Abstract
The kinetics of oxidation of various alcohols by purified rat liver alcohol dehydrogenase (ADH) were compared with the kinetics of elimination of the alcohols in rats in order to investigate the roles of ADH and other factors that contribute to the rates of metabolism of alcohols. Primary alcohols (ethanol, 1-propanol, 1-butanol, 2-methyl-1-propanol, 3-methyl-1-butanol) and diols (1,3-propanediol, 1,3-butanediol, 1,4-butanediol, 1,5-pentanediol) were eliminated in rats with zero-order kinetics at doses of 5–20 mmole/kg. Ethanol was eliminated most rapidly, at 7.9 mmole/kg•h. Secondary alcohols (2-propanol-d7, 2-propanol, 2-butanol, 3-pentanol, cyclopentanol, cyclohexanol) were eliminated with first order kinetics at doses of 5–10 mmole/kg, and the corresponding ketones were formed and slowly eliminated with zero or first order kinetics. The rates of elimination of various alcohols were inhibited on average 73% (55% for 2-propanol to 90% for ethanol) by 1 mmole/kg of 4-methylpyrazole, a good inhibitor of ADH, indicating a major role for ADH in the metabolism of the alcohols. The Michaelis kinetic constants from in vitro studies (pH 7.3, 37 °C) with isolated rat liver enzyme were used to calculate the expected relative rates of metabolism in rats. The rates of elimination generally increased with increased activity of ADH, but a maximum rate of 6 ± 1 mmole/kg•h was observed for the best substrates, suggesting that ADH activity is not solely rate-limiting. Because secondary alcohols only require one NAD+ for the conversion to ketones whereas primary alcohols require two equivalents of NAD+ for oxidation to the carboxylic acids, it appears that the rate of oxidation of NADH to NAD+ is not a major limiting factor for metabolism of these alcohols, but the rate-limiting factors are yet to be identified.
Keywords: Alcohol dehydrogenase, Alcohol metabolism, Inhibition, Rat metabolism, Enzyme specificity
1. Introduction
Alcohol dehydrogenases (ADH) are ubiquitous in higher organisms and participate in metabolizing a wide variety of alcohols and aldehydes, as an important “detoxification” mechanism [1]. ADHs catalyze the first step in alcohol oxidation, using NAD+ as a cofactor and producing NADH and the corresponding carbonyl compound. Various studies have suggested that ADH activity is a major rate-limiting factor in ethanol metabolism, because the amount of liver ADH is approximately sufficient to account for the rate of elimination in animals, and inhibitors of ADH act equivalently in vitro and in vivo in rats [2-8]. Fed rats eliminate ethanol at about 8 mmol/kg•h, and the total liver ADH activity could provide about a 1.4-fold higher rate, but the concentrations of coenzymes and acetaldehyde in vivo could limit the rate of ethanol metabolism to the observed value [7, 8]. Humans eliminate ethanol at about 2.2 mmol/kg•h, but the total ADH activity and the mass of the liver relative to body weight are each about one-half of that found in the rat, supporting the conclusion that ADH activity is a major rate limiting factor for ethanol metabolism in humans [2].
Kinetic simulation with estimated rate constants for alcohol and aldehyde dehydrogenases can approximately describe ethanol and acetaldehyde metabolism in humans, and it is significant that the rate of elimination of ethanol is directly related to ADH activity, whereas the steady-state level of acetaldehyde (almost a constant blood concentration) depends on levels of both alcohol and aldehyde dehydrogenases [9]. Although metabolism of ethanol in humans is complicated because humans have five different ADHs that can contribute to ethanol metabolism and the kinetic constants and the concentrations for these enzymes should be considered [10, 11], it is remarkable that the rate of metabolism can be described by a single set of kinetic constants [9]. However, in the steady-state of metabolism, it is likely that several steps, such as aldehyde dehydrogenase activity, transport of reducing equivalents from NADH into the mitochondria, and reoxidation of NADH in oxidative phosphorylation contribute to controlling the overall rate of metabolism, and more complete quantitative descriptions are required [2, 12]. Other enzymes, such as catalase and cytochrome P450 2E1, can also contribute to the oxidation of alcohols.
The specificities of ADHs for various alcohols and the kinetics of metabolism (elimination) in animals are also of fundamental interest because it is clear that ethanol is not the only substrate, and metabolism of ethanol can affect the metabolism of other alcohols and aldehydes, such as retinoids [13, 14]. Identification of endogenous substrates that may have physiological roles is a continuing challenge. The rat is a good model for these studies because of extensive prior use of this animal for studies of alcohol metabolism. Moreover, metabolism of various alcohols in rats should be studied as a prelude to any studies with humans.
Rats produce only four different active ADHs (see Ref. [15] for ADH nomenclature), but the single liver class 1 enzyme (ADH1, UniProt P06757) is the major ADH responsible for metabolism of common alcohols [16, 17]. The rodent ADH2 (UniProt Q64563) is much less active than the other ADHs [18]. The substrate specificities of three rat enzymes have been surveyed, and ADH3 (UniProt P12711) has no detectable activity on ethanol and butanol [19]. The “stomach” enzyme, ADH4 (UniProt P41682), has much lower catalytic efficiencies than ADH1, but may contribute to metabolism of high concentrations of alcohols and in metabolism of retinoids and lipid peroxidation products [16, 20-22]. ADH5 (UniProt Q5X195) is not expressed in an active form [23]. Having one major ADH makes it simpler to study the correlation of in vitro and in vivo activities. Studies with the rat are relevant for understanding metabolism of alcohols in humans, even though humans produce three class I ADHs with quantitatively-different substrate specificities, because the specificities of class I ADHs overlap and the metabolic pathways (chemical transformations) are probably similar in rats and humans. This work presents results on the specificities of rat liver ADH for alcohols, the kinetics of elimination of various alcohols in rats, and the correlation of in vitro ADH activities with the rates of metabolism.
2. Experimental Procedures
2.1. Substrate specificity of rat liver alcohol dehydrogenase
Rat liver alcohol dehydrogenase was partially purified by precipitation with (NH4)2SO4 (35–75% saturation), passage through DEAE-cellulose, and chromatography on Sephadex G-100 [24]. The specific activity was about 0.2 unit/mg, as assayed at 37 °C in a standard enzyme assay [25]. The enzyme was stabilized by adding 5% ethanol during the purification, but preparations gradually lost about 50% of the original activity during storage at 5 °C for a week. The insertion of Cys-112 may account for the lability [26]. (Adding NAD+ also stabilized the enzyme, but after a few days of storage, the kinetics did not fit simple Michaelis-Menten kinetics, as Km values for ethanol increased substantially, and it appeared that two forms of enzyme were present.) The ethanol was removed by gel filtration before kinetics studies with various alcohols, and a small reaction rate observed in the absence of added alcohol was subtracted from the rates with varied concentrations of alcohols. The reactions were also almost totally inhibited by 10 mM pyrazole, confirming the ADH activity. The kinetics of the ADH were studied in 83 mM potassium phosphate and 40 mM KCl buffer, pH 7.3, at 37 °C, with 0.5 mM NAD+, which is thought to resemble the conditions found in vivo [4]. The initial velocity data were fitted to the Michaelis-Menten equation [27]. Crystalline horse liver alcohol dehydrogenase, EE isoenzyme (ADH1E, UniProt P003327), was purchased from Boehringer Mannheim Co.
2.2. Elimination of alcohols in rats and effects of inhibitors
Male Sprague-Dawley rats (180–280 g) were fed certified rodent diet (equivalent to Harlan 8728C; 24% protein, 5% fat, 40% carbohydrate; tekladinfo@harlan.com) ad libitum, except for those that were fasted for 24 h. Lights were on from 5 a.m. to 5 p.m. during Central Standard Time and 6 a.m. to 6 p.m. during Daylight Savings Time. Experiments were begun mid-morning. Alcohols were diluted in physiological saline to 1 M or lower and administered intraperitoneally at sub-lethal doses. The doses were (usually) chosen to be within the concentration range used in the in vitro studies and to give measurable levels in the blood. Blood samples (10 μL) were drawn from the tail vein at regular intervals, such as 10 min, as shown in the figures, and mixed with an internal standard (an alcohol chosen to have a retention time close to the alcohol under investigation), and deproteinized with Ba(OH)2 and ZnSO4. (For 2,3-butanediol, methanol, and tert-butanol, which are slowly-eliminated, blood samples were taken about every 30 min.) The concentrations of ethanol determined in the tail vein are approximately the same as those in arterial blood [28]. The supernatant was analyzed on a Varian 3740 gas chromatograph with five different columns (usually 6 ft × 2 mm) and varied oven temperatures chosen to provide baseline resolution for the alcohols and their metabolites. Most primary and secondary alcohols and ketones were separated on 60/80 mesh Carbopack B/5% Carbowax PEG 20M or 60/80 mesh Carbopack C/0.2% Carbowax 1500 developed at 20 ml N2/min, with fixed column temperatures ranging from 60 to 130 °C. 1,2-Ethanediol, 1,5-pentanediol, and δ-valerolactone were separated on the Carbowax 20M column at 140 or 180 °C. Ethanol, 1-butanol, and 1,2-ethanediol were separated on 80/100 Chromasorb 102 at 30 ml N2/min, at 150, 200 and 200 °C. Ethanol and 1,2-ethanediol were also separated on 80/100 Porapak-S at 30 ml N2/min at 150 and 200 °C. The diols, benzyl alcohol and benzaldehyde were separated on 80/100 Carbopack C/0.8% THEED (3 ft × 2 mm ID) at 40 ml N2/min at 100 or 125 °C. The concentrations were calculated with a Hewlett-Packard 3388A integrator calibrated with standard curves.
The elimination data were fitted to the equations appropriate for the observed kinetics using the non-linear least squares Fortran program, NONLIN (C. M. Metzler, The Upjohn Co.). For zero-order (straight line dependence on time) elimination, the data were fitted to the equation [A]t = k1t + [A]o, and the rate constant was calculated by dividing the dose administered by the time required for complete elimination as determined by the linear extrapolation to zero concentration (mathematically, −[A]o/k1). The rate constants for three or more animals were averaged and the standard deviation calculated. The results for two animals were averaged and the range of values is reported. For non-linear (exponential) elimination, various equations were tested to determine the best fits. For the simplest reaction, A → B (A the alcohol and B an unidentified product), the equation d[A]/dt = −k1[A] was used. For sequential A → B → C reactions (B usually a ketone and C not identified), data for [A] and [B] were fitted simultaneously with various kinetic descriptions to find the best fit: irreversible, sequential first order, d[A]/dt = −k1[A] and d[B]/dt = k1[A] − k2[B]; sequential first order with reversible first step and irreversible second step, d[A]/dt = −k1[A] + k2[B] and d[B]/dt = k1[A] − (k−1+ k2)[B]; sequential, irreversible, with Michaelis-Menten kinetics for first step followed by first order reaction for second step, −d[A]/dt = k1[A]/(Km+ [A]) and d[B]/dt = k1[A]/(Km+ [A]) − k2[B]. The fits provided estimates of the rate constants and their standard errors; the average and standard deviation were calculated when three or more animals were used. For two animals, the average and range of values are reported. For single animals, the fitted value and standard error are reported.
The contribution of ADH to the elimination was assessed by administering inhibitors of ADH. A dose of 1 mmole/kg of 4-methylpyrazole (MP) or 10 mmole/kg of isobutyramide (IBA) were given 10 min before the i.p. dose of alcohol. 4-Methylpyrazole is a potent competitive (against alcohol) inhibitor of class I alcohol dehydrogenases, with an in vitro Ki of 0.11 μM for rat liver ADH and an effective, in vivo, competitive Ki value of 1.4 μmole/kg in the rat [8]. A dose of 1 mmole/kg inhibits elimination of a dose of ethanol of 20 mmole/kg by 87%. 4-Methylpyrazole has low toxicity and more specificity than pyrazole [29]. It does not appreciably inhibit rat liver catalase or cytochrome P-450, as the latter has an in vitro Ki of 5.7 mM [30, 31]. Likewise, 10 mmole/kg of isobutyramide, an uncompetitive inhibitor of ADH with an in vivo Ki of 1 mmole/kg, inhibited ethanol elimination comparably [8]. Using higher concentrations of 4-methylpyrazole and isobutryamide does not completely inhibit ethanol elimination, and the uninhibited rate depends on the dose of ethanol, as if the reaction were catalyzed by an enzyme with a Km of 21 mmole/kg and a Vmax of 2.1 mmole/(kg•h). The inhibitor-insensitive rate results from excretion and metabolism by other, unidentified enzymes [8].
3-Amino-1,2,4-triazole (AT) irreversibly inhibits catalase and was used to test the contribution of catalase to the metabolism [32] with a dose of 1 g/kg (12 mmole/kg) given 1 h before the alcohol. 3-Aminotriazole weakly inhibits or stimulates the activity of isolated ADH, depending upon the concentrations of substrates [32, 33]. 3-Aminotriazole modestly (~10%) inhibits ethanol metabolism in rats, but more strongly inhibits (~35%) methanol metabolism [32]. The rate of ethanol elimination observed in the presence of 4-methylpyrazole is not decreased significantly by co-administration of 3-aminotriazole, suggesting that the limiting rate of ethanol elimination is not due to catalase [8].
3. Results
3.1. Substrate specificities of liver alcohol dehydrogenases for oxidation of alcohols
Table 1 provides the kinetic constants for a variety of primary and secondary alcohols. The Km values and catalytic efficiencies (V/KmEt, turnover number per enzyme subunit divided by the Km) for many of these alcohols are similar to those found for horse liver ADH1E and monkey liver ADH1A, which were assayed under the same buffer conditions [34, 35]. Moreover, the catalytic efficiencies are similar to those for human liver ADHs 1A, 1B1, 1C, and 4 (Table 1S, Supplementary Data) even if the assay conditions differ from those used in this study. In contrast, human ADH2 and ADH3 have somewhat different specificities and probably different physiological roles (Table 1S). The class I enzymes have similar hydrophobic amino acid residues in the substrate binding sites, but rat ADH1 differs from human ADH1A in 6 out 9 residues, from ADH1B in 3 of 9, and from ADH1C in 2 of 9 residues [26, 35]. The specificities of the three human ADH1 enzymes may complement one another so that their combined physiological activities are similar to those of horse and rat ADH1 in those animals.
Table 1.
Kinetic constants for oxidation of alcohols by isolated rat liver alcohol dehydrogenase.a
| Alcohol | V relb | Km, mM | Vrel/K | Conc Range, mM |
|---|---|---|---|---|
| ethanol | (1.0) | 0.64 ± 0.22 | 1.6 | 0.4–10 |
| 1-propanol | 0.95 ± 0.07 | 0.22 ± 0.03 | 4.3 | 0.14–14 |
| 1-butanol | 1.3 ± 0.1 | 0.14 ± 0.03 | 9.3 | 0.1–10 |
| 2-methyl-1-propanol | 0.80 ± 0.04 | 0.19 ± 0.04 | 4.2 | 0.1–10 |
| 3-methyl-1-butanol | 0.54 ± 0.02 | 0.042 ± 0.013 | 13 | 0.3–15 |
| benzyl alcohol | 0.47 ± 0.03 | 0.036 ± 0.008 | 13 | 0.023–23c |
| 2-propanol | 0.40 ± 0.02 | 36 ± 7 | 0.011 | 10–200 |
| 2-propanol-d7 | 0.20 ± 0.02 | 83 ± 15 | 0.0024 | 10–200 |
| 2-butanold | 0.48 ± 0.03 | 12 ± 6 | 0.04 | 1.9–48 |
| 3-pentanol | 0.69 ± 0.03 | 4.8 ± 0.03 | 0.14 | 0.43–4.9 |
| cyclopentanol | 0.72 ± 0.04 | 0.32 ± 0.29 | 2.2 | 0.14–4.9 |
| cyclohexanol | 1.0 ± 0.1 | 0.54 ± 0.26 | 1.8 | 0.2–10 |
| 1,2-ethanediol | 0.99 ± 0.25 | 740 ± 120 | 0.0013 | 50–500 |
| 1,3-propanediol | 0.75 ± 0.04 | 24 ± 2 | 0.031 | 9–152 |
| 1,3-butanediol | 0.83 ± 0.10 | 4.4 ± 4.8 | 0.19 | 1.5–51 |
| 1,4-butanediol | 1.5 ± 0.3 | 9.5 ± 0.7 | 0.16 | 2.5–63 |
| 1,5-pentanediol | 3.6 ± 1.3 | 3.5 ± 1.6 | 1.0 | 1–20 |
The kinetic constants were determined in two or three separate experiments and the average values are reported. In each experiment, the data were fitted to the Michaelis-Menten equation, and the standard errors were less than 25% of the values, indicating a good estimation. Some of these constants were previously reported [20].
V rel, is the Vmax relative to ethanol as substrate; the kinetics with ethanol were determined for each batch of purified enzyme and used as the reference for the other substrates. Standard error on the estimation of Vmax with ethanol was about 5%, which was not propagated into the calculation of V rel for the other alcohols. To convert V rel to turnover number per enzyme subunit, multiply V rel by 2.4 s−1, which is based on the complete kinetic study [7].
Substrate inhibition at high concentration, Ki = 27 ± 13 mM
Some specificities should be further explained. Methanol is a very poor substrate for rat liver ADH1, with a Km of 380 mM and a V/Et of 12 min−1 [19], and human ADH1B1 and 1C1 have about 10-fold higher catalytic efficiency (Table 1S). Monkey and human ADH1A are about 10-fold less active on methanol, ethanol, and 1-propanol than the other class I enzymes because of the substitution of Phe-93 with Ala, but are about 10-fold more active on secondary alcohols, such as 2-propanol and cyclohexanol [34]. Human ADH1B is much less active than the other enzymes on cyclohexanol. A combination of the three human ADHs could function like the single rat ADH1, depending upon the concentrations of the enzymes in the liver and the concentrations of substrates.
The reactions of 1,5-pentanediol are complex, with several possible pathways. 1,5-Pentanediol would be oxidized by ADH on one primary hydroxyl group to form 5-hydroxypentanal, which might be further oxidized to produce the 1,5-dialdehyde and two NADHs per 1,5-pentanediol. 5-Hydroxypentanal may also cyclize to form the hemiacetal or be oxidized by ADH or aldehyde dehydrogenases to form 5-hydroxypentanoic acid, which can cyclize to form δ-valerolactone. (The oxidation of aldehydes by ADH has been described previously [36-40].) We studied these reactions using horse liver ADH1E, as this enzyme is more stable than the rat enzyme. Horse liver ADH1E is active on 1,5-pentanediol, with a Km of 21 mM and a Vmax relative to the activity on ethanol of 1.1. Analysis by GC of a reaction with 5 mM 1,5-pentanediol and 1 mM NAD+ at pH 7.0 and 25 °C with 0.01 mg/mL ADH over 14 h did not show a significant increase in 5-hydroxypentanal or δ-valerolactone concentrations, perhaps because the dialdehyde formed. Nevertheless, GC analysis of a reaction of 5 mM 5-hydroxypentanal and 1 mM NAD+ with 0.01 mg/mL of ADH showed that δ-valerolactone and 1,5-pentanediol formed slowly in approximately equal concentrations (~2 mM each at 12 h), in a dismutation reaction that consumed little NAD+.
3.2. Elimination of alcohols in rats by zero order kinetics
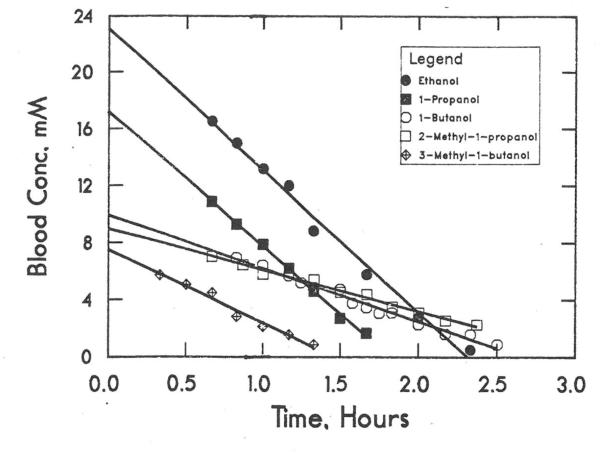
Fig. 1 shows the elimination of primary, aliphatic alcohols in the series from ethanol to 3-methyl-1-butanol. Data points for the initial increase in blood alcohol concentration are not shown, but the kinetics fit zero order kinetics after the absorption phase. Because the Km values for these alcohols are sub-millimolar and the concentrations in the rat are higher than mM, it appears that ADH would be saturated and be acting at maximum velocity. The elimination was substantially inhibited by 4-methylpyrazole and isobutyramide (Table 2), indicating that ADH has a major role in metabolism [8]. Although only one or two animals may have been tested with 4-methylpyrazole and isobutryamide, the extent of inhibition is similar with each inhibitor, supporting the significance of the result. In contrast, 3-aminotriazole did not significantly inhibit elimination, suggesting the catalase is not involved. The aldehydes are then presumably rapidly oxidized by aldehyde dehydrogenases to the carboxylic acids.
Fig. 1.
Elimination of primary alcohols. Doses were 20 mmole/kg ethanol (●), 10 mmole/kg 1-propanol (■), 8 mmole/kg 1-butanol (○), 10 mmole/kg 2-methyl-1-propanol (□), and 7 mmole/kg 3-methyl-1-butanol (◇). Data are for one rat.
Table 2.
Zero-order kinetics of elimination of alcohols in rats.a
| Alcohol | Animals, stateb |
Dose, mmole/kg |
Rate, mmole/kg•h |
r, mL/g |
|---|---|---|---|---|
| Ethanol | 29 | 20,33,43,65 | 7.9 ± 1.0 | 0.79 ± 0.09 |
| 2, MP | 20 | 0.92 ± 0.03 | 0.73 ± 0.01 | |
| 2, IBA | 20 | 1.7 ± 0.2 | 0.73 ± 0.01 | |
| 3, fasted | 43 | 5.7 ± 0.1 | 0.70 ± 0.01 | |
| 1-Propanol | 4 | 10 | 5.3 ± 0.5 | 0.67 ± 0.09 |
| 3, MP | 10 | 1.0 ± 0.2 | 0.72 ± 0.11 | |
| 3, IBA | 10 | 1.2 ± 0.2 | 0.71 ± 0.09 | |
| 3, AT | 10 | 5.0 ± 0.5 | 0.77 ± 0.20 | |
| 1-Butanol | 3 | 8 | 3.2 ± 0.4 | 0.82 ± 0.09 |
| 3, MP | 8 | 1.4 ± 0.1 | 0.90 ± 0.20 | |
| 1, IBA | 8 | 1.4 | 1.1 | |
| 2, AT | 8 | 4.0 ± 0.4 | 1.2 ± 0.2 | |
| 2-Methyl-1-propanol | 3 | 10 | 3.8 ± 0.6 | 1.1 ± 0.04 |
| 3, MP | 10 | 1.3 ± 0.2 | 0.92 ± 0.09 | |
| 1, IBA | 10 | 1.5 | 0.99 | |
| 2, AT | 10 | 4.5 ± 0.1 | 0.95 ± 0.13 | |
| 3-Methyl-1-butanol | 5 | 5, 7 | 4.6 ± 0.8 | 1.2 ± 0.2 |
| 2, MP | 5 | 1.8 ± 0.4 | 1.1 ± 0.1 | |
| 1, IBA | 5 | 1.5 | 1.0 | |
| 1, AT | 5 | 3.9 | 1.3 | |
| 1,3-Propanediol | 3 | 10 | 2.7 ± 0.8 | 0.9 ± 0.5 |
| 3, MP | 10 | 1.0 ± 0.2 | 0.6 ± 0.3 | |
| 1, IBA | 10 | 1.3 | 0.43 | |
| 1,3-Butanediol | 3 | 10 | 5.0 ± 0.3 | 0.82 ± 0.05 |
| 1, MP | 10 | 0.77 | 0.69 | |
| 1, IBA | 10 | 1.1 | 0.70 | |
| 1, AT | 10 | 4.9 | 0.98 | |
| 1,4-Butanediol | 2 | 12 | 6.7 ± 0.1 | 1.3 ± 0.2 |
| 2, MP | 12 | 0.95 ± 0.32 | 1.1 ± 0.1 | |
| 1, IBA | 12 | 1.2 | 0.68 | |
| 1, AT | 12 | 6.5 | 1.6 | |
| 1,5-Pentanediol | 5 | 10 | 5.0 ± 0.7 | 1.1 ± 0.2 |
| 2, MP | 10 | 1.05 ± 0.05 | 1.0 ± 0.3 | |
| 1, IBA | 10 | 1.5 | 0.87 | |
| 1, AT | 10 | 6.2 | 0.93 | |
| 1,2-Ethanediol | 2 | 50 | 5.6 ± 0.3 | 0.72 ± 0.08 |
| 2, MP | 50 | 5.0 ± 0.2 | 0.68 ± 0.06 | |
| 2, IBA | 50 | 5.2 ± 0.7 | 0.68 ± 0.06 | |
| 2, AT | 50 | 4.3 ± 0.3 | 0.76 ± 0.11 | |
| 2,3-Butanediol | 2 | 10 | 0.92 ± 0.08 | 0.89 ± 0.04 |
| 1, MP | 10 | 0.84 | 0.85 | |
| 1, AT | 10 | 0.86 | 0.93 | |
| Methanol | 2 | 10 | 0.64 ± 0.02 | 0.89 ± 0.04 |
| 2, MP | 10 | 0.56 ± 0.01 | 0.85 ± 0.05 | |
| 1, IBA | 10 | 0.62 | 0.79 | |
| 2, AT | 10 | 0.40 ± 0.05 | 0.82 ± 0.04 | |
| 1, MP+AT | 10 | 0.41 | 0.83 | |
| tert-Butanol | 2 | 7 | 0.39 ± 0.02 | 0.66 ± 0.01 |
| (2-Methyl-2-propanol) | 1,MP | 7 | 0.33 | 0.63 |
| 1, AT | 7 | 0.35 | 0.63 |
Rats (usually fed; fasted animals had no food for 24 h) were given the indicated doses of alcohol (i.p., diluted in saline) and the blood alcohol concentration was followed over time. Linear regression was used to calculate the parameters for the zero-order (straight line) fit to the data. The rates were calculated by dividing the dose given by the time required to completely eliminate the alcohol. The r value (fraction of body weight that is “water”, accessible to the alcohol or ketone) was calculated as the ratio of fitted concentration of alcohol in the blood at zero time divided by the dose given.
The number of animals, fed state usually (fasted animals had no food for 24 h), and treatments with inhibitors are given.
Elimination of ethanol is slower in fasted rats, consistent with the reduced content of liver ADH [41], and catalase may contribute in this state [42].
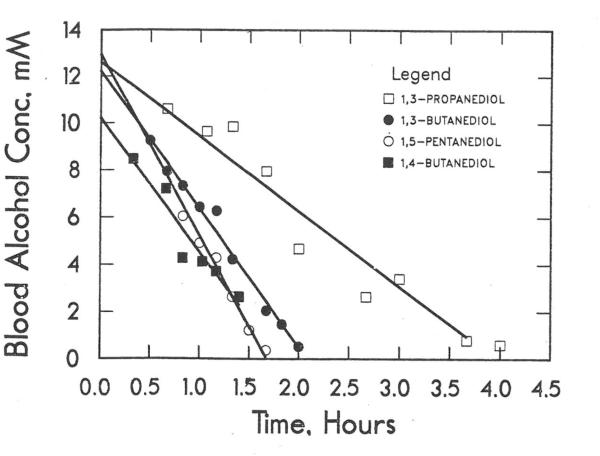
Diols with a primary hydroxyl group, but not vicinal diols, are also metabolized readily, with zero-order kinetics and involvement of ADH as demonstrated by significant inhibition by 4-methylpyrazole and isobutyramide (Fig. 2, Table 2). It is not clear why the alcohols are eliminated with zero-order kinetics, as the Km values from in vitro studies are comparable to, or larger than, the concentrations observed in the rat.
Fig. 2.
Elimination of diols with a primary hydroxyl group. All doses were 10 mmole/kg: 1,3-propanediol (□), 1,3-butanediol (●), 1,4-butanediol (■), 1,5-pentanediol (○).Data are for one rat.
1,3-Butanediol is metabolized almost quantitatively to β-hydroxybutyrate and acetoacetate [43]. 1,4-Butanediol is oxidized to γ-hydroxybutyrate, which can cyclize to γ-butyrolactone or be further oxidized to succinate [44].
1,5-Pentanediol is oxidized on one primary hydroxyl group to form 5-hydroxypentanal, and then is apparently oxidized by aldehyde dehydrogenases to form 5-hydroxypentanoic acid, which cyclizes to form the δ-valerolactone. With a dose of 10 mmole/kg 1,5-pentanediol, δ-valerolactone built up in the blood to levels of about 5 mM in one to two hours before clearing by six hours. It appears that aldehyde dehydrogenases are involved because the oxidation of 5-hydroxypentanal by ADH to δ-valerolactone in vitro is relatively slow.
The elimination of 1,2-ethanediol (ethylene glycol) is relatively rapid, but the inhibitors of ADH only slightly decreased the rate, whereas 3-aminotriazole had a larger effect, presumably because of inhibition of catalase. These results would suggest that ADH is not a major contributor to 1,2-ethanediol elimination. Excretion in the urine is significant [45], and other enzyme activities may contribute to metabolism. The low catalytic efficiency of ADHs on 1,2-ethanediol also indicates that ADH could only contribute slowly to the metabolism (Table 1, Table 1S). Nevertheless, even the slow oxidation of 1,2-ethanediol by ADH to glycoaldehyde and subsequent oxidation to glycolic acid causes toxic acidosis in primates [46, 47], which can be effectively inhibited by 4-methylpyrazole (Fomepizole) [48] and reduce mortality, in rats [45].
The elimination of 2,3-butanediol was relatively slow and was not inhibited by 4-methylpyrazole, suggesting that ADH is not involved. Previous studies showed that the 2R,3R and meso isomers were oxidized to R-acetoin and then cleaved to acetate [49].
The elimination of methanol was very slow, requiring 15 h to eliminate a dose of 10 mmole/kg (by strictly zero-order kinetics) and was only slightly inhibited by 4-methylpyrazole. Aminotriazole inhibited somewhat better, suggesting a role for catalase [32]. Methanol is eliminated significantly through lungs and kidneys [50]. Nevertheless, methanol is a substrate for rat liver alcohol dehydrogenase, with a Km of 340 mM and a Vmax relative to ethanol of 0.10 [32]. ADH oxidizes methanol to formaldehyde, which is rapidly oxidized by ADH3 to formic acid, which is slowly metabolized (especially in primates [51]) to CO2 by a folate-dependent pathway and builds up to produce metabolic acidosis, ocular toxicity, and potentially death in humans and monkeys [47]. Rodents metabolize formic acid more rapidly than humans and are not poisoned by doses of methanol that are toxic to humans [50]. As with 1,2-ethanediol, 4-methylpyrazole is useful for treatment of methanol toxicity in humans and in principle would be better to use than ethanol as a competitive inhibitor of ADH [47, 52, 53]. Selective uncompetitive inhibitors, such as isobutyramide, formamides and sulfoxides [54], offer an alternative, potentially more effective therapy, but studies in humans are required.
Tert-butanol cannot be oxidized by ADH, but its elimination serves as a reference for other metabolic and excretion pathways. As shown in Table 2, this alcohol is eliminated somewhat more slowly than methanol, and inhibitors of ADH or catalase did not substantially decrease the rate of elimination. Apparently tert-butanol is metabolized to 2-methyl-1,2-propanediol and subsequently oxidized to 2-hydroxy-iso-butyrate [55], perhaps by enzymes that also slowly act on ethanol.
3.3. Elimination of alcohols in rats by first order kinetics
Secondary alcohols typically are eliminated by a first order process that leads to the ketones, which form and are slowly metabolized to products (probably hydroxylated) that were not determined in the present studies. The exponential decay can also be described by the Michaelis-Menten equation when the apparent Km value is larger than the alcohol concentration because the equation then reduces to v = V[A]/Km, which is equivalent to −d[A]/dt = k[A]. Thus, elimination of 2-propanol (10, 17, and 30 mmole/kg doses) can be equally well described by simultaneous fitting of the progress curves to a first order process with k = 49 (± 1) × 10−4min−1(R2= 0.986) or the Michaelis-Menten equation with Vmax = 0.38 ± 0.10 mM/min and Km = 58 ± 20 mM (R2 = 0.981). (Similar Km values were determined previously in vivo and in vitro [5] and in this study (Table 1).) Because the single parameter is sufficient to describe the result, our choice is to fit the data for the secondary alcohols as first order, as described in Table 3.
Table 3.
First order kinetics of elimination of alcohols and ketones in rats.a
|
| ||||||
|---|---|---|---|---|---|---|
| Alcohol/ketone | Animals Stateb |
Dose, mmole/kg |
104
k1, min−1 |
104
k2, min−1 |
k2, mmole /kg•h |
r, mL/g |
| 2-Propanol | 7 | 10, 17, 30 | 49 ± 5 | 18 ± 2 | 0.84 ± 0.16 | 0.78 ± 0.07 |
| 1, MP | 10 | 22.6 ± 0.3 | 23.7 ± 1.1 | 0.33 | 0.75 | |
| 1, IBA | 10 | 31.7 ± 0.4 | 21.3 ± 1.0 | 0.44 | 0.76 | |
| 1, AT | 10 | 51.6 ± 0.5 | 27.3 ± 0.8 | 0.68 | 0.77 | |
| 2, fasted | 17 | 42 ± 4 | 14.3 ± 0.6 | 0.64 ± 0.02 | 0.75 ± 0.01 | |
| Acetone | 3 | 17, 34, 70 | -------- | 12 ± 4 | 1st orderc | 0.80 ± 0.05 |
| 2-Propanol-d7 | 9 | 15, 30 | 25 ± 2 | 19 ± 4 | 0.59 ± 0.08 | 0.74 ± 0.04 |
| 4, fasted | 15 | 18 ± 2 | 13 ± 2 | 0.44 ± 0.06 | 0.70 ± 0.03 | |
| 2-Butanol | 3 | 10 | 112 ± 17 | 37 ± 4 | 1.1 ± 0.2 | 0.74 ± 0.08 |
| 2, MP | 10 | 35 ± 1 | 55 ± 4 | 0.8 ± 0.1 | 0.73 ± 0.05 | |
| 2, IBA | 10 | 45 ± 1 | 56 ± 25 | 0.9 ± 0.3 | 0.75 ± 0.08 | |
| 3, AT | 10 | 110 ± 33 | 38 ± 5 | 1.2 ± 0.2 | 0.72 ± 0.04 | |
| 2-Butanone | 1 | 10 | -------- | 31 ± 2 | 1.3 | 0.81 |
| 3-Pentanol | 3 | 5 | 220 ± 50 | 51 ± 13 | 0.72 ± 0.24 | 0.71 ± 0.06 |
| 1, MP | 5 | 21.0 ± 0.4 | 62 ± 3 | 0.26 | 0.78 | |
| 1, AT | 5 | 220 ± 5 | 52 ± 2 | 0.92 | 0.88 | |
| 3-Pentanone | 1 | 5 | -------- | 59 ± 2 | 1.0 | 0.64 |
| 1, MP | 5 | -------- | 56 ± 2 | 0.96 | 0.66 | |
| Cyclopentanol | 4 | 5 | 280 ± 130 | 17 ± 9 | 0.30 ± 0.22 | 0.82 ± 0.08 |
| 2, 4MP | 5 | 68 ± 1 | 20 ± 8 | ---------- | 0.81 ± 0.05 | |
| 1, AT | 5 | 206 ± 6 | 5 ± 1 | 0.25 | 0.86 | |
| Cyclopentanone | 1 | 5 | -------- | 18 ± 0.4 | 0.37 | 0.72 |
| Cyclohexanol | 5 | 5 | 229 ± 70 | 50 ± 6 | 98 ± 46d | 0.75 ± 0.21 |
| 1, MP | 5 | 46 ± 1 | 380 ± 30 | Not fit | 0.68 | |
| 1, IBA | 5 | 59 ± 1 | 336 ± 33 | Not fit | 0.64 | |
| 1, AT | 5 | 142 ± 5 | 61 ± 1 | 56 ± 4d | 0.83 | |
| Cyclohexanone | 1 | 5 | 176 ± 13 | 67 ± 3 | 190 ± 22 | 0.53 |
| Benzyl alcohole | 2 | 4 | 560 ± 70 | -------- | -------- | 0.82 ± 0.07 |
| 2, MP | 4 | 150 ± 30 | -------- | -------- | 1.05 ± 0.06 | |
| 1, IBA | 4 | 340 ± 20 | -------- | -------- | 1.0 | |
| 1, AT | 4 | 300 ± 20 | -------- | -------- | 1.1 | |
The elimination data for each animal were fitted to the sequential mechanism for irreversible first order reactions (k1, k2) or an irreversible first order reaction (k1) followed by an irreversible zero order reaction (k2), which gave essentially equally good fits to the data. For cyclohexanol and cyclohexanone, the first step was reversible and the rate constant, k−1, was calculated instead of the zero order reaction. The r value was calculated as the ratio of the concentration of alcohol at zero time from the best fit to the differential equations divided by the dose given.
Number of animals, fed state usually (fasted animals had no food for 24 h) and inhibitors used.
The three (increasing) doses were eliminated (to 55, 67, 75% remaining, respectively) over 5.5 h with first order rate constants of 1.66 × 10−3 min−1, 1.15 × 10−3 min−1, and 0.87 × 10−3 min−1 , and thus the reaction is not strictly first order, but the estimated zero order rate constants also depended on the dose. The elimination is best described as first order, but previous studies show that a significant fraction (varies with dose) of acetone is excreted in breath and urine, thus yielding more complex kinetics [77]. Very small levels of 2-propanol (about 2% of the acetone levels) were also formed, but the reduction is negligible.
Fitted to reversible sequential mechanism with k−1, 104•min−1
Fitted to simple first order reaction, as no benzaldehyde was observed, and the data did not fit a zero order reaction.
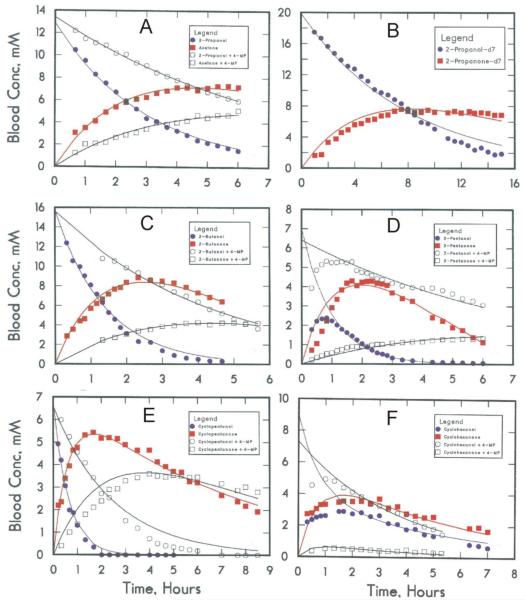
Metabolism of 2-propanol to acetone is somewhat more rapid than the subsequent elimination of acetone, and acetone levels became substantial (Fig. 3A). The rate of elimination of acetone was confirmed in separate experiments with three different doses of acetone. Acetone is very slowly reduced to 2-propanol, in accord with in vitro studies [56]. The elimination of 2-propanol is significantly inhibited (see Fig. 3A and k1 in Table 3) by 4-methylpyrazole and isobutyramide, but not by aminotriazole, suggesting a major role for ADH and not for catalase, consistent with previous studies [57]. Since acetone is hydroxylated and then reduced to 1,2-propanediol, the redox state of the liver is not altered substantially by 2-propanol metabolism [57]. Fully deuterated 2-propanol is eliminated at one-half the rate of protio 2-propanol (Fig. 3B, Table 3), consistent with the isotope effect for hydride transfer shown by the purified enzyme (Table 1). The in vitro oxidation and in vivo elimination kinetics of protio and deuterio 2-propanol in rats were previously analyzed by fitting the data to the Michaelis Menten equation, with the similar conclusion that ADH is a major limiting factor for 2-propanol metabolism [5]. An earlier study with mice also showed an isotope effect of 2.0 for in vivo metabolism of 2-propanol, but no isotope effect for metabolism of ethanol, because the rate-limiting step for the oxidation of ethanol with ADH is release of the product NADH [58].
Fig. 3.
Elimination of secondary alcohols. In each sub-figure, the concentration of alcohol is represented by filled circle (●), its corresponding ketone by a filled square (■), and the results when another rat was given 1 mmole/kg of 4-methylpyrazole are represented by the open symbols, alcohol (○) and ketone (□). (A) 2-propanol, 10 mmole/kg; (B) 2-propanol-d7, 15 mmole/kg; (C) 2-butanol (racemic mixture of isomers), 10 mmole/kg; (D) 3-pentanol, 5 mmole/kg; (E) cyclopentanol, 5 mmole/kg; (F) cyclohexanol, 5 mmole.kg. The lines represent the fitted values from simultaneous non-linear least squares fits of data for alcohol and ketone for the uninhibited (or inhibited) state to the differential equations for the first order, sequential reactions.
Our present study extends the series of alcohols to larger secondary alcohols (2-butanol, 3-pentanol, cyclopentanol and cyclohexanol), which are all eliminated with similar kinetics (Fig. 3C–F) and with relatively fast rates (Table 3), in accord with the good activity of ADH on these substrates (Table 1). The kinetics for cyclohexanol elimination are especially interesting because a quasi-equilibrium for [cyclohexanone]/[cyclohexanol] is rapidly reached during the steady-state elimination (Fig. 3F). When cyclohexanone is administered, it is also rapidly reduced to cyclohexanol, and the kinetic constants for the first order reactions are about the same as those determined for cyclohexanol elimination (Table 3). Moreover, simultaneous administration of cyclohexanol and ethanol shifts the equilibrium to greatly favor cyclohexanol, because of the effect on the redox state [9]. Ethanol metabolism can alter the metabolism of other alcohols and carbonyl compounds.
Benzyl alcohol is a primary alcohol, oxidized by ADH with a low Km, but it is eliminated, relatively rapidly, with first-order kinetics. The elimination is inhibited by 4-methylpyrazole and isobutyramide, but also by 3-aminotriazole, from which we conclude that ADH may be only one enzyme involved in the elimination. Perhaps the elimination is first-order because the other enzymatic systems that are involved have higher Kmvalues.
3.4. Correlation of activities of ADH and rates of elimination in rats
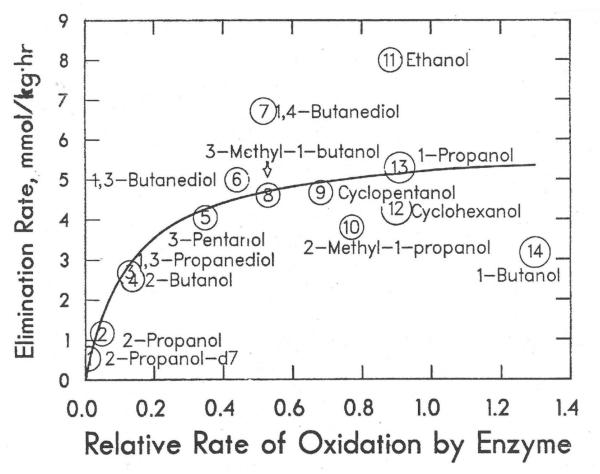
Table 4 compares the relative rates calculated with the Michaelis-Menten equation for the oxidation of the alcohols by ADH in vitro at a concentration of 5 mM with the rates of elimination in vivo that were observed or calculated for 5 mM concentrations. This concentration was chosen because such a concentration was observed in vivo at some point during the elimination in rats. (Choosing 10 mM for the calculation gives somewhat different results, but does not change the overall conclusions.) The contribution of ADH to the elimination, based on the extent of inhibition of the rate by 4-methylpyrazole (from Tables 2 and 3), is also listed. The correlation for 14 alcohols is shown in Fig. 4. (Benzyl alcohol, 1,2-ethanediol, 2,3-butanediol, and methanol are not included in the correlation because ADH is not the major contributor to their elimination. 1,5-Pentanediol is not included because the in vitro results may have overestimated the Vmaxdue to production of 2 NADH molecules per diol.) The data were fitted to the equation, rate = αk1• k2/(αk1+ k2), where k1is the relative rate for oxidation by ADH in vitro, α is a proportionality constant to adjust the relative rate with ADH to the rates in vivo, and k2is the limiting rate when αk1becomes large. The proportionality constant has units of mmoles/(kg•h•v-rel), and it represents the initial slope of the fit. This equation describes an irreversible, steady-state reaction, such as Alcohol → × → Y, where × is a carbonyl compound or NADH, and the second step could be various reactions. The fit is reasonable, yielding values of α = 43 (± 25) and k2= 6.0 (± 1.0) mmole/kg•h, with R2= 0.93, but there is considerable scatter. (For comparison, when the data are fitted to a straight line, the slope, equivalent to the proportionality factor α, is 6.0 (± 1.0), with R2 = 0.78, indicating a weak relationship.) The errors of the rate measurements have not been propagated. Although 2 or 3 determinations were made for each in vitro or in vivo parameter (and errors reported), a few values have errors of about ± 50%, and this would tend to disturb the correlation. Nevertheless, when 14 alcohols are used, there are sufficient data in the pattern to show that there is some overall limitation to the maximum rate of elimination.
Table 4.
Correlation of alcohol dehydrogenase activity and elimination of alcohols in rats
| Alcohol | in vitroa
v rel |
in vivob
mmole/kg/h |
Rate due to ADHc |
%ADHc |
|---|---|---|---|---|
| Ethanol | 0.88 | 7.9 ± 1.0 | 7.0 | 89 |
| 1-Propanol | 0.91 | 5.3 ± 0.5 | 4.3 | 81 |
| 1-Butanol | 1.3 | 3.2 ± 0.4 | 1.8 | 56 |
| 2-Methyl-1-propanol | 0.77 | 3.8 ± 0.5 | 2.5 | 66 |
| 2-Methyl-1-butanol | 0.53 | 4.6 ± 0.8 | 2.8 | 61 |
| 2-Propanol | 0.049 | 1.1 ± 0.1 | 0.6 | 55 |
| 2-Propanol-d7 | 0.011 | 0.55 ± 0.04 | N.D. | N.D. |
| 2-Butanol | 0.14 | 2.5 ± 0.4 | 1.7 | 68 |
| 3-Pentanol | 0.35 | 4.0 ± 1.0 | 3.6 | 90 |
| Cyclopentanol | 0.68 | 4.7 ± 3.2 | 3.0 | 64 |
| Cyclohexanol | 0.90 | 4.3 ± 1.5 | 3.2 | 74 |
| 1,3-Propanediol | 0.13 | 2.7 ± 0.8 | 1.7 | 63 |
| 1,3-Butanediol | 0.44 | 5.0 ± 0.3 | 4.1 | 82 |
| 1,4-Butanediol | 0.52 | 6.7 ± 0.1 | 5.8 | 86 |
Calculated from kinetic constants, v rel = Vrel(5)/Km + 5) for 5 mM alcohol.
Data for zero order elimination; or for first order elimination, where rate = k1(min−1 • (5 mmole/L)•r(L/kg)•60min/h).
Estimated from the observed rate without inhibitors minus the rate with 1 mmole/kg 4-methylpyrazole. “%ADH” is the estimated percentage of elimination due to ADH.
Fig. 4.
Correlation of rates of elimination of alcohols in rats with the rates of oxidation by isolated rat liver alcohol dehydrogenase. Rates are calculated for 5 mM alcohol, as shown in Table 4. The circles represent the data points, and the enclosed number is for the alcohol, which is also labeled.
The limiting rate for ethanol elimination does not appear to be due to aldehyde dehydrogenases, because acetaldehyde is eliminated rapidly, and acetaldehyde concentrations are relatively low after administration of ethanol [6]. NADH oxidation may be partially rate-limiting, especially for primary alcohols where the aldehyde dehydrogenases rapidly produce the corresponding acid and a second NADH. However, the secondary alcohols are typically oxidized by using one NAD+ in the first step, and the relative rates for the secondary alcohols fit the same trend in Fig. 4 as the primary alcohols. If only the six secondary alcohols are used for the analysis of the correlation, the fit to the sequential reaction provides α = 36 ± 7 and a limiting rate for k2= 5.4 (± 0.7) mmole/kg•h. Thus, there is still some factor (or factors) that limits the elimination. Because the limiting rate is about the same for primary and secondary alcohols, we would propose that NADH oxidation is not the limiting factor for elimination of these alcohols.
The data in Fig. 4 could also be interpreted as if the elimination of the alcohols (2-propanol, 2-butanol, and 1,3-propandiol) with the slowest rate of oxidation by ADH is limited by the activity of ADH, whereas the elimination of the ten alcohols with highest activity with ADH is limited by some other common factor. The average rate of elimination of these ten alcohols is 5.0 mmole/kg•h. The factors contributing to this limiting rate still need to be identified.
4. Discussion
4.1. Substrate specificities of ADH and endogenous substrates
The rat liver ADH is active on a variety of alcohols, with Kmvalues and relative catalytic efficiencies (V/Km) that are similar to those for the homologous horse liver ADH1E and mouse liver ADH1 [34, 35, 40]. Mouse liver ADH1 activity was determined on 28 alcohols and carbonyl compounds. The human ADHs have similar broad specificities (Table 1S). The specificities are generally consistent with the similar, hydrophobic substrate binding sites, although amino acid residues in the sites are unique for each enzyme. Three-dimensional structures for the horse and human ADHs have been determined, and we expect that the rat and mouse liver ADHs are similar. It remains for future studies to correlate substrate specificities with enzyme structure.
Because ADHs can oxidize many alcohols or reduce carbonyl compounds that are found in tissues of animals, defining the physiological roles of ADHs is of continuing interest [59]. ADH substrates may be ingested or be produced from various metabolic pathways (e.g., [60]), and the co-metabolism with ingested ethanol may cause alcoholic pathologies. Classic studies show that ethanol affects the metabolism of various alcohols [61, 62]. ADHs also oxidize 20-hydroxyeicosatetraenoic acid, which produces vasoconstriction in cerebral circulation, to the inactive dicarboxylic acid [39, 40]. This activity of ADH may be inhibited by ingested ethanol and thus cause hypertension.
Metabolism of other alcohols and carbonyl compounds may be physiologically important, but requires further investigation. Volatile compounds found in human serum include many potential substrates or products of ADH reactions: ethanol/acetaldehyde, 1-propanol/1-propanal, 2-propanol/acetone, 1-butanol/1-butanal, 2-butanol/2-butanone, 2-butenal, 2-methylpropanol/2-methylpropanal, 1-pentanol/1-pentanal, 2-methylbutanol/2-methylbutanal, 3-methylbutanol/3-methylbutanal, trans-2-methyl-2-butenal, 2-pentanone, 3-pentanone, 3-penten-2-one, 1-hexanol/1-hexanal, 2-hexanol/2-hexanone, 4-methyl-2-pentanone, 2-heptanone, 3-heptanone, 4-heptanone, octanal, 2-octanol/2-octanone, 4-octanone, 6-methyl-2-heptanone, 5-methyl-3-heptanone, nonanal, 2-nonenal, n-nona-2,4-dienal, 5-nonanone, 1-decanal, 1-undecanal, furfural, cyclohexanone, benzaldehyde, and 6-methyl-5-acetophenone [63-68]. Which substrates are metabolized by ADHs is thus of interest. Our study shows that 10 of these compounds are substrates or products of ADH activities in rats, and most of the rest probably are also because of the broad substrate specificities of the ADHs.
4.2. Contribution of ADH to the metabolism of alcohols in rats
The results show that ADH is a major contributor to the elimination of the alcohols listed in Table 4. This conclusion is based on the observation that 4-methylpyrazole and isobutyramide, which are potent inhibitors of ADH, significantly inhibited the elimination of the alcohols, whereas 3-aminotriazole did not. Inhibitors are not usually totally specific, however, and quantitative studies with other inhibitors are required to identify other enzymes that may be involved.
Liver ADHs have low activity on methanol and 1,2-ethanediol, even though metabolism of these alcohols by ADH leads to toxic metabolites. 2,3-Butanediol and 2-methyl-2-propanol are not substrates for ADH, and their elimination occurs by other pathways.
4.3. Contribution of other enzymes to the metabolism of alcohols in animals
The “microsomal ethanol oxidizing system” or NADPH-dependent cytochrome P-450 enzymes oxidize methanol, ethanol, propanol and butanol with Kmvalues of 22, 9.6, 5.5 and 4.9 mM, respectively, and could contribute 20–25% to the rate of ethanol metabolism [69, 70]. In deer mice lacking liver ADH, it was concluded that the activity of cytochrome P-450 was a minor contributor to ethanol metabolism, and that catalase-H2O2and non-hepatic ADHs were more involved [71]. Cytochrome P-450(CYP)2E1 is induced in animals by chronic treatment with ethanol, but knocking out CYP2E1 in mice did not significantly affect the rate of elimination of ethanol [72]. Although 4-methylpyrazole inhibits CYP2E1 [73, 74], it appears that the capacity of the P-450 system is relatively small.
4.4. Correlation of ADH activity and rates of elimination of alcohols in rats
A focus of this work was to determine if ADH activity is rate-limiting in the metabolism of the alcohols that are substrates for ADHs. The results in Table 4 and Fig. 4 show a relationship between the activity of rat liver ADH and the rate of elimination of the various alcohols. The rate of elimination reaches a maximum of about 6 mmole/kg•h for the 14 different alcohols, but there is considerable scatter, as ethanol is eliminated at a faster rate of 7.9 mmole/kg•h and 1-butanol at the slower rate of 3.2 mmole/kg•h. That there is a limiting rate might be taken as evidence for some rate limitation for reoxidation of NADH via oxidative phosphorylation, which has been investigated with rat liver cells and slices, producing differing conclusions [4, 12, 75]. The fact that the redox state becomes 2–3-fold more reduced during ethanol oxidation clearly indicates that NADH reoxidation is partially rate-limiting for the overall rate. During ethanol metabolism in the rat, acetaldehyde levels also increase – to variable extents – indicating partial rate limitation for aldehyde dehydrogenase [6]. In our calculations of ADH activity (for Table 4), we only used the initial velocity for alcohol oxidation, without consideration of the full kinetic equation that includes product inhibition by NADH or the carbonyl compound. Such calculations for rat ADH suggest that for ethanol oxidation, product inhibition might decrease the rate of ethanol metabolism by about 45%, but ADH activity would still be the major rate-limiting factor for ethanol oxidation [4, 7].
In principle, the reduction of the carbonyl products of ADH1 activity by medium and short chain alcohol dehydrogenases and aldo-keto reductases [76] could decrease the overall net rate of metabolism by reversing the oxidation of the alcohols. Reduction of the aldehydes back to the alcohols probably is not very significant because the aldehyde dehydrogenases are very active, and the aldehyde levels are low. Furthermore, our experiments show that the ketones are not being reduced in vivo because the corresponding alcohols are not detected at significant levels in the blood when the ketones are administered. An exception is cyclohexanone, which is illustrative because it is reduced to cyclohexanol in vivo, and the kinetics of elimination of cyclohexanol involves the reversible dehydrogenation. Nevertheless, as can be shown by simulations, the rate calculated for the initial phase of elimination is almost the same whether the reduction of cyclohexanone is included or not. As noted above, the kinetics of elimination of the secondary alcohols, which produce just one NADH for the first step, also show a limiting rate of elimination.
Other factors could influence the relationship shown in Fig. 4. Enzyme systems, such as cytochrome P-450 (CYP)2E1, could be contributing to the elimination. The quantitative contribution of CYP2E1 for metabolism of various alcohols needs to be studied. A criterion for including the data in Fig. 4 was that the rate of elimination of the alcohol should be inhibited by at least 50% by 4-methylpyrazole, but the uninhibited rates could be due to CYP2E1 or other activities. The alcohols are probably also excreted at different rates. The alcohols may be distributed to different extents in fat, muscle, blood, etc., although the body water parameters, r, are similar for the various alcohols. Further studies are required to evaluate these factors.
A major conclusion of this study is that rat liver ADH contributes significantly to the metabolism of a broad variety of alcohols. Because the specificities of the various Class I human ADHs are similar in an aggregate, complementary manner to that of rat ADH1, we suggest that metabolism of these alcohols in humans and rats would be similar. Whether the metabolism of particular alcohols by ADHs, and co-metabolism with ethanol, has biological effects remains an important question for further studies.
Supplementary Material
Highlights for Plapp, et al.
Rat liver alcohol dehydrogenase is active on a wide variety of alcohols. Rats eliminate primary alcohols and diols with zero order kinetics.
Rats metabolize secondary alcohols to the ketones with first order kinetics.
Alcohol dehydrogenase is a major contributor to metabolism of various alcohols.
Elimination of alcohols is limited by alcohol dehydrogenase and unknown factors.
Acknowledgments
This work was supported by the U. S. Public Health Service, National Institute on Alcohol Abuse and Alcoholism, Grant AA00279.
Abbreviations
- ADH
alcohol dehydrogenase
- MP
4-methylpyrazole
- IBA
isobutyramide
- AT
3-amino-1,2,4-triazole
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest statement
The authors declare that there are no conflicts of interest.
Appendix A. Supplementary data
A table comparing substrate specificities of horse, rat, and human alcohol dehydrogenases can be found in the on-line version, at http://dx.doi.org/10.1016/j.cbi.2014.12.040.
References
- [1].Jörnvall H, Hedlund J, Bergman T, Kallberg Y, Cederlund E, Persson B. Origin and evolution of medium chain alcohol dehydrogenases. Chem. Biol. Interact. 2013;202:91–96. doi: 10.1016/j.cbi.2012.11.008. [DOI] [PubMed] [Google Scholar]
- [2].Plapp BV. Rate-limiting steps in ethanol metabolism and approaches to changing these rates biochemically. In: Majchrowicz E, editor. Biochemical Pharmacology of Ethanol. Vol. 56. Plenum; New York: 1975. pp. 77–109. Adv. Exp. Med. Biol. [DOI] [PubMed] [Google Scholar]
- [3].Havre P, Abrams MA, Corrall RJM, Yu LC, Szczepanik PA, Feldman HB, Klein P, Kong MS, Margolis JM, Landau BR. Quantitation of pathways of ethanol metabolism. Arch. Biochem. Biophys. 1977;182:14–23. doi: 10.1016/0003-9861(77)90278-8. [DOI] [PubMed] [Google Scholar]
- [4].Cornell NW, Crow KE, Leadbetter MG, Veech RL, Li T-K, Schenker S, Lumeng L. Alcohol and Nutrition. U.S. Government Printing Office; Washington,D.C.: 1979. Rate determining factors for ethanol oxidation in rats in vivo and in isolated rat hepatocytes; pp. 315–330. [Google Scholar]
- [5].Chen WS, Plapp BV. Kinetics and control of alcohol oxidation in rats. Adv. Exp. Med. Biol. 1980;132:543–549. doi: 10.1007/978-1-4757-1419-7_56. [DOI] [PubMed] [Google Scholar]
- [6].Braggins TJ, Crow KE. The effects of high ethanol doses on rates of ethanol oxidation in rats. A reassessment of factors controlling rates of ethanol oxidation in vivo. Eur. J. Biochem. 1981;119:633–640. doi: 10.1111/j.1432-1033.1981.tb05654.x. [DOI] [PubMed] [Google Scholar]
- [7].Crabb DW, Bosron WF, Li T-K. Steady-state kinetic properties of purified rat liver alcohol dehydrogenase: Application to predicting alcohol elimination rates in vivo. Arch. Biochem. Biophys. 1983;224:299–309. doi: 10.1016/0003-9861(83)90213-8. [DOI] [PubMed] [Google Scholar]
- [8].Plapp BV, Leidal KG, Smith RK, Murch BP. Kinetics of inhibition of ethanol metabolism in rats and the rate-limiting role of alcohol dehydrogenase. Arch. Biochem. Biophys. 1984;230:30–38. doi: 10.1016/0003-9861(84)90083-3. [DOI] [PubMed] [Google Scholar]
- [9].Plapp BV. Control of alcohol metabolism. In: Jansson HJB, Rydberg U, Terenius L, Vallee BL, editors. Toward a Molecular Basis of Alcohol Use and Abuse. Birkhäuser Verlag; Basel: 1994. pp. 311–322. [Google Scholar]
- [10].Lee SL, Chau GY, Yao CT, Wu CW, Yin SJ. Functional assessment of human alcohol dehydrogenase family in ethanol metabolism: Significance of first-pass metabolism. Alcohol. Clin. Exp. Res. 2006;30:1132–1142. doi: 10.1111/j.1530-0277.2006.00139.x. [DOI] [PubMed] [Google Scholar]
- [11].Yin S-J, Lee S-L, Yao C-T, Lai C-L. Functional roles of alcohol dehydrogenases in human ethanol metabolism. In: Weiner H, Lindahl R, Plapp B, editors. Enzymology and Molecular Biology of Carbonyl Metabolism 13. Purdue University Press; West Lafayette, IN: 2007. pp. 134–143. [Google Scholar]
- [12].Cederbaum AI, Dicker E, Rubin E. Transfer and reoxidation of reducing equivalents as the rate-limiting steps in the oxidation of ethanol by liver cells isolated from fed and fasted rats. Arch. Biochem. Biophys. 1977;183:638–646. doi: 10.1016/0003-9861(77)90398-8. [DOI] [PubMed] [Google Scholar]
- [13].Plapp BV, Mitchell JL, Berst KB. Mouse alcohol dehydrogenase 4: Kinetic mechanism, substrate specificity and simulation of effects of ethanol on retinoid metabolism. Chem. Biol. Interact. 2001;130-132:445–456. doi: 10.1016/s0009-2797(00)00284-2. [DOI] [PubMed] [Google Scholar]
- [14].Plapp BV, Berst KB. Human alcohol dehydrogenase 4: Mechanism, specificity and effects of ethanol on retinoid metabolism. In: Weiner H, Plapp BV, Lindahl R, Maser E, editors. Enzymology and Molecular Biology of Carbonyl Metabolism: Aldehyde Dehydrogenase, Aldo-Keto Reductase and Alcohol Dehydrogenase 12. West Lafayette, IN; Purdue University Press: 2005. pp. 190–199. [Google Scholar]
- [15].Duester G, Farrés J, Felder MR, Holmes RS, Höög J-O, Parés X, Plapp BV, Yin SJ, Jörnvall H. Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem. Pharmacol. 1999;58:389–395. doi: 10.1016/s0006-2952(99)00065-9. [DOI] [PubMed] [Google Scholar]
- [16].Julià P, Boleda MD, Parés X. Kinetic properties and physiological significance of the ADH-1 isoenzyme of rat stomach alcohol dehydrogenase. Prog. Clin. Biol. Res. 1987;232:189–201. [PubMed] [Google Scholar]
- [17].Boleda MD, Julià P, Moreno A, Parés X. Role of extrahepatic alcohol dehydrogenase in rat ethanol metabolism. Arch. Biochem. Biophys. 1989;274:74–81. doi: 10.1016/0003-9861(89)90416-5. [DOI] [PubMed] [Google Scholar]
- [18].Svensson S, Strömberg P, Höög J-O. A novel subtype of class II alcohol dehydrogenase in rodents. Unique Pro-47 and Ser-182 modulates hydride transfer in the mouse enzyme. J. Biol. Chem. 1999;274:29712–29719. doi: 10.1074/jbc.274.42.29712. [DOI] [PubMed] [Google Scholar]
- [19].Julià P, Farrés J, Parés X. Characterization of three isoenzymes of rat alcohol dehydrogenase. Tissue distribution and physical and enzymatic properties. Eur. J. Biochem. 1987;162:179–189. doi: 10.1111/j.1432-1033.1987.tb10559.x. [DOI] [PubMed] [Google Scholar]
- [20].Plapp BV, Parsons M, Leidal KG, Baggenstoss BA, Ferm JR, Wear SS. Characterization of alcohol dehydrogenase from cultured rat hepatoma (HTC) cells. Prog. Clin. Biol. Res. 1987;232:203–215. [PubMed] [Google Scholar]
- [21].Boleda MD, Saubi N, Farrés J, Parés X. Physiological substrates for rat alcohol dehydrogenase classes: Aldehydes of lipid peroxidation, omega-hydroxyfatty acids, and retinoids. Arch. Biochem. Biophys. 1993;307:85–90. doi: 10.1006/abbi.1993.1564. [DOI] [PubMed] [Google Scholar]
- [22].Allali-Hassani A, Martínez SE, Peralba JM, Vaglenova J, Vidal F, Richart C, Farrés J, Parés X. Alcohol dehydrogenase of human and rat blood vessels. Role in ethanol metabolism. FEBS Lett. 1997;405:26–30. doi: 10.1016/s0014-5793(97)00151-8. [DOI] [PubMed] [Google Scholar]
- [23].Östberg LJ, Strömberg P, Hedberg JJ, Persson B, Höög J-O. Analysis of mammalian alcohol dehydrogenase 5 (ADH5): Characterisation of rat ADH5 with comparisons to the corresponding human variant. Chem. Biol. Interact. 2013;202:97–103. doi: 10.1016/j.cbi.2012.11.002. [DOI] [PubMed] [Google Scholar]
- [24].Markovič O, Theorell H, Rao S. Rat liver alcohol dehydrogenase - Purification and properties. Acta Chem. Scand. 1971;25:195–205. doi: 10.3891/acta.chem.scand.25-0195. [DOI] [PubMed] [Google Scholar]
- [25].Plapp BV. Enhancement of the activity of horse liver alcohol dehydrogenase by modification of amino groups at the active sites. J. Biol. Chem. 1970;245:1727–1735. [PubMed] [Google Scholar]
- [26].Crabb DW, Edenberg HJ. Complete amino acid sequence of rat liver alcohol dehydrogenase deduced from the cDNA sequence. Gene. 1986;48:287–291. doi: 10.1016/0378-1119(86)90087-9. [DOI] [PubMed] [Google Scholar]
- [27].Cleland WW. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- [28].Levitt MD, Furne J, DeMaster EG. Magnitude, origin, and implications of the discrepancy between blood ethanol concentrations of tail vein and arterial blood of the rat. Alcohol. Clin. Exp. Res. 1994;18:1237–1241. doi: 10.1111/j.1530-0277.1994.tb00111.x. [DOI] [PubMed] [Google Scholar]
- [29].Deis FH, Lester D. Biochemical pharmacology of pyrazoles. In: Majchrowicz E, Noble EP, editors. Biochemistry and Pharmacology of Ethanol. Vol. 2. Plenum; New York: 1979. pp. 303–323. [Google Scholar]
- [30].Makar AB, Tephly TR. Inhibition of monkey liver alcohol dehydrogenase by 4-methylpyrazole. Biochem. Med. 1975;13:334–342. doi: 10.1016/0006-2944(75)90172-6. [DOI] [PubMed] [Google Scholar]
- [31].Damgaard SE. The D(V/K) isotope effect of the cytochrome P-450-mediated oxidation of ethanol and its biological applications. Eur. J. Biochem. 1982;125:593–603. doi: 10.1111/j.1432-1033.1982.tb06724.x. [DOI] [PubMed] [Google Scholar]
- [32].Feytmans E, Leighton F. Effects of pyrazole and 3-amino-1,2,4-triazole on methanol and ethanol metabolism by the rat. Biochem. Pharmacol. 1973;22:349–360. doi: 10.1016/0006-2952(73)90416-4. [DOI] [PubMed] [Google Scholar]
- [33].Theorell H, Yonetani T, Sjöberg B. On the effects of some heterocyclic compounds on the enzymic activity of liver alcohol dehydrogenase. Acta Chem. Scand. 1969;23:255–260. doi: 10.3891/acta.chem.scand.23-0255. [DOI] [PubMed] [Google Scholar]
- [34].Light DR, Dennis MS, Forsythe IJ, Liu CC, Green DW, Kratzer DA, Plapp BV. Alpha-isoenzyme of alcohol dehydrogenase from monkey liver. Cloning, expression, mechanism, coenzyme, and substrate specificity. J. Biol. Chem. 1992;267:12592–12599. [PubMed] [Google Scholar]
- [35].Green DW, Sun HW, Plapp BV. Inversion of the substrate specificity of yeast alcohol dehydrogenase. J. Biol. Chem. 1993;268:7792–7798. [PubMed] [Google Scholar]
- [36].Dalziel K, Dickinson FM. Aldehyde mutase. Nature. 1965;206:255–257. doi: 10.1038/206255a0. [DOI] [PubMed] [Google Scholar]
- [37].Henehan GT, Oppenheimer NJ. Horse liver alcohol dehydrogenase-catalyzed oxidation of aldehydes: Dismutation precedes net production of reduced nicotinamide adenine dinucleotide. Biochemistry. 1993;32:735–738. doi: 10.1021/bi00054a001. [DOI] [PubMed] [Google Scholar]
- [38].Shearer GL, Kim K, Lee KM, Wang CK, Plapp BV. Alternative pathways and reactions of benzyl alcohol and benzaldehyde with horse liver alcohol dehydrogenase. Biochemistry. 1993;32:11186–11194. doi: 10.1021/bi00092a031. [DOI] [PubMed] [Google Scholar]
- [39].Collins XH, Harmon SD, Kaduce TL, Berst KB, Fang X, Moore SA, Raju TV, Falck JR, Weintraub NL, Duester G, Plapp BV, Spector AA. ω -Oxidation of 20-hydroxyeicosatetraenoic acid (20-HETE) in cerebral microvascular smooth muscle and endothelium: Role of alcohol dehydrogenase 4. J. Biol. Chem. 2005;280:33157–33164. doi: 10.1074/jbc.M504055200. [DOI] [PubMed] [Google Scholar]
- [40].Plapp BV, Thulasiram HV, Berst KB, Kaduce TL. Recombinant mouse alcohol dehydrogenase 1: Kinetic mechanism, substrate specificity and oxidation of 20-hydroxyeicosatetraenoic acid. In: Weiner H, Maser E, Lindahl R, Plapp B, editors. Enzymology and Molecular Biology of Carbonyl Metabolism 13. Purdue University Press; West Lafayette, IN: 2007. pp. 123–133. [Google Scholar]
- [41].Lumeng L, Bosron WF, Li T-K. Quantitative correlation of ethanol elimination rates in vivo with liver alcohol dehydrogenase activities in fed, fasted and food-restricted rats. Biochem. Pharmacol. 1979;28:1547–1551. doi: 10.1016/0006-2952(79)90471-4. [DOI] [PubMed] [Google Scholar]
- [42].Handler JA, Thurman RG. Redox interactions between catalase and alcohol dehydrogenase pathways of ethanol metabolism in the perfused rat liver. J. Biol. Chem. 1990;265:1510–1515. [PubMed] [Google Scholar]
- [43].Tate RL, Mehlman MA, Tobin RB. Metabolic fate of 1,3-butanediol in the rat: Conversion to β-hydroxybutyrate. J. Nutr. 1971;101:1719–1726. doi: 10.1093/jn/101.12.1719. [DOI] [PubMed] [Google Scholar]
- [44].Bessman SP, McCabe E., III 1,4-Butanediol–A substrate for rat liver and horse liver alcohol dehydrogenases. Biochem. Pharmacol. 1972;21:1135–1142. doi: 10.1016/0006-2952(72)90107-4. [DOI] [PubMed] [Google Scholar]
- [45].Chou JY, Richardson KE. The effect of pyrazole on ethylene glycol toxicity and metabolism in the rat. Toxicol. Appl. Pharmacol. 1978;43:33–44. doi: 10.1016/s0041-008x(78)80030-1. [DOI] [PubMed] [Google Scholar]
- [46].Clay KL, Murphy RC. On the metabolic acidosis of ethylene glycol intoxication. Toxicol. Appl. Pharmacol. 1977;39:39–49. doi: 10.1016/0041-008x(77)90175-2. [DOI] [PubMed] [Google Scholar]
- [47].Jacobsen D, McMartin KE. Methanol and ethylene glycol poisonings. Mechanism of toxicity, clinical course, diagnosis and treatment. Med. Toxicol. 1986;1:309–334. doi: 10.1007/BF03259846. [DOI] [PubMed] [Google Scholar]
- [48].Brent J, McMartin K, Phillips S, Burkhart KK, Donovan JW, Wells M, Kulig K, Methylpyrazole for Toxic Alcohols Study Group Fomepizole for the treatment of ethylene glycol poisoning. New Eng. J. Med. 1999;340:832–838. doi: 10.1056/NEJM199903183401102. [DOI] [PubMed] [Google Scholar]
- [49].Montgomery JA, David F, Garneau M, Brunengraber H. Metabolism of 2,3-butanediol stereoisomers in the perfused rat liver. J. Biol. Chem. 1993;268:20185–20190. [PubMed] [Google Scholar]
- [50].Röe O. The metabolism and toxicity of methanol. Pharmacol. Rev. 1955;7:399–412. [PubMed] [Google Scholar]
- [51].McMartin KE, Martin-Amat G, Makar AB, Tephly TR. Methanol poisoning. V. Role of formate metabolism in the monkey. J. Pharm. Exp. Ther. 1977;201:564–572. [PubMed] [Google Scholar]
- [52].Jacobsen D, McMartin KE. Antidotes for methanol and ethylene glycol poisoning. J. Toxicol. Clin. Toxicol. 1997;35:127–143. doi: 10.3109/15563659709001182. [DOI] [PubMed] [Google Scholar]
- [53].Jacobsen D, Sebastian CS, Barron SK, Carriere EW, McMartin KE. Effects of 4-methylpyrazole, methanol/ethylene glycol antidote, in healthy humans. J. Emerg. Med. 1990;8:455–461. doi: 10.1016/0736-4679(90)90176-v. [DOI] [PubMed] [Google Scholar]
- [54].Plapp BV, Chadha VK, Leidal KG, Cho H, Scholze M, Schindler JF, Berst KB, Ramaswamy S. Uncompetitive inhibitors of alcohol dehydrogenases. Adv. Exp. Med. Biol. 1999;463:295–303. doi: 10.1007/978-1-4615-4735-8_36. [DOI] [PubMed] [Google Scholar]
- [55].McGregor D. Tertiary-butanol: A toxicological review. Crit. Rev. Toxicol. 2010;40:697–727. doi: 10.3109/10408444.2010.494249. [DOI] [PubMed] [Google Scholar]
- [56].Cook PF, Cleland WW. pH variation of isotope effects in enzyme-catalyzed reactions. 2. Isotope-dependent step not pH dependent. Kinetic mechanism of alcohol dehydrogenase. Biochemistry. 1981;20:1805–1816. doi: 10.1021/bi00510a015. [DOI] [PubMed] [Google Scholar]
- [57].Nordmann R, Ribiere C, Rouach H, Beauge F, Giudicelli Y, Nordmann J. Metabolic pathways involved in the oxidation of isopropanol into acetone by the intact rat. Life Sci. 1973;13:919–932. doi: 10.1016/0024-3205(73)90082-9. [DOI] [PubMed] [Google Scholar]
- [58].Gershman H, Abeles RH. Deuterium isotope effects in the oxidation of alcohols in vitro and in vivo. Arch. Biochem. Biophys. 1973;154:659–674. doi: 10.1016/0003-9861(73)90021-0. [DOI] [PubMed] [Google Scholar]
- [59].Edenburg HJ, Bosron WF. Alcohol Dehydrogenases. In: Guengerich FP, editor. Comprehensive Toxicology Vol. 3 Biotransformation. Pergamon Elsevier Scence; New York: 1997. pp. 119–131. [Google Scholar]
- [60].Dickinson JR, Lanterman MM, Danner DJ, Pearson BM, Sanz P, Harrison SJ, Hewlins MJ. A 13C nuclear magnetic resonance investigation of the metabolism of leucine to isoamyl alcohol in Saccharomyces cerevisiae. J. Biol. Chem. 1997;272:26871–26878. doi: 10.1074/jbc.272.43.26871. [DOI] [PubMed] [Google Scholar]
- [61].Tank AW, Weiner H. Ethanol-induced alteration of dopamine metabolism in rat liver. Biochem. Pharmacol. 1979;28:3139–3147. doi: 10.1016/0006-2952(79)90624-5. [DOI] [PubMed] [Google Scholar]
- [62].Weiner H, Coker FG, Vrbanac JJ. Application of metabolic profiling to study the effects of ethanol on metabolism in rats. Alcohol. 1984;1:105–109. doi: 10.1016/0741-8329(84)90064-8. [DOI] [PubMed] [Google Scholar]
- [63].Zlatkis A, Bertsch W, Bafus DA, Liebich HM. Analysis of trace volatile metabolites in serum and plasma. J. Chromatogr. 1974;91:379–383. doi: 10.1016/s0021-9673(01)97916-6. [DOI] [PubMed] [Google Scholar]
- [64].Liebich HM, Woll J. Volatile substances in blood serum: profile analysis and quantitative determination. J. Chromatogr. 1977;142:505–516. doi: 10.1016/s0021-9673(01)92063-1. [DOI] [PubMed] [Google Scholar]
- [65].Zlatkis A, Poole CF, Brazeli R, Bafus DA, Spencer PS. Volatile metabolites in sera of normal and diabetic patients. J.Chromatogr. 1980;182:137–145. doi: 10.1016/s0378-4347(00)81617-5. [DOI] [PubMed] [Google Scholar]
- [66].Zlatkis A, Brazell RS, Poole CF. The role of organic volatile profiles in clinical diagnosis. Clin. Chem. 1981;27:789–797. [PubMed] [Google Scholar]
- [67].Goldberg EM, Blendis LM, Sandler S. A gas chromatographic--mass spectrometric study of profiles of volatile metabolites in hepatic encephalopathy. J. Chromatogr. 1981;226:291–299. doi: 10.1016/s0378-4347(00)86063-6. [DOI] [PubMed] [Google Scholar]
- [68].Liebich HM, Buelow HJ, Kallmayer R. Quantification of endogenous aliphatic alcohols in serum and urine. J. Chromatogr. 1982;239:343–349. doi: 10.1016/s0021-9673(00)81993-7. [DOI] [PubMed] [Google Scholar]
- [69].Lieber CS, DeCarli LM. The Role of the hepatic microsomal ethanol oxidizing system (MEOS) for ethanol metabolism in vivo. J. Pharmacol. Exp. Ther. 1972;181:279–287. [PubMed] [Google Scholar]
- [70].Teschke R, Hasumura Y, Lieber CS. Hepatic microsomal alcohol-oxidizing system. Affinity for methanol, ethanol, propanol, and butanol. J. Biol. Chem. 1975;250:7397–7404. [PubMed] [Google Scholar]
- [71].Norsten C, Cronholm T, Ekström G, Handler JA, Thurman RG, Ingelman-Sundberg M. Dehydrogenase-dependent ethanol metabolism in deer mice (Peromyscus maniculatus) lacking cytosolic alcohol dehydrogenase. Reversibility and isotope effects in vivo and in subcellular fractions. J. Biol. Chem. 1989;264:5593–5597. [PubMed] [Google Scholar]
- [72].Kono H, Bradford BU, Yin M, Sulik KK, Koop DR, Peters JM, Gonzalez FJ, McDonald T, Dikalova A, Kadiiska MB, Mason RP, Thurman RG. CYP2E1 is not involved in early alcohol-induced liver injury. Am. J. Physiol. 1999;277:G1259–1267. doi: 10.1152/ajpgi.1999.277.6.G1259. [DOI] [PubMed] [Google Scholar]
- [73].Feierman DE, Cederbaum AI. Increased sensitivity of the microsomal oxidation of ethanol to inhibition by pyrazole and 4-methylpyrazole after chronic ethanol treatment. Biochem. Pharmacol. 1987;36:3277–3283. doi: 10.1016/0006-2952(87)90645-9. [DOI] [PubMed] [Google Scholar]
- [74].Spracklin DK, Hankins DC, Fisher JM, Thummel KE, Kharasch ED. Cytochrome P450 2E1 is the principal catalyst of human oxidative halothane metabolism in vitro. J. Pharm. Exp. Ther. 1997;281:400–411. [PubMed] [Google Scholar]
- [75].Svanas GW, Weiner H. Aldehyde dehydrogenase activity as the rate-limiting factor for acetaldehyde metabolism in rat liver. Arch. Biochem. Biophys. 1985;236:36–46. doi: 10.1016/0003-9861(85)90603-4. [DOI] [PubMed] [Google Scholar]
- [76].Persson B, Kallberg Y. Classification and nomenclature of the superfamily of short-chain dehydrogenases/reductases (SDRs) Chem. Biol. Interact. 2013;202:111–115. doi: 10.1016/j.cbi.2012.11.009. [DOI] [PubMed] [Google Scholar]
- [77].Haggard HW, Greenberg LA, Turner JM. The physiological principles governing the action of acetone together with determination of toxicity. J. Indust. Hyg.Toxicol. 1944;26:133–151. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.