWA studies may be statistically underpowered. In TB, this can be explained by the strong selective pressure exerted by M. tuberculosis, an ancient and wide-spread pathogen. Nevertheless, our results demonstrate that when associated variants are found, they can highlight novel molecular mechanisms implicated in infection and host responses. Future studies will further clarify how the ASAP1-mediated pathways are involved in mycobacterial infection and predispose to TB.
Online Methods
Ethics
Blood samples from all participants have been collected and studied with the written informed consent according to the Declaration of Helsinki and with approvals from the local Ethics Committees in Russia (St. Petersburg and Samara), and the UK (Human Biological Resource Ethics Committee of the University of Cambridge and the National Research Ethics Service Cambridgeshire 1 REC, ref 10/H0304/71).
TB patients and controls
Pulmonary TB patients attending civilian TB dispensaries and TB clinics along with healthy subjects attending the blood transfusion service were recruited in two Russian cities, St. Petersburg and Samara, as described previously24. TB patients have been diagnosed using information about TB contact, medical history and clinical symptoms (cough, haemoptysis, chest pain, fever, weight loss), presence of acid fast bacilli in sputum smear and symptoms characteristic of pulmonary TB on chest X rays. For all patients in this study diagnosis has been confirmed by culture of M. tuberculosis from sputum; otherwise patients were excluded. We also excluded from the study patients with extra-pulmonary TB and all HIV-positive subjects. Controls were healthy adult blood bank donors with no history of TB. M. tuberculosis infection status of these controls was unknown. We studied genotypes of 15,087 subjects, including 6,615 TB patients (4,074 from St. Petersburg and 2,541 from Samara) and 8,472 controls (5,555 from St. Petersburg and 2,917 from Samara). Average age was 45 years for patients and 31 years for controls. There were 73% of males among patients and 71% of males among controls. This sample size provides >80% statistical power to detect association with P = 5 × 10−8 for polymorphisms with minor allele frequencies (MAF) > 20% and OR > 1.2.
Genotyping, quality control and association analyses
We extracted genomic DNA from whole blood of the participating subjects using a standard chloroform/proteinase K protocol. We checked DNA quality using 1% agarose gel electrophoresis, determined DNA concentration using Picogreen assay and then normalized concentration for genotyping.
In GWAS, genotyping was done using the Affymetrix Genome-Wide Human SNP Array 6.0. Genotypes were called in 5,914 TB cases and 6,022 controls using Birdseed25. Individuals were excluded if they had more than 2% missing genotype data or showed excess of heterozygous genotypes (±3.5 standard deviations, Supplementary Fig. 1). For each pair of individuals, we calculated identity-by-state (IBS) and excluded samples with IBS > 80% as likely duplicates or close relatives. Finally, IBS was also calculated between each sample from this study and 1,397 samples from the International HapMap project. The IBS relationships were converted to distance, and projected onto two axes of multidimensional scaling. We removed non-European ancestry outliers based on these projections (Supplementary Fig. 2a). After removing these individuals, we excluded SNPs with a call rate of less than 98%, Hardy-Weinberg P-value < 10−6 (in controls), missing rate per SNP difference in cases and controls > 0.02 or a minor allele frequency less than 1%. In total, 799 individuals and 175,385 SNPs were excluded, leaving for analysis 707,452 SNPs in 5,530 cases and 5,607 controls, which we call Set 1. Principal component analysis was performed across autosomal SNPs within Set 1 (Supplementary Figs. 2b and 2c). We computed the first 10 principal components and decided to include the top 4 in our downstream analysis to account for population structure remaining in our dataset.
We imputed SNPs from the 1000 Genomes Phase I (interim) release into our Set 1 samples using IMPUTE226,27. Imputed SNPs were excluded if the imputation quality score r2 was < 0.5 and minor allele frequency was < 1%. After filtering, 7,614,862 SNPs were left for further association analyses.
TB association was tested with logistic regression implemented in SNPTEST28 on genotype likelihoods from imputation, with 4 PCs as covariates. The genomic control inflation factor λGC for SNPs before and after imputation was 1.10 (Supplementary Fig. 3), indicating that we have successfully controlled for any residual population structure between cases and controls.
SNPs from the 1000 Genomes Phase I (interim) release were imputed into Ghanaian and Gambian subjects using IMPUTE2 and TB association of the seven ASAP1 SNPs was tested with logistic regression implemented in SNPTEST.
We genotyped seven ASAP1 SNPs using custom Taqman assays (Supplementary Table 6) and 7900HT system from Applied Biosystems. We visually checked all genotype clusters, assigned calls and extracted genotypes using SDS 2.3 software. TB association was tested in STATA11 using logistic regression, with the city of the sample origin (St. Petersburg or Samara) as a covariate.
To conduct Bayesian fine-mapping we calculated marginal likelihoods for each SNP and Bayes Factors using a normal prior with mean 0 and variance 0.2. Posterior probabilities were assigned using the relative contribution of each SNP’s Bayes Factor to the sum of Bayes Factors across all seven SNPs29.
We analyzed the seven ASAP1 SNPs in the Ghanaian and Gambian TB GWAS datasets that were published previously4,5. The Gambian dataset is available in the The European Genome-phenome Archive (accession number EGAS00000000027). Both Ghanaian and Gambian datasets have been QCed by filtering individuals with discordant sex information, elevated missing data rates (>2%), outlying heterozygosity rate (±3.5 standard deviations), duplicated or related individuals with IBS >80% and divergent ancestry based on principal component projections using the HapMap dataset. In total, 101 (13 cases and 88 controls) and 316 (192 cases and 124 controls) individuals were excluded from Ghanaian and Gambian datasets, respectively. After individual quality control, we imputed the seven ASAP1 SNPs from the 1000 Genomes Phase I (interim) release into Ghanaian and Gambian samples using IMPUTE226,27.
Analysis of association between SNPs and levels of ASAP1 mRNA expression in DCs
We analyzed genome-wide expression data from DCs isolated from 65 Caucasian subjects23. These DCs were either non-infected or infected in vitro for 18 hours with M. tuberculosis and were studied using Illumina HT-12 expression arrays23. The raw data can be obtained from the GEO database (accession numbers GSE34588 and GSE34151). For our analysis, we used a cleaned dataset provided by L. Barreiro (personal communications) in which expression levels were regressed using the first 5 principal components for the data from non-infected cells and the first 8 principal components for the data from infected cells. We used the R package snpStats (D. Clayton, snpStats: snpMatrix and XSnpMatrix classes and methods, R package version 1.12.10) to fit generalized linear models, with SNP genotypes as predictor variables and expression levels of ASAP1 as responses. We report P-values for the two-tailed test where the null hypothesis was that the true value of the generalized linear model coefficient is zero. We imputed genotypes of the ASAP1 SNPs rs2033059, rs17285138, rs1469288, rs12680942, rs1017281 with the MaCH and minimac software packages30 using genotype data from 1000 Genomes.
Cells and Antibodies
Human PBMC were isolated from fresh peripheral blood of healthy adult volunteers participating in the Cambridge BioResource cohort by centrifugation of peripheral blood with a gradient of Ficoll-paque Plus (GE Healthcare) at 860 g for 23 minutes at room temperature. Buffy coat layer containing PBMC was used for sequential isolation of monocytes, CD8+ T cells, CD4+ T cells and B cells. Red blood cells (RBCs) and polymorphonuclear cells sediment was used for neutrophil isolation. Monocytes were purified from PBMC using the MACS CD14+ Isolation kit (Miltenyi, UK Cat. No 130-050-201). CD8+ and CD4+ T cells were isolated from PBMCs using immunomagnetic positive selection kits (Stemcells Technologies Cat. No. 18053 and 18052). B cells were isolated from PBMCs using an immunomagnetic negative selection kit (Miltenyi Biotec 130-091-151). To isolate neutrophils, RBCs and granulocytes sediments were incubated with the same volume of fresh cold RBC lysis buffer on ice for 30 minutes and centrifuged for 5 minutes at 200 g. Cell pellets were then suspended with 10 ml of cold RBC lysis buffer and incubated on ice for 10 minutes. Neutrophil pellets were then washed with cold PBS 3 times.
To derive dendritic cells (DCs), the monocytes were cultured for 6 days in RPMI-1640 (Life technologies) with gentamycin (50 μg/ml), 5% FCS, GM-CSF (50 U/ml, Miltenyi, UK) and IL-4 (1000 U/ml, R&D Systems). To derive macrophages, the monocytes were cultured for 5 days in RPMI-1640 with HEPES (0.02 M), gentamycin (50 μg/ml), 10% FCS and 50 ng/ml M-CSF (Miltenyi, UK). Where indicated DCs were infected with live M. bovis BCG genetically modified to express mCherry protein (provided by Dr Brian Robertson, Imperial College, UK) at multiplicity of infection 3:1 and then lysed for RNA or protein extraction.
For immunofluorescence the following antibodies were used: anti-ASAP1 antibody (Sc-81896, Santa Cruz Biotechnology), anti-Vinculin antibody (700062, Invitrogen) and Alexa Fluor 568-coupled phalloidin (A12380, Molecular Probes). The secondary antibodies were Alexa Fluor 633-conjugated goat anti-rabbit IgG (Molecular Probes), and Alexa Fluor 488-conjugated donkey anti-mouse IgG (Molecular Probes). For western blots the following antibodies were used: anti-ASAP1 antibody (ab11011, Abcam) and anti-Actin antibody (A5441, Sigma).
qRT-PCR
Two-step qRT-PCR was used for analysis of the ASAP1 mRNA expression in neutrophils, monocytes, CD4+ T cells, CD8+ T cells, B cells, monocyte-derived macrophages and monocyte-derived DCs. Total RNA was isolated from cells using Isolate RNA Mini Kit as per the manufacturer’s instructions (Bioline UK, Cat No: BIO-52044). Then the first strand cDNA was generated from 200 ng of total RNA using Thermo Scientific Maxima Reverse Transcriptase (Thermo Scientific, UK, Cat. No. EP0741) and diluted 1:5. Then 2 μl were taken for qPCR using KAPA SYBR FAST Universal qPCR kit (Kapa Biosystems, UK, Cat. No. KK4602), ASAP1 primers and primers for the beta-2 microglobulin gene (Supplementary Table 7). qPCR reactions were done in triplicate. The data were analyzed using delta Ct values, providing ASAP1 mRNA expression levels relative to the levels of beta-2 microglobulin mRNA.
siRNA transfection
DCs were transfected at day 1 and day 4 using Hiperfect (Qiagen) with either a pool of three ASAP1-specific siRNAs (Supplementary Table 7) or with a control, scrambled, siRNA (1027281, Qiagen). Western blotting with the anti-ASAP1 antibody (ab11011, Abcam) was done in each case to assess knockdown efficiency. Results were analysed only if in cells treated with the ASAP1 siRNA expression of the ASAP1 protein was reduced to below 60% of that in cells treated with the control siRNA.
DC matrix degradation assay
Cover slips were coated with 0.2 mg/ml FITC-coupled gelatine (G13187, Molecular Probes) and subsequently blocked using complete media. The siRNA-treated cells were seeded on the cover slips and incubated at 37°C for 4 hours and fixed with 4% PFA. The cells were labelled with Alexa Fluor 568-coupled phalloidin (A12380, Molecular Probes) and mounted in anti-fade with DAPI (P36931, Molecular Probes). Cells were imaged on a Leica SP5 confocal microscope. The images were processed using ImageJ (Wayne Rasband, NIH). Matrix degradation was quantified by measuring the area where the FITC labelled-gelatine was below the threshold and counting the number of cells using ImageJ, as described before31.
DC migration assay
The siRNA-treated cells were seeded on cover slips coated with 0.025 mg/ml fibronectin (F1141, Sigma) and incubated at 37°C for 4 hours in their usual media without GM-CSF or IL-4. Cover slips were then inverted onto Dunn chambers containing GM-CSF (100 U/ml). The cells were imaged every 2-4 minutes for a minimum of 2 hours in the 37°C chamber of Leica SPE. Images were processed and analyzed using the manual track plugin of ImageJ and velocity was calculated.
Supplementary Material
Acknowledgements
The study was supported by the Wellcome Trust grants 088838/Z/09/Z and 095198/Z/10/Z, the EU Framework Programme 7 Collaborative grant 201483, the European Research Council Starting grant 260477, and the Royal Society grants UF0763346 and RG090638. S.N. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science and is also supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113, 085475 and 090355.
Footnotes
Author Information The authors declare no competing financial interests.
References
- 1.Zumla A, Raviglione M, Hafner R, von Reyn CF. Tuberculosis. N Engl J Med. 2013;368:745–755. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]
- 2.Moller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis (Edinb) 2010;90:71–83. doi: 10.1016/j.tube.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Apt A, Kramnik I. Man and mouse TB: contradictions and solutions. Tuberculosis (Edinb) 2009;89:195–198. doi: 10.1016/j.tube.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thye T, et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet. 2010;42:739–741. doi: 10.1038/ng.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thye T, et al. Common variants at 11p13 are associated with susceptibility to tuberculosis. Nat Genet. 2012;44:257–259. doi: 10.1038/ng.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chimusa ER, et al. Genome-wide association study of ancestry-specific TB risk in the South African Coloured population. Hum Mol Genet. 2013;23:796–809. doi: 10.1093/hmg/ddt462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang FR, et al. Genomewide association study of leprosy. N Engl J Med. 2009;361:2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 8.Zhang F, et al. Identification of two new loci at IL23R and RAB32 that influence susceptibility to leprosy. Nat Genet. 2011;43:1247–1251. doi: 10.1038/ng.973. [DOI] [PubMed] [Google Scholar]
- 9.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nie Z, Randazzo PA. Arf GAPs and membrane traffic. J Cell Sci. 2006;119:1203–1211. doi: 10.1242/jcs.02924. [DOI] [PubMed] [Google Scholar]
- 11.Randazzo PA, Inoue H, Bharti S. Arf GAPs as regulators of the actin cytoskeleton. Biol Cell. 2007;99:583–600. doi: 10.1042/bc20070034. [DOI] [PubMed] [Google Scholar]
- 12.Murphy DA, Courtneidge SA. The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function. Nat Rev Mol Cell Biol. 2011;12:413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bharti S, et al. Src-dependent phosphorylation of ASAP1 regulates podosomes. Mol Cell Biol. 2007;27:8271–8283. doi: 10.1128/MCB.01781-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin D, et al. ASAP1, a gene at 8q24, is associated with prostate cancer metastasis. Cancer Res. 2008;68:4352–4359. doi: 10.1158/0008-5472.CAN-07-5237. [DOI] [PubMed] [Google Scholar]
- 15.Onodera Y, et al. Expression of AMAP1, an ArfGAP, provides novel targets to inhibit breast cancer invasive activities. EMBO J. 2005;24:963–973. doi: 10.1038/sj.emboj.7600588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehlers JP, Worley L, Onken MD, Harbour JW. DDEF1 is located in an amplified region of chromosome 8q and is overexpressed in uveal melanoma. Clin Cancer Res. 2005;11:3609–3613. doi: 10.1158/1078-0432.CCR-04-1941. [DOI] [PubMed] [Google Scholar]
- 17.Muller T, et al. ASAP1 promotes tumor cell motility and invasiveness, stimulates metastasis formation in vivo, and correlates with poor survival in colorectal cancer patients. Oncogene. 2010;29:2393–2403. doi: 10.1038/onc.2010.6. [DOI] [PubMed] [Google Scholar]
- 18.Lin PL, et al. Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nat Med. 2014;20:75–79. doi: 10.1038/nm.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst JD. The immunological life cycle of tuberculosis. Nat Rev Immunol. 2012;12:581–591. doi: 10.1038/nri3259. [DOI] [PubMed] [Google Scholar]
- 20.Wolf AJ, et al. Mycobacterium tuberculosis infects dendritic cells with high frequency and impairs their function in vivo. J Immunol. 2007;179:2509–2519. doi: 10.4049/jimmunol.179.4.2509. [DOI] [PubMed] [Google Scholar]
- 21.Wolf AJ, et al. Initiation of the adaptive immune response to Mycobacterium tuberculosis depends on antigen production in the local lymph node, not the lungs. J Exp Med. 2008;205:105–115. doi: 10.1084/jem.20071367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts LL, Robinson CM. Mycobacterium tuberculosis infection of human dendritic cells decreases integrin expression, adhesion and migration to chemokines. Immunology. 2013;141:39–51. doi: 10.1111/imm.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barreiro LB, et al. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc Natl Acad Sci U S A. 2012;109:1204–1209. doi: 10.1073/pnas.1115761109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional References
- 24.Szeszko JS, et al. Resequencing and association analysis of the SP110 gene in adult pulmonary tuberculosis. Hum Genet. 2007;121:155–160. doi: 10.1007/s00439-006-0293-z. [DOI] [PubMed] [Google Scholar]
- 25.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2012;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 29.Maller JB, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gotz A, Jessberger R. Dendritic cell podosome dynamics does not depend on the F-actin regulator SWAP-70. PLoS One. 2013;8:e60642. doi: 10.1371/journal.pone.0060642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen CY, et al. Improved ancestry inference using weights from external reference panels. Bioinformatics. 2013;29:1399–1406. doi: 10.1093/bioinformatics/btt144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.