Abstract
Following light-induced nuclear translocation, phytochrome photoreceptors interact with and induce rapid phosphorylation and degradation of bHLH transcription factors, such as PHYTOCHROME-INTERACTING FACTOR 3 (PIF3), to regulate gene expression. Concomitantly this interaction triggers feedback reduction of phytochrome B (phyB) levels. Light-induced phosphorylation of PIF3 is necessary for the degradation of both proteins. We report that this PIF3 phosphorylation induces, and is necessary for, recruitment of LRB (Light-Response Bric-a-Brack/Tramtrack/Broad (BTB)) E3 ubiquitin ligases to the PIF3-phyB complex. The recruited LRBs promote concurrent polyubiqutination and degradation of both PIF3 and phyB in vivo. These data reveal a linked signal-transmission and attenuation mechanism involving mutually assured destruction of the receptor and its immediate signaling partner.
The mechanisms by which cells perceive and adapt to external signals remains an area of central interest in the biosciences. The capacity to respond rapidly and robustly to such signals is frequently coupled to a capacity to subsequently modulate the intensity of that response by feedback attenuation of the signaling process (1). Whereas the initial robust burst of signaling activity is presumably necessary for the induction of the consequent adaptational or developmental switch, unfettered prolongation of the elevated activity can be harmful to the organism. A plethora of attenuation mechanisms have been identified (1, 2). Defects in these mechanisms are under increasing scrutiny as evidence increases that they are major causes of human malignancies (3).
The initial emergence of seedlings from subterranean darkness into sunlight triggers a rapid and extensive redirection of gene expression that drives a developmental switch from skotomorphogenic to photomorphogenic development, observed as production of normal green seedlings (4, 5). The red and far-red wavelengths inducing this switch are perceived by the phytochrome family of sensory photoreceptors (phyA-E) by virtue of a capacity to convert reversibly between biologically-inactive Pr (red-absorbing), and biologically-active Pfr (far-red-absorbing), conformers (6). Photoactivation of the cytoplasmically-localized photoreceptors triggers their rapid translocation into the nucleus where they interact directly with a subfamily of bHLH transcription factors, termed PIFs (Phytochrome-Interacting Factors; PIF1-8 (4, 7)). This interaction induces multisite phosphorylation, ubiquitination and degradation of the PIFs via the 26S proteasome system (4, 8-14), altering the transcription of target genes within minutes (4, 5, 15). The PIFs promote skotomorphogenic development in the dark (16), but their red-light-induced degradation represses this activity, inducing the switch to the photomorphogenic pathway (4).
In addition to their direct function in phytochrome signal transduction, the PIFs also de-sensitize cells to red light through negative feedback regulation of phyB levels (17-19). We showed recently, that Pfr-induced multisite phosphorylation of PIF3 is not only necessary for the degradation of PIF3, but also for concomitant PIF-interaction-induced degradation of phyB (8). Although there is evidence that E3 ligases assembled with COP1 are involved in ubiquitylation and degradation of phyB, in a PIF-promoted manner (20), the signaling mechanism has remained unclear.
To address this question, we performed mass-spectrometric analysis of affinity-purified PIF3 to identify components associated with PIF3 in a light-dependent manner. Dark-grown seedlings transgenically-expressing YFP-PIF3 were either kept in the dark or irradiated with 10 min of red light before protein extraction and affinity purification using a GFP antibody. Quantitative spectral count analysis, not unexpectedly, identified all five phytochromes (phyA-E) specifically in the red light-treated samples (Table S1). In addition, components of a Bric-a-Brack/Tramtrack/Broad (BTB)-Cullin3-type E3 ubiquitin ligase were also identified as red-light-specific, PIF3-interacting proteins in three biological replicates (Table S1 and Fig. S1). BTB proteins are substrate-specific proteins that bridge target proteins to Cullin3 for ubiquitin ligation (21, 22). The two highly conserved BTB proteins (Light-Response-BTB1 (LRB1) and LRB2) identified here were shown previously to be nuclear-localized and required genetically for proteasome-mediated phyB protein degradation in the light, but whether the involvement is direct or indirect was not determined (23). A third LRB homolog (LRB3) was not detected in our mass spectrometric analysis, possibly due to the lower expression level than the other two (23).
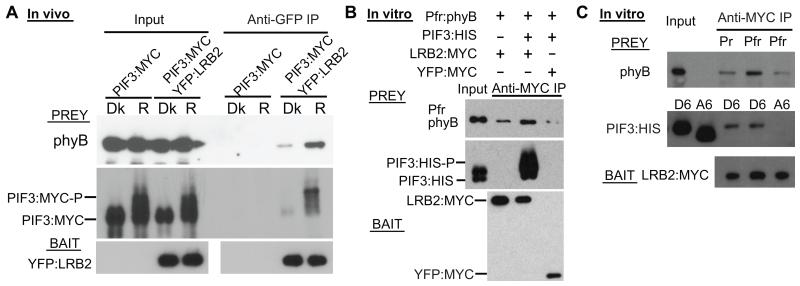
To further investigate the light dependent interaction of PIF3 with the LRBs in vivo, we generated transgenic lines expressing both PIF3:MYC and YFP:LRB2. Immunoprecipitation using YFP:LRB2 as bait showed considerably higher levels of PIF3:MYC co-precipitation from red-treated than dark-control seedlings (Fig. 1A). The red-treated samples also displayed enrichment of the mobility-shifted forms of PIF3:MYC in the immunoprecipitates compared to the input (Fig.1A), suggesting preferential binding of in vivo-phosphorylated PIF3 to the LRB protein. Together with previous evidence that red light induces LRB-Cullin3 interaction in vivo (23), our present findings indicate that this interaction is promoted by light-induced binding of phospho-PIF3 to the LRBs.
Fig. 1.
Light-induced phosphorylation of PIF3 promotes interaction with Cullin E3-ligase substrate-recognition subunit LRB2.
(A) Light induces LRB2 and PIF3 interaction in vivo. Dark-grown seedlings from either PIF3:MYC or PIF3:MYC/YFP:LRB2 double-transgenic lines were pretreated with MG132, then either kept in the dark (Dk) or irradiated with a pulse of red light (Rp) before protein extraction and co-immunoprecipitation (Co-IP) with an anti-GFP antibody. The proteins were analyzed by Western blot with either anti-MYC antibody (Prey, Top and middle panels), or anti-GFP antibody (Bait, Bottom panel). (B –D) Phosphorylation or phosphomimic mutations of the light-induced phosphosite residues in PIF3 promote its association with LRB2 in vitro. (B) and (C) In vitro-expressed recombinant LRB2 or YFP-control (Bait) proteins were precipitated with a MYC antibody, then incubated with normal (WT) PIF3 or the indicated light-induced-phosphosite mutant variants (A6, A20, D6, D19) of PIF3 (Prey). Bait and prey proteins were detected by Western blot using anti-MYC and anti-His antibody, respectively. All proteins were expressed in Hela cell lysate. A: Ser/Thr to Ala substitutions; D: Ser/Thr to Asp substitutions. A6/D6: PIF3 with substitutions in the six strongly light-induced phosphosite residues (8) (see Fig. S4A). A20/D19: PIF3 with substitutions in the majority of the light-induced phosphosite residues. Co-IP products were treated with alkaline phosphatase (CIAP) in panel (B). (D) PIF3-A6, PIF3-WT or PIF3-D6 expressed in Hela cell lysates were either treated (+) or not (−) with CIAP or with heat inactivated CIAP before Western blot analysis using anti-His antibody. (E) High affinity interaction of LRB2 and PIF3 in vitro requires multisite light-induced phosphorylation of PIF3. Immunoprecipitations and Western blots as in (C). PIF3 mutant-variant substitutions: D1: S88D, D3: S58/S88/S102D. See Fig. S4A for all S to A or D substitutions.
In vitro pulldown assays using recombinant LRB2:MYC as bait were also performed to further probe the molecular mechanism of the interaction with PIF3. Because phyB does not display autonomous light-activated protein kinase activity toward PIF3 in our hands, and no other kinase has been identified, we examined the LRB-binding behavior of a set of mutant PIF3 variants. Previously, using mass spectrometric analysis and transgenic expression of targeted missense mutants of PIF3, we identified twenty light-induced phosphorylated residues in the protein that are functionally necessary for its rapid degradation in vivo (8). When expressed in a Hela cell lysate, the phospho-dead PIF3 mutant protein (A20) showed no apparent binding to LRB2, whereas PIF3 with phospho-mimic mutations in the majority of the light-induced residues (D19), or simply the six most strongly light-induced residues (D6), exhibited high binding affinity towards LRB2 (Fig. 1B). The absence of binding to PIF3-A6 and YFP-MYC further supports the specificity of phosphorylation-dependent binding of LRB2 to PIF3 (Fig. 1C). The in vitro-expressed wild-type PIF3 intrinsically displayed multiple bands, reminiscent of the pattern from light-treated seedlings, and only the slower migrating form showed high affinity for LRB2 by co-immunoprecipitation (Fig.1B and 1C). Moreover, the slower migrating band of wild-type PIF3 was sensitive to alkaline phosphatase treatment (Fig. 1B and 1D). These results suggest that recombinant wild-type PIF3 expressed in Hela cell lysates is phosphorylated by an unknown kinase in the lysate. Moreover, it appears that one or more of the strongly light-induced sites (8) in PIF3 is phosphorylated by this kinase, since the PIF3-A6 variant, mutated at six of these sites, did not have a similarly slower migrating form (Fig. 1C and 1D). In addition, in vitro pulldown assays using LRB2 as prey also indicated that PIF3-WT, D6 and D19, but not A20, bind LRB2 with high affinity (Fig. S2), and that both LRB1 and LRB3 are able to bind PIF3-D6 with high affinity in vitro, similarly to LRB2 (Fig. S3).
Since the six most strongly light-induced phosphosites in PIF3 are predominantly responsible for rapid degradation in vivo (8), we tested here whether binding to LRB2 in vitro also requires these multiple phospho-sites. Phosphomimic substitutions in individual (variants 1, D1 or 3) or three (variants 4 or D3) of these sites were introduced into the PIF3-A6 sequence (Fig. S4A). In vitro pulldown assays showed that none of these mutant variants has LRB2-binding-affinity comparable to PIF3-D6 (Fig. 1E and S4B), suggesting that high-affinity binding to LRB2 requires between 3 and 6 light-induced phosphosites in PIF3. In addition, PIF3-D6 showed relatively low binding to either the N- or C-terminal domain of LRB2 (Fig. S5B), suggesting that determinants in both domains are required for its strong binding to PIF3. These data favor the model that the cooperative action of multiple low-affinity sites between PIF3 and LRBs enable light-induced degradation of PIF3, similar to that proposed for yeast Sic1 degradation (24).
Because light-induced phosphorylation of PIF3 is also required for its negative feedback-regulation of phyB levels, despite not being required for phyB-PIF3 binding per se (8), we examined the possibility that phosphorylated PIF3 bridges the interaction of LRBs and phyB-Pfr in vivo. Pull-down assays from seedling extracts using YFP:LRB2 as bait showed light-dependent co-immunoprecipitation of phyB and PIF3 with LRB2 (Fig. 2A). In vitro binding assays indicated that LRB2 itself has some apparent affinity for phyB-Pfr alone, but that PIF3 binding significantly enhances LRB2-phyB-Pfr association (Fig. 2B and 2C).
Fig. 2.
Light-induced phosphorylation of PIF3 promotes phyB-LRB2 association.
(A) Light induces phyB-LRB2 association in vivo. Proteins were extracted and immunoprecipitated using the same lines and procedure as in Fig. 1A, and then subjected to Western blot analysis using anti-phyB antibody (Prey, top panel), anti-MYC antibody (Prey, middle panel), or anti-GFP antibody (Bait, bottom panel). (B) Phosphorylated PIF3 promotes phyB-LRB2 association in vitro. In vitro-expressed recombinant LRB2:MYC or YFP:MYC-control bait proteins were incubated with phyB-Pfr in the presense (+) or absence (−) of PIF3:His prey proteins, and immunoprecipitated with anti-MYC antibody. Western blots were probed with anti-phyB (Top), anti-His-epitope (middle) or anti-MYC (bottom) antibodies. (C) PIF3-D6 phosphomimic (8) (see Fig. S4A) promotes Pfr dependent phyB-LRB2 association in vitro. In vitro-expressed recombinant LRB2:MYC bait protein was incubated with phyB (as Pr or Pfr) and PIF3:His (D6- or A6-mutant (8) (see Fig. S4A)) prey proteins, and immunoprecipitated with anti-MYC antibody. Western blots as in (B).
To examine the functional role of the LRBs in PIF3 and PIF3-mediated-phyB degradation, we utilized a set of single, double and triple lrb mutants (23). Dark-grown seedlings of both the lrb3 single and lrb1lrb2 double mutants have normal PIF3 protein levels, and normal light-induced PIF3 degradation (Fig. S6A). By contrast, the lrb1lrb2lrb3 (lrb123) triple mutant exhibits a significantly reduced rate of light-induced endogenous PIF3 degradation (Fig. 3A and Fig. S6B, C). In addition, the light-induced, slower-migrating forms of PIF3 display accumulation in the lrb123 triple mutant, compared to rapid degradation in the wild type (Fig. 3A and Fig. S6B). A similar pattern was observed for a PIF3:GFP fusion protein transgenically expressed under control of the constitutive CaMV 35S promoter in the lrb123 mutant background (Fig. 3B, top two panels, and Fig. S6C), indicating that LRB regulation is exerted at the post-translational level. Phosphatase treatment reversed the mobility shift of the accumulated slower migrating PIF3:GFP bands (Fig. 3B, third panel). These data indicate that PIF3 accumulates in the phosphorylated form in the light when the LRBs are mutated. The reason for the retention of higher relative levels of PIF3:GFP than endogenous PIF3 in prolonged red light (Fig.S6C) is undetermined, but could reflect the higher absolute levels of PIF3:GFP than endogenous PIF3.
Fig. 3.
LRBs function in both PIF3 and PIF3-mediated-phyB degradation in the light.
(A) Endogenous PIF3 degradation is reduced in the lrb1lrb2lrb3 triple-mutant (lrb123) compared to Col wild-type (WT) seedlings in the light. Dark-grown seedlings of Col and lrb123 were irradiated with red light (R) for the period indicated before protein extraction and Western blot analysis using anti-PIF3 antibody (Left panel). PIF3: unphosphorylated PIF3; PIF3-P: phosphorylated PIF3; NS: nonspecific bands. Right panel shows quantification of the Western blot results from both the Left panel (blue and red curves) and Fig. S6B (green and purple curves) after normalization to the Tubulin loading control. The dark PIF3 level was set as 100%. (B) PIF3:GFP accumulates in the phosphorylated form in the lrb123 triple mutant in the light. Seedling growth was as in panel (A). Extracted proteins were either analyzed directly (Top two panels), or after phosphatase (CIAP) treatment (Third panel) by Western blot using anti-GFP antibody. (C) LRBs are essential for light-induced, PIF3-promoted feedback degradation of phyB. Light-induced degradation of phyB (accelerated by PIF3:GFP overexpression) is absent in the lrb123 mutant background. Dark-grown seedlings of the indicated genotypes were irradiated with 3 (R3h), 24 (R24h) or 96 (Rc96h) hours of continuous red light before protein extraction and Western blot analysis using an anti-phyB antibody (Left and middle panels). Tubulin was used as a loading control. Right panel shows quantification of the relative phyB/Tubulin protein levels from left and middle panels. Dark levels were set as 100%. (D) PIF3:GFP over-expression does not complement the short-hypocotyl phenotype of the lrb123 mutant in the light. Seedlings of the indicated genotypes were grown for 4 days in the dark (Dk) or continuous red light (Rc). Visible phenotypes (left); hypocotyl lengths (right). Error bars represent standard errors. (E) Endogenous PIF3 levels are strongly reduced in dark-grown seedlings of the cop1 and spa123 mutants.
We showed previously that overexpression of PIF3 markedly accelerates the rate of phyB degradation in response to light (8). This finding indicates that the absolute rate of PIF3 degradation is rate-limiting to the concurrent degradation of phyB. Because the relative rate of phyB degradation is about 50-fold slower than that of PIF3 in the wild type seedling (8, 18, 19, 25), this observation suggests in turn that phyB is substantially more abundant than PIF3. The data for phyB in the Col wild type and the Col transgenic line overexpressing PIF3:GFP in Fig. 3C support this notion, showing the accelerated degradation of phyB in the overexpressor line. However, the light-induced degradation of phyB is absent in both the lrb12 double (Fig. S6D) and lrb123 triple (Fig. 3C) mutants, with or without overexpression of PIF3:GFP, in the latter case. These data are consistent with previous evidence for the lrb mutants (23), and establish that LRB1 and LRB2 are essential for the PIF3-mediated, light-induced degradation of phyB. The absence of accelerated phyB degradation by overexpression of the PIF3-A20 variant (8) that cannot bind LRB2 (Fig. 1B) is similarly in agreement with this conclusion.
The morphogenic phenotypes of these lines are consistent with these findings. While seedlings over-expressing PIF3 in wild-type Col show an etiolated-like phenotype in the light, seedlings over-expressing a similarly high or higher level of PIF3:GFP or endogenous PIF3 in the lrb123 mutant (Figs. 3A, 3B and S6C) remain hypersensitive to red light, like the lrb123 mutant itself (23) (Fig. 3D). These data are in accord with the levels of phyB in these lines under prolonged red light (Fig. 3C), whereby the depletion of phyB in the wild-type Col PIF3-overexpressor results in hyposensitivity to the light signal, and the stability of phyB in the lrb123 background (23) results in hypersensitivity, despite high levels of PIF3 (Figs. 3A, 3B and S6C). These results indicate that the suppressive activity of high phyB levels overrides the antagonistic promotive activity of PIF3 on hypocotyl growth (17). This effect could result from phyB activity independent of PIF3, reduced intrinsic activity of PIF3, or phyB-induced dissociation of PIF3 from its DNA binding sites.
COP1-assembled E3 ligases were reported to be an important negative regulator of phyB levels (20). However, in agreement with other data (14, 16), we observed that PIF3 levels are reduced in dark-grown seedlings of both the cop1 and spa123 mutants, and no accumulation is observed in the light (Fig. 3E). These data suggest that the reduction in light-induced phyB degradation in the cop1 mutant is partially due to an indirect effect of the reduced levels of PIF3 (and possibly other PIFs) initially present in the dark. Regardless, the absence of detectable light-induced phyB degradation in the lrb123 mutant (Fig 3C) indicates that COP1 is not sufficient for phyB degradation in the absence of the LRBs. Consistent with previous data that LRB1 and LRB2 are not directly involved in phyA degradation (23), we found that LRB3 also has no detectable role (Fig. S7).
Regulated protein degradation through the 26S proteasome requires E3 ligase-mediated poly-ubiquitination of target proteins (21, 22). In support of the LRBs being components of an E3 ligase responsible for PIF3 polyubiqutination, light-induced ubiquitination of PIF3:GFP (Fig. S8) is significantly reduced in the lrb123 mutant (Fig. 4A). Moreover, photoactivated phyB, which co-immunoprecipitates with PIF3:GFP from extracts of light-treated seedlings, also displays high molecular weight-shifted bands in wild-type but not in lrb123 mutant seedlings (Fig. 4B), suggestive of a sub-fraction of light-dependent, LRB-generated polyubiquitinated phyB molecules. More definitively, precipitation of total ubiquitinated proteins from the seedling extracts, followed by immunoblot detection of either phyB, PIF3:GFP or endogenous PIF3, indicates that both phyB and PIF3 are polyubiquitinated in the wild-type in the light, and that this concomitantly-induced, bimolecular ubiquitination is undetectable (Fig. 4C) or markedly reduced (Fig. S9) in the lrb123 mutant.
Fig. 4.
LRB E3 ligases function in polyubiquitination of both PIF3 and phyB. (A) Light-induced poly-ubiquitination of PIF3 is reduced in the lrb123 mutant. Dark-grown wild-type (col) or lrb123-mutant seedlings, transgenically, or not, expressing PIF3:GFP (PIF3:GFP/col and PIF3:GFP/lrb123, respectively), were irradiated with 10 min of red light (R), or not (Dk), before protein extraction and immunoprecipitation (IP) using anti-GFP antibody. The IP products were then analyzed by Western blot with anti-GFP antibody (left panel) or anti-ubiquitin antibody (right panel). (B) Light-induced mobility shift of PIF3-bound phyB (phyB-ubi) is absent in the lrb123 mutant. Co-IP products prepared as in panel A were analyzed with anti-phyB antibody. (C) Light-induced poly-ubiquitination of both phyB and PIF3 is strongly reduced in the lrb123 mutant. Seedling growth and extraction was as in panel A. Total ubiquitinated proteins were immunoprecipitated with TUBEs (Tandem Ubiquitin Binding Entities), then analyzed by Western blot using antibody against ubiquitin (left panel), phyB (middle panel), or GFP (right panel). (D) A Cullin3-LRB2 E3 ubiquitin-ligase complex ubiquitinates phosphomimic PIF3 (D6) in vitro. A Cullin3 E3 ubiquitin ligase complex was assembled with recombinant LRB2 protein and incubated with D6 or A20 PIF3:MYC (8) (see Fig. S4A for D6) variants in vitro (see Methods) before western blot analysis with anti-MYC antibody.
Finally, to examine whether LRB2 has the capacity to directly catalyze ubiquitination of PIF3, we assembled a Cullin3-based E3 ligase complex in vitro using recombinant LRB2 and human Cullin3/RBX1, and performed an in vitro ubiquitination assay. The data show that this complex can indeed ubiquitinate PIF3 in a phosphomimic-dependent manner, consistent with its PIF3 recognition mechanism and confirming its biochemical function (Fig. 4D, Fig. S10). Although the evidence for LRB-dependent, light-induced ubiquitination of phyB in vivo is compelling (Fig. 4C), for undetermined reasons we were unable to detect such activity using the above in vitro assay.
We propose a mechanistic model (Fig. S11), whereby light-activated phyB induces multisite phosphorylation of PIF3 upon direct binding, thereby enhancing the affinity of PIF3 for the LRBs, which then bind to Cul3, forming an active E3 ligase complex, which ubiquitinates both PIF3 and phyB, targeting them for degradation by the 26S proteasome. This mechanism of attenuation thus embodies a mutually-assured destruction configuration of bidirectional signaling, directly at the receptor-primary-signaling-partner interface, that is unusual among reported mechanisms (1-3). It is notable, however, that while light-induced phyB ubiquitination and degradation is essentially eliminated in the lrb123 mutant, light-induced PIF3 ubiquitination and degradation is slowed but not eliminated (Fig. 3, 4 and S6C and D). This observation suggests the existence of partial functional redundancy between the LRBs and yet another unknown E3 ligase(s) for PIF3 degradation, and uncovers a dichotomy between PIF3 and phyB regulation.
Supplementary Material
Acknowledgments
We thank R. J. Chalkley, A. Pfeiffer, M. Zhuang, and B. A. Schulman for materials and technical assistance, and S. McCormick for comments on the manuscript. We also thank the Salk Institute and ABRC for the Arabidopsis T-DNA insertion lines. The plasmid pGEX-CUL3 (split ‘n’ coexpress)-Rbx1 used in the in vitro ubiquitination assays, was provided by Dr. B. A. Schulman under an MTA with St. Jude Children’s Research Hospital. For access to this reagent, contact Dr. B. A. Schulman. Supplement contains additional data. This work was supported by NIH (2R01 GM-047475), DOE (DEFG03-87ER13742), and USDA ARS Current Research Information System (5335-21000-032-00D) to P.H.Q; by NIH (5R01GM066258) and DOE (DEFG02-08ER15973) to Z.Y.W.; by NIH (8P41GM103481) to A.L.B; by NIH (P50 GM082250) to J.D.G.
Footnotes
Supplementary Materials:
Materials and Methods
Figures S1-S11
Tables S1-S2
References (26-27)
References and Notes
- 1.Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12:104. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 2.Yarden Y. The biological framework: translational research from bench to clinic. Oncologist. 2010;15:1. doi: 10.1634/theoncologist.2010-S5-01. [DOI] [PubMed] [Google Scholar]
- 3.Amit I, et al. A module of negative feedback regulators defines growth factor signaling. Nat Genet. 2007;39:503. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- 4.Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 6.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400:781. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- 8.Ni W, et al. Multisite Light-Induced Phosphorylation of the Transcription Factor PIF3 Is Necessary for Both Its Rapid Degradation and Concomitant Negative Feedback Modulation of Photoreceptor phyB Levels in Arabidopsis. Plant Cell. 2013;25:2679. doi: 10.1105/tpc.113.112342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen H, et al. Light-Induced Phosphorylation and Degradation of the Negative Regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis Depend upon Its Direct Physical Interactions with Photoactivated Phytochromes. Plant Cell. 2008;20:1586. doi: 10.1105/tpc.108.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 11.Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol. 2008;59:281. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- 12.Shen Y, Khanna R, Carle CM, Quail PH. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007;145:1043. doi: 10.1104/pp.107.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Bauer D, et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004;16:1433. doi: 10.1105/tpc.021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, et al. A Quartet of PIF bHLH Factors Provides a Transcriptionally Centered Signaling Hub That Regulates Seedling Morphogenesis through Differential Expression-Patterning of Shared Target Genes in Arabidopsis. PLoS Genet. 2013;9:e1003244. doi: 10.1371/journal.pgen.1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leivar P, et al. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leivar P, Monte E, Cohn MM, Quail PH. Phytochrome signaling in green Arabidopsis seedlings: impact assessment of a mutually negative phyB-PIF feedback loop. Mol Plant. 2012;5:734. doi: 10.1093/mp/sss031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Al-Sady B, Kikis EA, Monte E, Quail PH. Mechanistic duality of transcription factor function in phytochrome signaling. Proc Natl Acad Sci U S A. 2008;105:2232. doi: 10.1073/pnas.0711675105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khanna R, et al. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell. 2007;19:3915. doi: 10.1105/tpc.107.051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH. Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell. 2010;22:2370. doi: 10.1105/tpc.109.072520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua Z, Vierstra RD. The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol. 2011;62:299. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- 22.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 23.Christians MJ, Gingerich DJ, Hua Z, Lauer TD, Vierstra RD. The light-response BTB1 and BTB2 proteins assemble nuclear ubiquitin ligases that modify phytochrome B and D signaling in Arabidopsis. Plant Physiol. 2012;160:118. doi: 10.1104/pp.112.199109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang X, et al. Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase. Proc Natl Acad Sci U S A. 2012;109:3287. doi: 10.1073/pnas.1116455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monte E, et al. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci U S A. 2004;101:16091. doi: 10.1073/pnas.0407107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhuang M, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.