Abstract
The diaphragm is an essential mammalian skeletal muscle, and defects in diaphragm development are the cause of congenital diaphragmatic hernias (CDH), a common and often lethal birth defect. The diaphragm is derived from multiple embryonic sources, but how these give rise to the diaphragm is unknown and, despite the identification of many CDH-associated genes, the etiology of CDH is incompletely understood. Using mouse genetics, we show that the pleuroperitoneal folds (PPFs), transient embryonic structures, are the source of the diaphragm’s muscle connective tissue, regulate muscle development, and their striking migration controls diaphragm morphogenesis. Furthermore, Gata4 mosaic mutations in PPF-derived muscle connective tissue fibroblasts result in the development of localized amuscular regions that are biomechanically weaker and more compliant and lead to CDH. Thus the PPFs and muscle connective tissue are critical for diaphragm development and mutations in PPF-derived fibroblasts are a source of CDH.
INTRODUCTION
The muscularized diaphragm is not only a unique and defining character of all mammals1, but is an essential skeletal muscle. Diaphragm contraction drives inspiration and is critical for respiration2. In addition, the diaphragm has a significant passive functional role, since it serves as a barrier between the thoracic and abdominal cavities1. These important functions are carried out by the costal diaphragm: a radial array of myofibers, surrounded by muscle connective tissue, that extend from the ribs to the central tendon (Fig. 1y). Development of a functional diaphragm therefore requires the coordinated morphogenesis of muscle, muscle connective tissue, and tendon, and these tissues have been suggested to develop from multiple embryonic sources3. However, our knowledge of how this muscle develops has been limited by the inaccessibility of mammalian embryos to classic experimental embryological techniques and lack of genetic reagents to manipulate key embryonic sources.
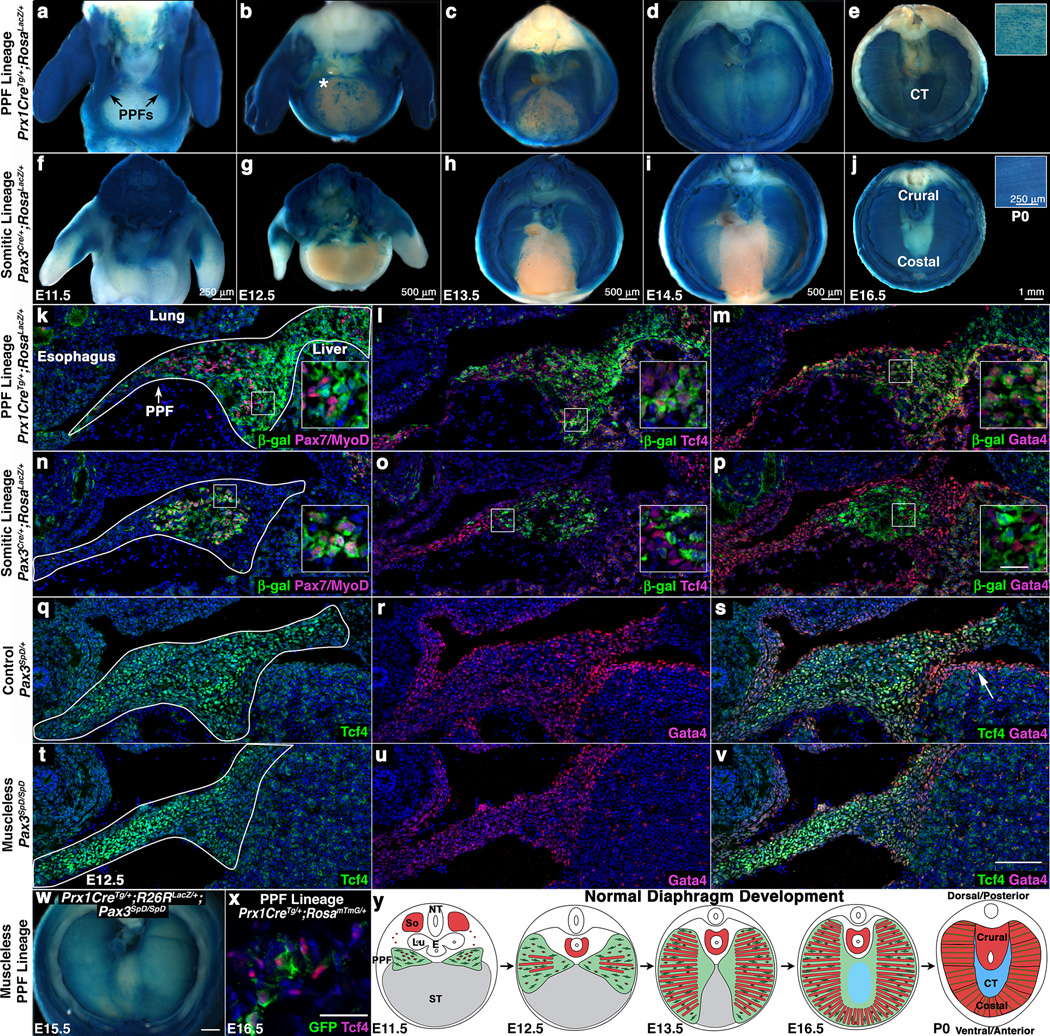
Fig. 1. PPFs contains Gata4+ muscle connective tissue fibroblasts that migrate independently and in advance of myogenic cells.
(a–e) Prx1CreTg labels PPF cells (a) that expand throughout diaphragm (d) and contribute to the central tendon (e) and costal muscle connective tissue fibroblasts (inset e) As expected, Prx1CreTg labels limb and body wall lateral plate mesoderm. (f–j) Pax3Cre labels myogenic cells (f) that give rise to crural and costal muscles (j). (k–m) Prx1CreTg does not label Pax7+ muscle progenitors or MyoD+ myoblasts (k), but labels Tcf4+ (l) and Gata4+ (m) fibroblasts in the PPFs. (n–p) Pax3Cre labels Pax7+/MyoD+ myogenic cells (n), but not Tcf4+ (o) or Gata4+ cells (p). n > 3/time point. (q–v) Gata4+ cells are Tcf4+ muscle connective tissue fibroblasts (q–s) and present in the absence of muscle (t–v). w–x, Prx1-derived cells expand in absence of muscle (w, n = 4/4) and are Tcf4+ (x). (y) Model of diaphragm development. (a–j). n > 5 for each time point and genotype. Whole-mount β-galactosidase staining. (k–v) Sections through E12.5 PPFs. (x) Section through E16.5 diaphragm. Asterisk in b shows location of Video S1. Arrow in s shows Gata4+ septum transversum. (a–j, w) Dorsal at top. Heart and lungs removed. (k–v) Dorsal at top, medial at left. Scale bar = 100 µm (k–v), 20 µm (insets k–p), 500 µm (w), 50 µm (x). CT, central tendon; E, esophagus; Lu, lung; NT, neural tube; PPFs, pleuroperitoneal folds; So, somite, ST, septum transversum.
Defects in diaphragm development are the cause of congenital diaphragmatic hernias (CDHs), common (1/3,000 of total births) and costly (exceeding $250 million/year in the USA) birth defects4,5. In CDH, weaknesses in the developing diaphragm allow abdominal contents to herniate into the thoracic cavity and impede lung development. The associated lung hypoplasia is the cause of the 50% neonatal mortality and long-term morbidity associated with CDH6. Despite the prevalence and severity of CDH, the genetic and cellular etiology of this birth defect is incompletely understood. The majority of CDH cases appear as isolated defects6, and the low incidence of sibling/familial recurrence suggests that CDH generally arises from de novo genetic mutations7. Molecular cytogenetic analyses of CDH patients have identified copy number variants (CNVs) in multiple chromosomal regions strongly associated with CDH6,8,9, and detailed analyses of these regions and a limited number of mouse studies have identified over 50 candidate CDH-causative genes8,10. A region that contains recurrent CDH-associated CNVs is 8p23.1 (OMIM 22240)8, and within this region variants in the GATA binding protein 4 (Gata4) transcription factor have recently been shown to strongly correlate with CDH11–13. Nevertheless, how these candidate CDH genes mechanistically cause CDH is not clear. Functional analyses of mouse mutants have been severely limited by early embryonic lethality14,15, low or variable incidence of CDH16–18, and lack of conditional mutants19,20. Furthermore, the notable incomplete penetrance and variable expressivity of CDH-associated CNVs and genetic mutations6,11–13 suggest that the genetic architecture underlying CDH is complex.
RESULTS
Pleuroperitoneal folds regulate diaphragm development
The diaphragm has been proposed to develop primarily from two embryonic sources. The somites are well documented to be the source of the diaphragm’s muscle21–23. Less understood, the pleuroperitoneal folds (PPFs) are two pyramidal-shaped mesodermal structures located between the thoracic (pleural) and abdominal (peritoneal) cavities and hypothesized to be critical for diaphragm development22. However, without genetic reagents to label and manipulate the PPFs, it has been unclear what function they play in diaphragm development. A third embryonic structure, the septum transversum, has been suggested to be a source of the central tendon22,24.
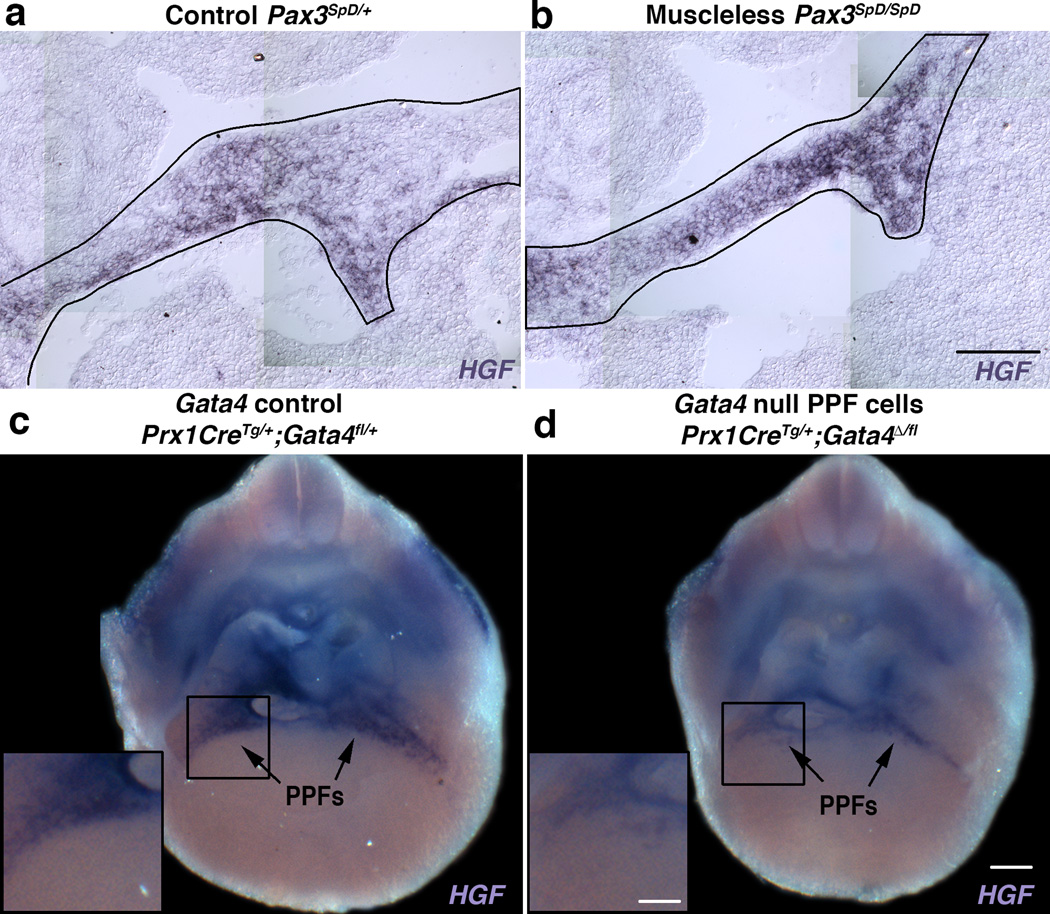
To test the contribution of the PPFs to diaphragm development and structure, we identified the first genetic reagent to label the PPFs. Based on the suggestion that the PPFs are of lateral plate origin25, we tested Prx1Cre26, a transgene composed of a Prx1-regulatory element that drives Cre-mediated recombination in the flank and limb lateral plate mesoderm (including the limb muscle connective tissue, tendons and bones). We found that when crossed with the Cre-responsive reporter RosaLacZ27 it robustly labels the PPFs, but not the septum transversum (Fig. 1a, k–m, s, and data not shown). The PPFs are present at embryonic day (E) 11.5 and expand across the surface of the liver to give rise to cells throughout the diaphragm at E14.5 (Fig. 1a–d). Although Prx1Cre labels body wall non-muscle tissues (Fig. 1a–e), these cells do not appear to contribute to the diaphragm (based on two-photon studies, see below). PPF cells are distinct from somite-derived muscle progenitors, myoblasts, and myofibers (Fig. 1e, k). Instead, the PPFs give rise to two non-myogenic tissues. First, in contrast to the previous hypothesis that the central tendon derives from the septum transversum22,24, the PPFs give rise to the central tendon (Fig. 1e). Second, the PPFs give rise to muscle connective tissue fibroblasts. Prx1-derived cells express the muscle connective tissue marker Tcf4 (Tcf7l2)28,29 in the E12.5 PPF (Fig. 1l) and in the fully developed diaphragm (Fig. 1x) and ultimately reside interstitial to costal (but not crural) myofibers (Fig. 1e). Importantly, the PPF cells also strongly express the CDH-implicated gene Gata4; at E12.5 most Prx1-derived cells express Gata4 (Fig. 1m), and all Tcf4+ fibroblasts are Gata4+ (Fig. 1q–s). Thus, we demonstrate that the PPFs are not simply transient developmental structures, but ultimately give rise to the central tendon and the muscle connective tissue fibroblasts (Fig. 1y). Furthermore, the expression in PPF cells of CDH-implicated Gata4 (as also noted in12,16,30) suggests that the PPFs may be important in the etiology of CDH.
To trace the contribution of myogenic cells to diaphragm morphogenesis and compare their spatiotemporal relationship to PPF cells, we genetically labeled myogenic cells using Pax3Cre/+;RosaLacZ/+ mice31, in which Cre-mediated recombination in the somite labels all myogenic cells. Myogenic cells migrate from the somite and enter the PPFs by E11.5 (Fig. 1f, n), then spread ventrally and dorsally (Fig. 1g–i), differentiate into myofibers (Fig. 1h–i), and form a completely muscularized diaphragm by E16.5 (Fig. 1j). Comparison of muscle and PPF morphogenesis reveals that the PPFs expand ventrally in advance of the muscle (Fig. 1d, i). Labeled myogenic cells do not express either Tcf4 or Gata4, but instead are surrounded by non-myogenic Tcf4+Gata4+ cells (Fig. 1o–p). The myogenic cells’ migration into the PPFs and their subsequent expansion within, but behind the leading edge of the PPF cells suggest that the PPFs may guide the morphogenesis of the diaphragmatic muscle (Fig. 1y).
If PPFs are critical for morphogenesis of the diaphragm’s muscle, we expected that formation and morphogenesis of the PPFs should occur independently of muscle. To test this, we analyzed Prx1CreTg/+;RosaLacZ/+;Pax3SpD/SpD mice32, which have a muscleless diaphragm. We found that indeed, even in the absence of muscle, PPF cells are present and express Tcf4 and Gata4 (Fig. 1t–v) at E12.5 and subsequently undergo normal morphogenetic expansion (Fig. 1w).
The expansion of PPF cells, even in the absence of muscle, suggests that the morphogenetic movement of the PPF cells drives normal diaphragm morphogenesis. While the static images of PPF cells suggest that these cells migrate across the surface of the liver, it is formally possible that this simply reflects the dynamic activation of Prx1Cre and not cell movement. To test this, we cultured ex-vivo E12.5 diaphragms (leaving ribs and underlying liver intact) and imaged genetically labeled PPF cells via 2-photon microscopy. We found that PPF cells actively migrate across the liver surface (at the rate of 5 µm/hour); although a few cells migrate singly, most appear to migrate collectively (Fig. 1b, Supplementary Fig. 1 and Videos 1–2). This collective PPF cell migration, which occurs independent of muscle, likely drives diaphragm morphogenesis.
CDH originates with defects in PPF-derived fibroblasts
Human genetic studies have shown non-coding variants and missense mutations of Gata4 are correlated with CDH11–13, but explicit tests of the function of Gata4 in development of CDH have been limited by the early embryonic lethality of Gata4 null mice14,15 and the reported low penetrance of CDH in Gata4 heterozygous mice16. The strong Gata4 expression in the PPFs (Fig. 1 and12,16,30), suggests that Gata4 functions in this tissue. To test this, we generated Prx1CreTg/+;Gata4Δ/fl mice (using Gata4fl mice33), in which Gata4 is specifically deleted in the PPFs. Strikingly, we found that 100% of mutant mice (n > 33/33) develop multiple hernias throughout the diaphragm (Fig. 2a, b). Similar to CDH patients6,34, in Prx1CreTg/+;Gata4Δ/fl mice the size and location of hernias varies: most (68%) form in the dorsal lateral diaphragm (Bochdalek hernias) and the rest (32%) develop in the ventral diaphragm (Morgagni hernias, Fig. 2b). We found that hernias only occur in muscle-associated regions and not in the central tendon. Thus although the PPFs give rise to both the muscle connective tissue and the central tendon, hernias never arise in the central tendon and so it is defects in the PPF-derived muscle connective tissue that cause CDH in these mice. Unlike previous reports16, we never observed diaphragm defects in heterozygous Gata4Δ/+ mice (n > 66/66, Fig. 2a). In addition, we tested whether Gata4 was required in myogenic cells for diaphragm development by analyzing Pax3Cre/+;Gata4Δ/fl mice. As predicted based on the lack of Gata4 expression in myogenic cells, diaphragms develop normally in these mice (n = 28/28, Fig. 2c). Because Pax3 is also expressed in neural crest cells, this rules out neural crest as a source of CDH from Gata4 mutations. Thus we definitively demonstrate that Gata4 null mutations cause CDH. Moreover, defects in the PPFs and muscle connective tissue are a source of CDH.
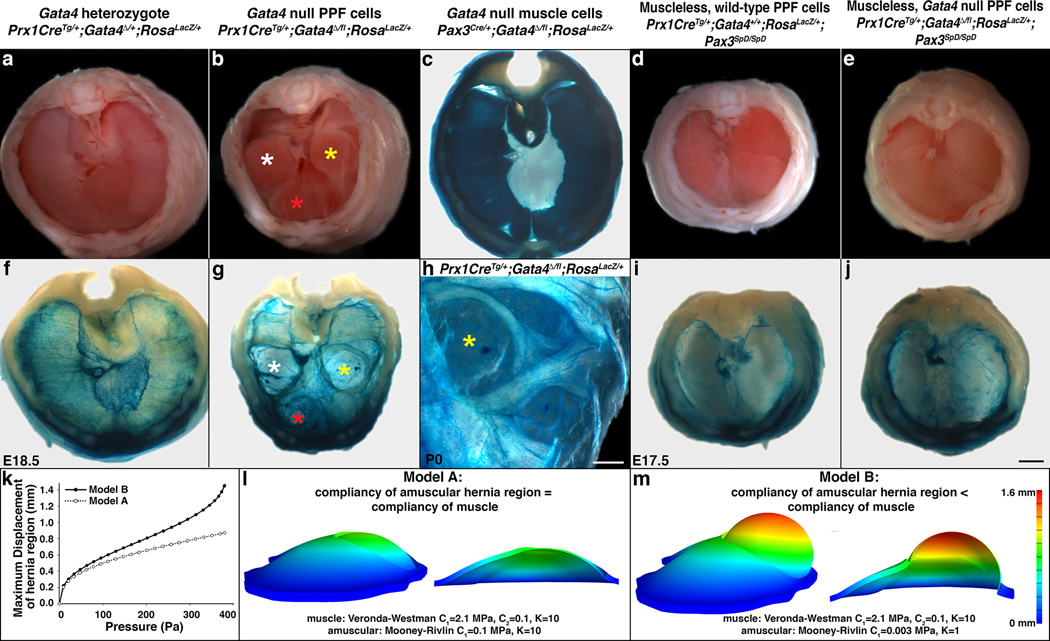
Fig. 2. Deletion of Gata4 in the PPFs produces localized amuscular regions that are weaker than juxtaposed muscular regions and results in CDH.
(a–d) CDH develops in mice with Gata4 null PPF cells (n > 33/33; Bochdalek hernias labeled with white and yellow asterisks and Morgagni hernia labeled with red asterisk in b), but not in Gata4 heterozygotes (n > 66/66; a), mice with Gata4 null muscle (n = 28/28; c), or mice with muscleless diaphragms (n > 10/10; d). (e, j) Loss of muscle in diaphragms with Gata4 null PPF cells rescues herniation, indicating that juxtaposition of amuscular with muscular regions is required for CDH (n = 3/3). (g–h) In herniated regions PPF cells are present, but muscle is not (n = 7/7). (k–m), Finite element modeling shows that a hernia only develops when the amuscular region is more compliant than the muscular region. Models are lateral views (c, f–j) Whole-mount β-galactosidase staining. (a–j) Dorsal is at top. Scale bar = 1 mm (a–g, i–j), 0.5 mm (h).
Localized weaker amuscular regions give rise to hernias
Our results demonstrate that, with complete penetrance, loss of Gata4 in PPF-derived muscle connective tissue fibroblasts results in diaphragmatic hernias, but what mechanistically causes herniation? The most commonly proposed mechanism is that hernias are caused by a morphogenetic defect in the PPFs; this defect results in a localized loss of PPF-derived tissue, a consequent absence of muscle normally associated with this tissue, formation of a hole in the diaphragm, and herniation of growing abdominal tissue through this hole24,35. If this hypothesis is true, PPF-derived cells, and associated muscle, should not be present in herniated regions. To test this, we examined Prx1CreTg/+;Gata4Δ/fl;RosaLacZ/+ mice, in which PPF-derived Gata4 null fibroblasts are β-galactosidase+ (β-gal+) and muscle is β-gal-. Surprisingly, β-gal+ fibroblasts are present as a sac covering the herniated regions, and thus these regions are not simply holes in this tissue (n = 7/7; Fig. 2g–h). Yet, the sacs are devoid of muscle, and muscle surrounding the sacs is aberrantly patterned (Fig. 2g–h). Therefore, although muscle is absent in herniated regions, hernias in Prx1CreTg/+;Gata4Δ/fl mice are not caused by a failure of the morphogenetic expansion of the PPFs and the formation of holes in the diaphragm.
A second hypothesis for how CDH develops is that PPF-derived muscle connective tissue alone, without muscle, is weak and allows herniation through the weaker tissue. We tested this by examining Prx1CreTg/+;RosaLacZ/+;Pax3SpD/SpD diaphragms, in which muscle is absent but PPF-derived muscle connective tissue is present (Fig. 1w). However, in these mice with muscleless diaphragms, hernias never formed (n > 10/10, Fig. 2d, i). This demonstrates that PPF-derived muscle connective tissue alone, even in the absence of muscle, is sufficiently strong to prevent herniation of abdominal tissues.
These data suggest two additional hypotheses for the mechanism underlying herniation. First, the muscle connective tissue produced by Gata4 null fibroblasts may be weaker than connective tissue made by wild-type fibroblasts and this weaker tissue allows for herniation of abdominal tissue. Alternatively, formation of relatively weak regions of amuscular connective tissue juxtaposed to stronger muscularized regions may allow abdominal contents to herniate through the localized weak regions. To test this, we generated Prx1CreTg/+;Gata4Δ/fl;Pax3SpD/SpD mice, in which diaphragms are muscleless and the PPF-derived fibroblasts are Gata4 null. If the first hypothesis is correct, then hernias should form, while if the second hypothesis is correct, hernias should be absent. Strikingly, no hernias develop in these mice (n = 3/3), and the diaphragms are indistinguishable from Pax3SpD/SpD mice (Fig. 2d–e, i–j). Thus the loss of muscle rescues the herniation phenotype of Prx1CreTg/+;Gata4Δ/fl mice. This demonstrates that the muscle connective tissue produced by Gata4 null fibroblasts is not inherently weaker than wild-type connective tissue. Instead, hernias only develop when localized regions of amuscular connective tissue develop in juxtaposition with muscularized regions; through these amuscular regions that are relatively weaker than muscularized regions, abdominal tissue herniates.
To gain further insight into the biomechanics governing herniation, we turned to finite element modeling. The amuscular regions of E16.5 diaphragms, prior to overt herniation of abdominal tissue, are 25% of the thickness of the muscular regions (see Fig. 4c, g). This suggests that the relative weakness of the amuscular compared to the muscular regions could simply be due to its decreased thickness. Alternatively, the amuscular regions may be both thinner and composed of a more compliant material than muscle. We tested this by creating a finite element model. The geometry of the diaphragm was based on the dimensions of an E16.5 mutant diaphragm (see Fig. 4c), and a uniform physiologically reasonable pressure was applied to the diaphragm to simulate the pressure of the growing liver. Using the FEBio nonlinear finite element solver36, when the muscle material behavior is represented by an isotropic hyperelastic constitutive equation (based on37) and the amuscular region is made significantly more compliant (deforms more in response to an applied force) than the muscle, a pressure of 380 Pa induced a bulge in the amuscular region that matched the geometry of the hernias in experimental mice (Model B, Fig. 2 k, m, Supplementary Videos 3–4). In contrast, when the thinner amuscular region was assigned the same material properties as the muscle, pressures up to 380 Pa were unable to generate a bulge in the amuscular region (Model A, Fig. 2k–l). Together these results indicate that the weakness in the amuscular regions is due to both its decreased thickness and increased compliance as compared with the muscularized regions.
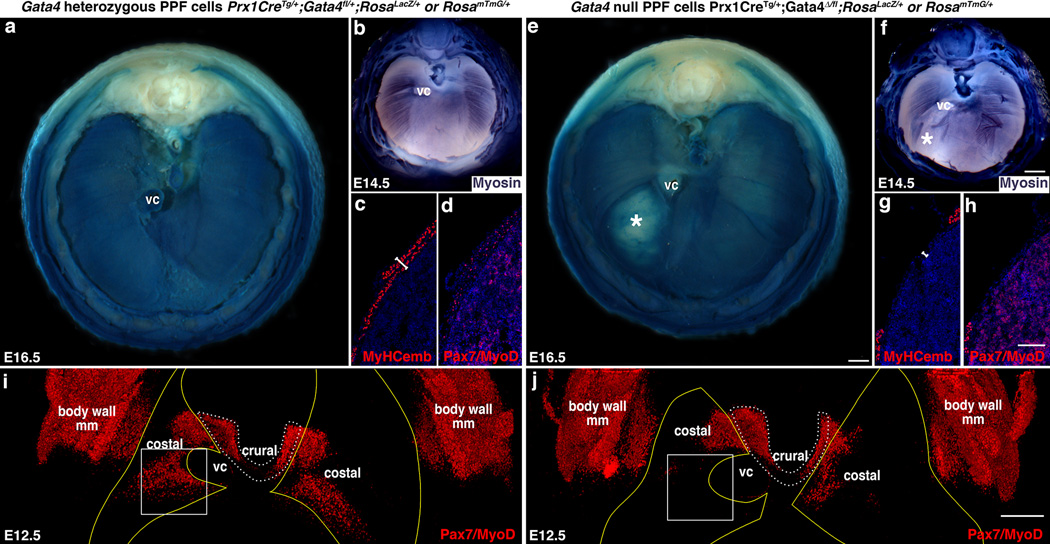
Fig. 4. CDH results from early defects in the localization of muscle progenitors.
(a, e) Overt liver herniation through diaphragms with Gata4 null PPF cells first appears at E16.5 (* in e, n = 3/3). (b–d, f–h) At E14.5 differentiating myofibers are aberrant (* in f; n = 7/7; b, f) and myofibers (c, g) and Pax7+/MyoD+ muscle progenitors (d, h) are absent in localized regions (n = 1/1). (i–j) At E12.5 Pax7+/MyoD+ muscle progenitors are absent in localized regions, particularly in the region (box) that consistently gives rise to hernias (n = 7/7). Pleuroperitoneal folds are outlined in yellow (outline derived from GFP immunofluorescence shown in Fig. 6a and b) and costal muscle is outlined in white dashed lines. (a, b) Whole-mount β-galactosidase staining. (b, f) Whole-mount myosin-alkaline phosphatase staining. (c–d, g-–h) Section immunofluorescence. (i-j) Whole-mount immunofluorescence (same diaphragms shown in Fig. 6a and b). (a–b, e–f, i–j) Dorsal is at the top. Scale bar = 500 µm (a, e), 500 µm (b, f), 100 µm (c–d, g–h), 200 µm (i–j). VC, vena cava; body wall mm, body wall muscles.
Herniated tissue physically impedes lung development
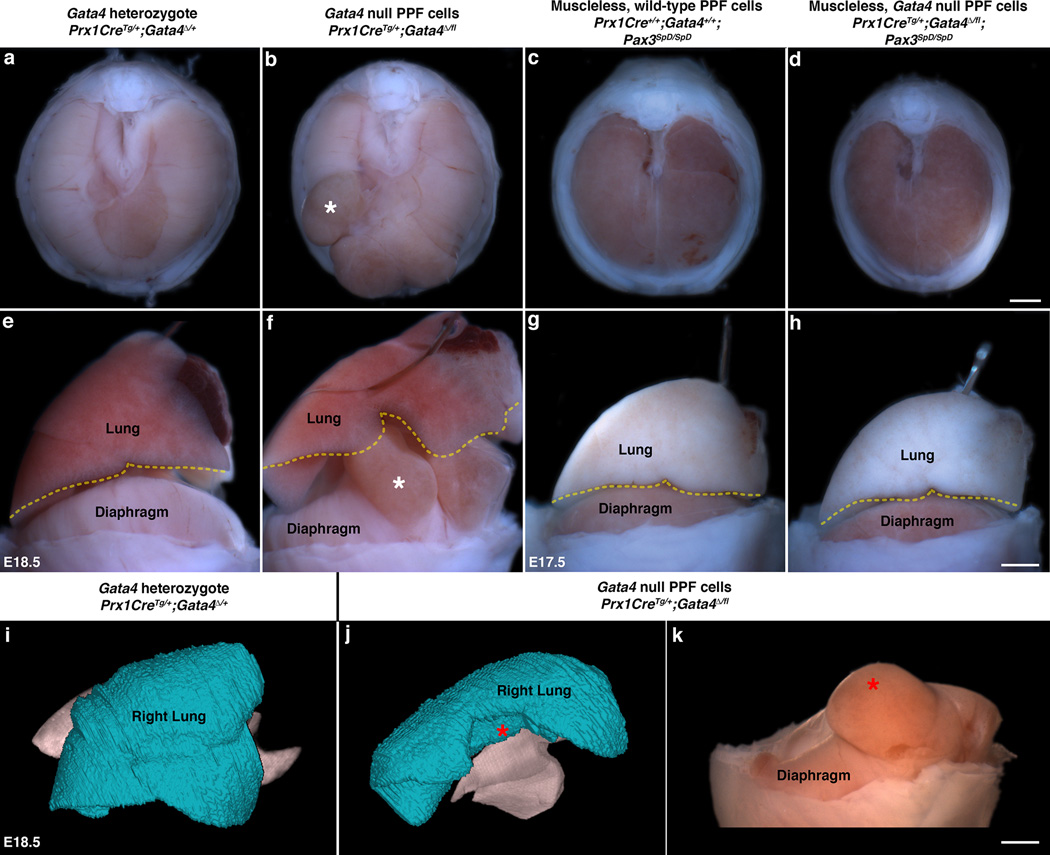
The neonatal mortality and long-term morbidity of CDH patients is caused by secondary lung hypoplasia, which is thought to arise from the physical impedance of lung growth by the herniated abdominal tissue. However, a “dual-hit hypothesis” has also been proposed38, whereby lung hypoplasia can result both from physical impedance by herniated tissue and from cell-autonomous effects on lung development by CDH-associated genes. Consistent with this hypothesis, mutations in Gata4 can cell-autonomously affect lung development39. Similar to CDH patients, Prx1CreTg/+;Gata4Δ/fl mice have low O2 blood saturation (as measured by pulse oximetry; data not shown) and most die within a few hours of birth. Defects in the lung lobar structure were found in all mice examined (n = 8/8, Fig. 3a–b, e–f, i–j) and resulted in up to a 34% reduction in lung volume (as quantified by microCT) of lobes adjacent to hernias (Fig. 3i–k; Supplementary Videos 5–6). The tight correlation of lung defects with hernias in all mice strongly suggests that the herniated tissue physically impedes lung growth in these mice. Furthermore, the absence of lung defects in Prx1CreTg/+;Gata4Δ/fl;Pax3SpD/SpD mice (Fig. 3c–d, g–h) argues that herniation precedes and causes lung hypoplasia. Thus Prx1CreTg/+;Gata4Δ/fl mice not only develop CDH, but also the lung defects and neonatal lethality typically accompanying CDH, and impedance of lung growth by herniated tissue is sufficient to induce lung lobar defects. It is also likely that Gata4 mutations have additional cell-autonomous effects on lung development (as shown by39), although we could not directly evaluate this, as Prx1Cre has limited recombination in the lungs and the alveolar structure is largely unaffected in these mutant mice (Supplementary Fig. 2a–c).
Fig. 3. In CDH, physical impedance by herniated tissue causes lung hypoplasia.
(a–b, e–f) As compared with lungs in mice with a normal diaphragm (n > 50; a, e), development of lung lobar structure is impeded by liver herniated through a defective diaphragm (n = 8/8; white asterisks show equivalent herniated region in b, f). (c–d, g–h) Lungs develop normally in mice with a muscleless diaphragm (n > 5/5; c, g) or with a muscleless diaphragm with Gata4 null PPF cells (n = 2/2; d, h). (i–k) MicroCT scans show that lungs (i–j) are greatly reduced in presence of hernia (red asterisks show herniated region j–k; n = 3). (i–j) Lateral right view of lungs, right lungs shown in blue. (k) Lateral view of diaphragmatic hernia associated with malformed lungs (j). (a–b, e–f, i–k) E18.5. (c–d, g–h) E17.5. Scale bar = 1 mm (a–d), 1 mm (e–h, k).
Aberrant HGF and muscle proliferation and apoptosis in CDH
Our experiments demonstrate that hernias form when localized amuscular regions develop within the muscular diaphragm. To determine when hernias and amuscular regions arise, we examined a developmental time series of mutant Prx1CreTg/+;Gata4Δ/fl embryos. Overt herniation of liver through the diaphragm first occurs at E16.5 (Fig. 4a, e). However, defects in muscle are present by E14.5 (Fig. 4b, f), and neither differentiated myofibers nor muscle progenitors are present in amuscular regions (Fig. 4c–d, g–h). Even at E12.5 there is a profound defect in the number and localization of muscle progenitors (Fig. 4i–j).
A critical early regulator of muscle progenitors migrating into the limb and diaphragm is Hepatocyte Growth Factor (HGF)23,40,41 and therefore an attractive candidate downstream target of Gata4. We found in control mice that the PPF cells, independent of the presence of muscle, robustly express the ligand HGF E12-13.5 (Fig. 5a–c), while the muscle progenitors express the HGF receptor, Met (data not shown). In Prx1CreTg/+;Gata4Δ/fl E12.5 diaphragms, HGF is markedly down-regulated and diminished in regions that consistently give rise to hernias (n = 11/11, Fig. 5d).
Fig. 5. HGF is strongly expressed in PPF cells and down-regulated in Gata4 null fibroblasts.
(a–c) HGF is expressed in PPF cells independent of muscle. PPFs are outlined in black. (d) Deletion of Gata4 in PPF cells leads to HGF down-regulation, particularly in the region (box) that consistently give rise to hernias. (n = 11/11). Dorsal is at top of all panels. Scale bar = 100 µm (a, b), 250 µm (c, d) and 125 µm (insets c, d).
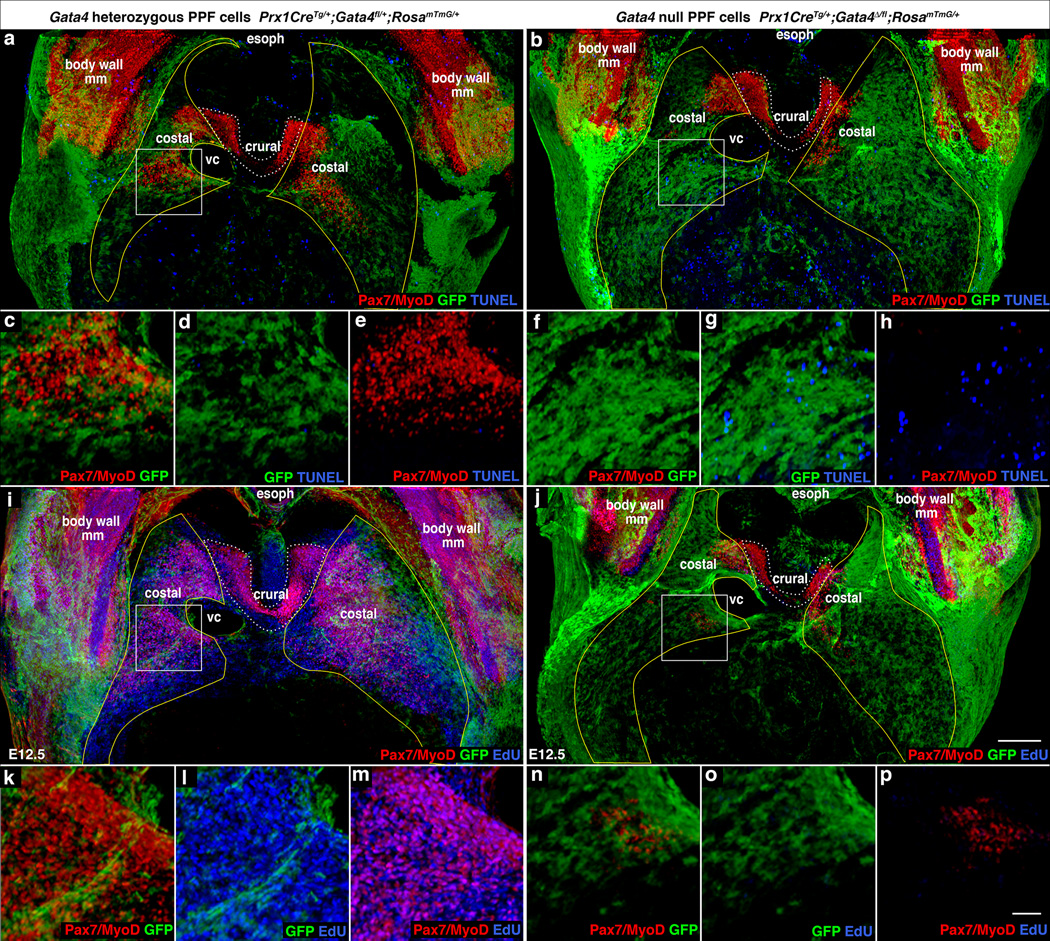
HGF functions as a mitogen and a cell survival factor42, and therefore we examined whether changes in apoptosis and proliferation determine why fewer muscle progenitors are present in mutant embryos. At E12.5 in the developing diaphragm of control embryos, few TUNEL+ apoptotic cells are present (Fig. 6a, c–e, Supplementary Video 7), while nearly all muscle progenitors are actively proliferating (Fig. 6i, k–m, Supplementary Video 9). In contrast, in mutant embryos there is a marked increase in apoptotic cells, many of which are present in regions that are abnormally devoid of muscle and consistently give rise to hernias (Fig. 6b, f–h, Supplementary Video 8). In addition, there is a profound decrease in EdU+ proliferative cells (Fig. 6j, n–p, Supplementary Video 10). Similar to the heart43,44, we find that loss of Gata4 leads to decreased levels of cell cycle regulators Cyclin D2 and Cdk4 in PPF cells (Supplementary Fig. 3a–l). In culture, isolated Gata4 null PPF fibroblasts proliferate at less than half the level of wild-type fibroblasts (Supplementary Fig. 4a). Moreover, when diaphragm myogenic cells are cultured with PPF cells, Gata4 null fibroblasts (as compared with wild-type fibroblasts) fail to support the growth of myogenic cells (Supplementary Fig. 4b–c).
Fig. 6. Early defects in proliferation, apoptosis, and localization of muscle progenitors lead to CDH.
(a–h) At E12.5 there is a marked increase in apoptotic cells in diaphragms with Gata4 null PPF cells (b, f–h versus a, c–e), particularly in the region (boxes a and b, magnified c–e, f–h) that consistently gives rise to hernias (n = 3/3). (i–p) There also is a marked decrease in EdU+ proliferating cells in mutant diaphragms (n = 7/7). (j, n–p versus i, k–m). In the region (boxes i and j, magnified in k–m, n–p) destined to give rise to hernias the few myogenic cells are EdU−. (a–p) Costal muscle progenitors are surrounded by GFP+ PPF cells in control diaphragms but are excluded from regions with GFP+ Gata4 null PPF cells in mutant diaphragms, particularly in destined hernia regions (boxes b, j magnified f–h, n–p). (a–p) Whole-mount immunofluorescence. (a–b, i–j) Pleuroperitoneal folds are outlined in yellow and costal muscle is outlined in white dashed lines. (a–p) Dorsal is at top. Scale bar = 200 µm (a–b, i–j), 50 µm (c–h, k–p). PPF, pleuroperitoneal fold; NT, neural tube; E or esoph, esophagus; So, somite; ST, septum transversum, VC, vena cava, body wall mm, body wall muscles.
In mutant diaphragms, the number of muscle progenitors is greatly reduced by increased cell death and decreased proliferation, but how do localized amuscular regions develop? In control Prx1CreTg/+;Gatafl/+;RosamTmG/+ embryos, muscle progenitors migrate into and develop completely surrounded by GFP+ PPF cells (Fig. 6a, i; Supplementary Videos 7, 9). As the PPF cells expand they carry with them the proliferating and differentiating myogenic cells. In contrast, in Prx1CreTg/+;Gata4Δ/fl;RosamTmG/+ mutants, at E12.5 myogenic cells are largely excluded from GFP+Gata4− regions (Fig. 6b, j; Supplementary Videos 8,10). These localized amuscular regions likely result from the mosaic deletion of one Gata4 allele by the Prx1Cre transgene which does not efficiently recombine in all PPF cells at E11.5–12.5 (Supplementary Fig. 2d–i; the other Gata4 allele is deleted in the germline); muscle is excluded from the PPF regions where Gata4 has been deleted during this early time window. Only during this early time are myogenic cells sensitive to Gata4, as deletion of Gata4 via Tcf4Cre29, which causes recombination in PPF fibroblasts primarily after E12.5, does not result in hernias (n = 16/16, data not shown). Altogether these data indicate that early mosaic deletion of Gata4 in PPF cells leads to increased apoptosis and decreased proliferation of muscle progenitors and the development of localized amuscular regions, which ultimately allow herniation (Fig. 7).
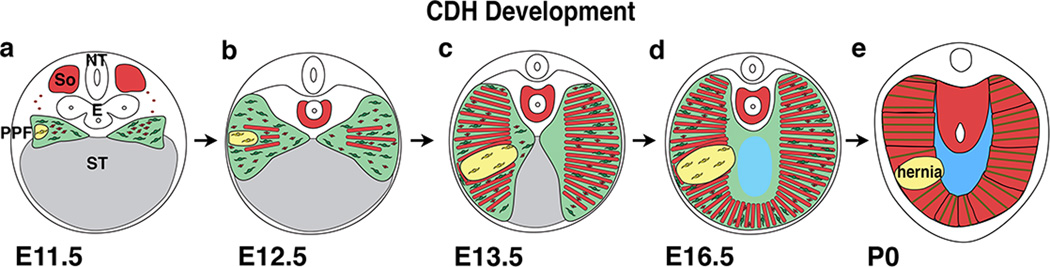
Fig. 7. Model of CDH development.
Our data supports a model whereby CDH arises from early genetic mutations in a subset of PPF-derived muscle connective tissue fibroblasts (a; mutant fibroblasts are yellow, wild-type fibroblasts are green). Mutant fibroblasts clonally expand and inhibit muscle progenitors from developing in these regions (via decreased proliferation and increased apoptosis of muscle progenitors) and results in local regions (shown in yellow) that are amuscular, but contain connective tissue fibroblasts and their associated extracellular matrix (b–d). Amuscular regions are thinner and more compliant than surrounding thicker and stiffer muscularized diaphragm and allow herniation of abdominal contents into the thoracic cavity (d–e).
DISCUSSION
Our study establishes that the PPFs and muscle connective tissue fibroblasts, although previously underappreciated, are critical for the development of the diaphragm and CDH. PPF cells not only give rise to the diaphragm’s muscle connective tissue and the central tendon, but the connective tissue fibroblasts control the morphogenesis of the diaphragm’s muscle. Our finding that the connective tissue regulates muscle development has precedence, as muscle connective tissue fibroblasts regulate the pattern of limb muscles28 and the fiber-type of limb and diaphragm muscles29. Completely novel is our finding that the active, apparently collective, cell migration of connective tissue fibroblasts across the liver’s surface controls the expansion of muscle progenitors and overall diaphragm morphogenesis.
We also definitively demonstrate for the first time that defects in the muscle connective tissue fibroblast component of the PPFs are a cellular source of CDH and Gata4 null mutations in these cells cause CDH. Surprisingly, we show that hernias do not result from defects in PPF cell migration and the formation of holes in the diaphragm. Instead, PPF deletion of Gata4 leads to the development of localized amuscular regions through which the growing liver and intestines herniate. In humans, such hernias with connective tissue surrounding the herniated tissue are classified as “sac hernias”6, and we hypothesize that many hernias are covered by connective tissue early in development. Mechanistically, CDH arises when thinner and more compliant amuscular regions develop within the thicker and stiffer muscularized diaphragm and allow herniation. Development of the amuscular regions in mutants results from a marked decrease in cell proliferation, an increase in cell death and, most notably, the localized exclusion of myogenic cells from regions with early PPF deletion of Gata4 and down-regulation of HGF expressed by the muscle connective tissue fibroblasts. Thus we elucidate how defects in the muscle connective tissue are a potent source of CDH. CDH has been associated with over 50 candidate genes and recently several alternative mechanisms of CDH have been described45,46. An important area of future research will be to determine whether CDH is of heterogeneous origin or if any cellular or molecular defect is common to the etiology of CDH.
Our data suggests a novel genetic hypothesis for the origin of CDH. Our finding that CDH derives from localized, weaker amuscular regions that develop specifically where Gata4 has been mutated early in muscle connective tissue fibroblasts suggests that somatic mosaic mutations in fibroblasts may be a genetic feature of some CDH patients. A potential role for somatic mosaicism in CDH has previously been suggested by the largely discordant occurrence of CDH in monozygotic twins47, including twins with an 8p23.1 deletion48, and the finding of genetic mosaicism in CDH patients49,50. Our hypothesis that CDH can arise from somatic mosaic mutations may explain the notable incomplete penetrance and variable expressivity of many CDH-associated CNVs and genetic mutations, particularly Gata411–13,48. Interestingly, in humans Gata4 haploinsufficiency and in mice 70% reduction in Gata4 expression leads with nearly complete penetrance to heart defects12,48,51. In contrast, in humans Gata4 haploinsufficiency is incompletely penetrant for CDH12,48, and in mice Gata4 heterozygotes either do not develop CDH (our data) or do so with low penetrance16,52. In addition, for CDH patients with shared Gata4 missense mutations13 or intronic variants11, the location and severity of hernias is highly variable. Our data suggest the hypothesis that loss of one Gata4 allele confers susceptibility, but is not sufficient to cause CDH; development of CDH requires somatic loss of a second allele in a subset of the connective tissue fibroblasts early during diaphragm morphogenesis. When and where this second allele is deleted determines the size and location of the amuscular region and hernia (i.e. CDH expressivity). In humans, somatic mosaicism in Gata4 mutations and 8p23.1 polymorphisms are well documented53–56. Alternatively, the second somatic mosaic, CDH-causative mutation may not be loss of the second Gata4 allele, but another CDH-associated gene (resulting in non-allelic non-complementation); multiple CDH-associated genes are strongly expressed in the PPF cells, including Zfpm219 and Nr2f218 which are known to genetically interact with Gata4. Thus the genetic architecture underlying CDH may be complex.
In summary, although muscle connective tissue has often been relegated to a role supporting muscle structure and function, we demonstrate that muscle connective tissue fibroblasts dynamically control diaphragm morphogenesis and their interactions with muscle progenitors critically regulate the development of the diaphragm’s muscle and are a source of CDH. The role of mosaicism in human disease has received increasing attention in recent years57. Here we show in mice that mosaic mutations in muscle connective tissue have profound cellular and biomechanical consequences and lead to hernias. We hypothesize that early somatic mosaic mutations are critical for the etiology of CDH in humans, and this hypothesis will be tested in future experiments.
Online Methods
Materials and Methods
Mice
All mice have been previously published. We used Prx1Cre26, Pax3Cre31, and HPRTCre58 Cre alleles; RosaLacZ27 and RosamTmG59 Cre-responsive reporter alleles; and Pax3SpD32, Gata4fl33 mutant alleles. Gata4del/+ mice were generated by breeding Gata4fl mice to HPRTCre mice. Mice were back-crossed on to C57/Bl6J background. No statistical method was used to predetermine sample size; all animals were included; and the experiments were not randomized. Animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee at the University of Utah.
Immunofluorescence, β-Galactosidase staining, and microscopy
For section immunofluorescence, embryos were fixed, embedded, cryosectioned, and immunostained as described previously29. EdU (Life Technologies) was detected per manufacturer’s directions. For whole mount immunofluorescence, embryos were fixed 24 h in 4% PFA, dissected, incubated 24 hours in Dent’s bleach (1:2 30% H2O2:Dent’s fix), and stored in Dent’s fix (1:4 DMSO:MeOH) for at least 5 days. Embryos were washed in PBS, blocked 1h in 5% serum + 20% DMSO, incubated in primary antibody at room temperature 48 h, washed in PBS, incubated in secondary antibody 48 h, washed in PBS, and subjected to EdU reaction 1 h. Diaphragms were cleared in BABB (33% Benzyl Alcohol, 66% Benzyl Benzoate). AP-conjugated mouse IgG1 α My32 was incubated for 48 h and detected with 250 µg/ml NBT and 125 µg/ml BCIP (Sigma) in NTMT.
Cultured cells were fixed for 20 minutes in 4% PFA, blocked in 5% serum in PBS with 0.1% Triton X-100 for 1 h, incubated in primary antibody at 4°C overnight, washed with PBS, and incubated for two hours in secondary antibody. Following antibody staining, EdU was detected as above. Cells were incubated in 0.4 ug/ml Hoechst for 5 minutes and mounted with Fluoromount-G.
Antibodies are listed in Supplementary Table 1.
For whole-mount β-galactosidase staining, embryos were fixed 1.5 hours in 4% PFA + 2mM MgCl2. Diaphragms were dissected, washed in PBS and rinse buffer (100mM Sodium Phosphate, 2mM MgCl2, 0.01% Na Deoxycholate, 0.02% Ipegal) and stained 16 h at 37°C in X-gal staining solution (5mM potassium ferricyanide, 5 mM potassium ferrocyanide, 1 mg/ml Xgal).
Fluorescent images were taken on a Nikon A1 confocal. Optical stacks of whole-mounts were rendered using FluoRender60. β-galactosidase stained embryos were imaged with a Qimaging camera. All analyses were performed blinded.
Cell Culture
Fibroblasts were isolated from E15.5 Gata4fl/fl;RosamTmG/+ or RosamTmG/+ diaphragms, cultured, and expanded 3 weeks. 4 µM TAT-Cre (M Hockin, MR Capecchi) was added to induce recombination. EdU was given 4 h prior to fixation and immunofluorescence. For co-cultures, fibroblasts and myoblasts were isolated from E15.5 Prx1CreTg/+;Gata4del/fl;RosamTmG/+ or Prx1Cre+/+;Gata4del/fl;RosamTmG/+ diaphragms, cultured 2 days, and given EdU 1 h prior to fixation and immunofluorescence. Statistical significance was performed with 2-tailed Student’s t test.
Diaphragm Explants
E12.5 Prx1CreTg/+;RosamTmG/+ mice were harvested and trimmed to only include the diaphragm with attached ribs, hind limbs, and liver. Explanted diaphragms were cultured 2–6 h in 100% horse serum, at 37°C, in the presence of 5% CO2. Explants were imaged on a Bruker (Prairie) 2 Photon microscope, and 4-dimensional datasets were rendered and visualized using FluoRender60. Individual cells were tracked using FluoRender.
Modeling
To gain insights into the biomechanics of herniation, we used finite element modeling, a computational method for assembling the response of a complex system from individual contributions of discrete elements36. The finite element model of hernia development was based on an E16.5 Prx1CreTg/+;Gata4Δ/fl herniated mouse diaphragm. The geometry of the diaphragm was based on surfaces created from segmenting microscopy images. Surfaces were discretized with quadratic-tetrahedral elements, and adequacy of the spatial discretization was confirmed with a mesh convergence study. Meshes were created with ANSA software (BETA CAE Systems USA) and analyzed with FEBio36. For the simulations, the rib cage surrounding the diaphragm was rigidly constrained in the model (kept immobile), while a uniform pressure of 380 Pa was applied to simulate the pressure applied by the liver. The pressure was chosen so that the bulge height of the muscle region predicted by the model matched the height measured in the experiment. The muscle was represented with an isotropic hyperelastic Veronda-Westmann constitutive equation61 with coefficients (C1 = 2.1 MPa, C2 = 0.1, K = 10) based on published data37. The connective tissue of the hernia was represented with an isotropic hyperelastic Mooney-Rivlin constitutive equation62 using either the coefficients C1 = 0.003 MPa, K = 1 or C1 = 0.01 MPa, K = 10.
MicroCT Analysis of Lungs
Embryos were fixed in 4% PFA in PBS overnight. Lungs were removed, pre-treated with 25% Lugol’s solution for 1 hour, and then soaked in PBS for 1 hour prior to imaging. CT images consisting of 360 degrees and 600 projections were acquired by using an Inveon trimodality PET/SPECT/CT scanner (Siemens Preclinical Solutions, Knoxville, TN). The exposure time was 2.9 seconds with detector settings at 80 kVp and 150 µA. Data was reconstructed onto a 1792×1792×2688 image matrix using the COBRA software package (Exxim Computing Corporation, Pleasanton, CA). The effective image voxel size was 33.6 µm isotropic. Reconstructed images were analyzed using Osirix software.
Supplementary Material
ACKNOWLEDGEMENTS
We thank C. Rodesch at the U of Utah Imaging Core for help with microscopy, Samer Merchant for help with microCT imaging, Y. Wan and C. Hansen for Fluorender analysis, M Hockin and MR Capecchi for Cre protein, and M Colasanto, N Elde, LB Jorde, A Keefe, A Letsou, LC Murtaugh, CJ Tabin for critical comments on the manuscript. AJM was supported by U of Utah Graduate Fellowship, FEBio is supported by NIH R01GMo83925 to JA Weiss and GA Ateshian and Fluorender is supported by NIH R01-GM098151 to C Hansen. This research was supported by NIH R01 HD053728 and March of Dimes FY12-405 to GK.
Footnotes
AUTHOR CONTRIBUTIONS
A. J. M. conducted experiments, analyzed data, and wrote the manuscript. Z. J. F. conducted 2 photon experiments. B. J. E. and J. A. W. contributed FEM analysis. J. A. L. managed mouse colony. G.K. conducted experiments, analyzed data, and wrote the manuscript.
The authors declare no competing financial interests.
REFERENCES
- 1.Perry SF, Similowski T, Klein W, Codd JR. The evolutionary origin of the mammalian diaphragm. Respir Physiol Neurobiol. 2010;171:1–16. doi: 10.1016/j.resp.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Campbell EJM, Agostoni E, Newsom Davis J. The Respiratory Muscles: Mechanics and Neural Control. London: Lloyd-Luke; 1970. [Google Scholar]
- 3.Merrell AJ, Kardon G. Development of the diaphragm -- a skeletal muscle essential for mammalian respiration. FEBS J. 2013;280:4026–4035. doi: 10.1111/febs.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raval MV, Wang X, Reynolds M, Fischer AC. Costs of congenital diaphragmatic hernia repair in the United States-extracorporeal membrane oxygenation foots the bill. J Pediatr Surg. 2011;46:617–624. doi: 10.1016/j.jpedsurg.2010.09.047. [DOI] [PubMed] [Google Scholar]
- 5.Torfs CP, Curry CJ, Bateson TF, Honore LH. A population-based study of congenital diaphragmatic hernia. Teratology. 1992;46:555–565. doi: 10.1002/tera.1420460605. [DOI] [PubMed] [Google Scholar]
- 6.Pober BR. Overview of epidemiology, genetics, birth defects, and chromosome abnormalities associated with CDH. Am J Med Genet C Semin Med Genet. 2007;145C:158–171. doi: 10.1002/ajmg.c.30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pober BR, et al. Infants with Bochdalek diaphragmatic hernia: sibling precurrence and monozygotic twin discordance in a hospital-based malformation surveillance program. Am J Med Genet A. 2005;138A:81–88. doi: 10.1002/ajmg.a.30904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holder AM, et al. Genetic factors in congenital diaphragmatic hernia. Am J Hum Genet. 2007;80:825–845. doi: 10.1086/513442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veenma DC, de Klein A, Tibboel D. Developmental and genetic aspects of congenital diaphragmatic hernia. Pediatr Pulmonol. 2012;47:534–545. doi: 10.1002/ppul.22553. [DOI] [PubMed] [Google Scholar]
- 10.Russell MK, et al. Congenital diaphragmatic hernia candidate genes derived from embryonic transcriptomes. Proc Natl Acad Sci U S A. 2012;109:2978–2983. doi: 10.1073/pnas.1121621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arrington CB, et al. A family-based paradigm to identify candidate chromosomal regions for isolated congenital diaphragmatic hernia. Am J Med Genet A. 2012;158A:3137–3147. doi: 10.1002/ajmg.a.35664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longoni M, et al. Congenital diaphragmatic hernia interval on chromosome 8p23.1 characterized by genetics and protein interaction networks. Am J Med Genet A. 2012;158A:3148–3158. doi: 10.1002/ajmg.a.35665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu L, et al. Variants in GATA4 are a rare cause of familial and sporadic congenital diaphragmatic hernia. Hum Genet. 2013;132:285–292. doi: 10.1007/s00439-012-1249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo CT, et al. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 15.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 16.Jay PY, et al. Impaired mesenchymal cell function in Gata4 mutant mice leads to diaphragmatic hernias and primary lung defects. Dev Biol. 2007;301:602–614. doi: 10.1016/j.ydbio.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendelsohn C, et al. Function of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development. 1994;120:2749–2771. doi: 10.1242/dev.120.10.2749. [DOI] [PubMed] [Google Scholar]
- 18.You LR, et al. Mouse lacking COUP-TFII as an animal model of Bochdalek-type congenital diaphragmatic hernia. Proc Natl Acad Sci U S A. 2005;102:16351–16356. doi: 10.1073/pnas.0507832102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ackerman KG, et al. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 2005;1:58–65. doi: 10.1371/journal.pgen.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coles GL, Ackerman KG. Kif7 is required for the patterning and differentiation of the diaphragm in a model of syndromic congenital diaphragmatic hernia. Proc Natl Acad Sci U S A. 2013;110:E1898–E1905. doi: 10.1073/pnas.1222797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allan DW, Greer JJ. Embryogenesis of the phrenic nerve and diaphragm in the fetal rat. J Comp Neurol. 1997;382:459–468. doi: 10.1002/(sici)1096-9861(19970616)382:4<459::aid-cne3>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 22.Babiuk RP, Zhang W, Clugston R, Allan DW, Greer JJ. Embryological origins and development of the rat diaphragm. J Comp Neurol. 2003;455:477–487. doi: 10.1002/cne.10503. [DOI] [PubMed] [Google Scholar]
- 23.Dietrich S, et al. The role of SF/HGF and c-Met in the development of skeletal muscle. Development. 1999;126:1621–1629. doi: 10.1242/dev.126.8.1621. [DOI] [PubMed] [Google Scholar]
- 24.Greer JJ, et al. Structure of the primordial diaphragm and defects associated with nitrofen-induced CDH. J Appl Physiol. 2000;89:2123–2129. doi: 10.1152/jappl.2000.89.6.2123. [DOI] [PubMed] [Google Scholar]
- 25.Ackerman KG, Greer JJ. Development of the diaphragm and genetic mouse models of diaphragmatic defects. Am J Med Genet C Semin Med Genet. 2007;145C:109–116. doi: 10.1002/ajmg.c.30128. [DOI] [PubMed] [Google Scholar]
- 26.Logan M, et al. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 27.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 28.Kardon G, Harfe BD, Tabin CJ. A Tcf4-positive mesodermal population provides a prepattern for vertebrate limb muscle patterning. Dev Cell. 2003;5:937–944. doi: 10.1016/s1534-5807(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 29.Mathew SJ, et al. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development. 2011;138:371–384. doi: 10.1242/dev.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clugston RD, Zhang W, Greer JJ. Gene expression in the developing diaphragm: significance for congenital diaphragmatic hernia. Am J Physiol Lung Cell Mol Physiol. 2008;294:L665–L675. doi: 10.1152/ajplung.00027.2008. [DOI] [PubMed] [Google Scholar]
- 31.Engleka KA, et al. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev Biol. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Vogan KJ, Epstein DJ, Trasler DG, Gros P. The splotch-delayed (Spd) mouse mutant carries a point mutation within the paired box of the Pax-3 gene. Genomics. 1993;17:364–369. doi: 10.1006/geno.1993.1333. [DOI] [PubMed] [Google Scholar]
- 33.Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ackerman KG, et al. Congenital diaphragmatic defects: proposal for a new classification based on observations in 234 patients. Pediatr Dev Pathol. 2012;15:265–274. doi: 10.2350/11-05-1041-OA.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clugston RD, Greer JJ. Diaphragm development and congenital diaphragmatic hernia. Semin Pediatr Surg. 2007;16:94–100. doi: 10.1053/j.sempedsurg.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Maas SA, Ellis BJ, Ateshian GA, Weiss JA. FEBio: finite elements for biomechanics. J Biomech Eng. 2012;134:011005. doi: 10.1115/1.4005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strumpf RK, Humphrey JD, Yin FC. Biaxial mechanical properties of passive and tetanized canine diaphragm. Am J Physiol. 1993;265:H469–H475. doi: 10.1152/ajpheart.1993.265.2.H469. [DOI] [PubMed] [Google Scholar]
- 38.Keijzer R, Liu J, Deimling J, Tibboel D, Post M. Dual-hit hypothesis explains pulmonary hypoplasia in the nitrofen model of congenital diaphragmatic hernia. Am J Pathol. 2000;156:1299–1306. doi: 10.1016/S0002-9440(10)65000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ackerman KG, et al. Gata4 is necessary for normal pulmonary lobar development. Am J Respir Cell Mol Biol. 2007;36:391–397. doi: 10.1165/rcmb.2006-0211RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376:768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- 41.Maina F, et al. Uncoupling of Grb2 from the Met receptor in vivo reveals complex roles in muscle development. Cell. 1996;87:531–542. doi: 10.1016/s0092-8674(00)81372-0. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura K, Hongo A, Kodama J, Hiramatsu Y. The role of hepatocyte growth factor activator inhibitor (HAI)-1 and HAI-2 in endometrial cancer. Int J Cancer. 2011;128:2613–2624. doi: 10.1002/ijc.25606. [DOI] [PubMed] [Google Scholar]
- 43.Rojas A, et al. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol. 2008;28:5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamak A, et al. Cyclin D2 rescues size and function of GATA4 haplo-insufficient hearts. Am J Physiol Heart Circ Physiol. 2012;303:H1057–H1066. doi: 10.1152/ajpheart.00250.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Domyan ET, et al. Roundabout receptors are critical for foregut separation from the body wall. Dev Cell. 2013;24:52–63. doi: 10.1016/j.devcel.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang B, et al. Heparan sulfate deficiency disrupts developmental angiogenesis and causes congenital diaphragmatic hernia. J Clin Invest. 2014;124:209–221. doi: 10.1172/JCI71090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veenma D, et al. Copy number detection in discordant monozygotic twins of Congenital Diaphragmatic Hernia (CDH) and Esophageal Atresia (EA) cohorts. Eur J Hum Genet. 2012;20:298–304. doi: 10.1038/ejhg.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wat MJ, et al. Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am J Med Genet A. 2009;149A:1661–1677. doi: 10.1002/ajmg.a.32896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kantarci S, et al. Characterization of the chromosome 1q41q42.12 region, and the candidate gene DISP1, in patients with CDH. Am J Med Genet A. 2010;152A:2493–2504. doi: 10.1002/ajmg.a.33618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veenma D, et al. Comparable low-level mosaicism in affected and non affected tissue of a complex CDH patient. PLoS One. 2010;5:e15348. doi: 10.1371/journal.pone.0015348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pu WT, Ishiwata T, Juraszek AL, Ma Q, Izumo S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol. 2004;275:235–244. doi: 10.1016/j.ydbio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 52.Wat MJ, et al. Mouse model reveals the role of SOX7 in the development of congenital diaphragmatic hernia associated with recurrent deletions of 8p23.1. Hum Mol Genet. 2012;21:4115–4125. doi: 10.1093/hmg/dds241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bosch N, et al. Nucleotide, cytogenetic and expression impact of the human chromosome 8p23.1 inversion polymorphism. PLoS One. 2009;4:e8269. doi: 10.1371/journal.pone.0008269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giglio S, et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet. 2001;68:874–883. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reamon-Buettner SM, Borlak J. GATA4 zinc finger mutations as a molecular rationale for septation defects of the human heart. J Med Genet. 2005;42:e32. doi: 10.1136/jmg.2004.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reamon-Buettner SM, Cho SH, Borlak J. Mutations in the 3'-untranslated region of GATA4 as molecular hotspots for congenital heart disease (CHD) BMC Med Genet. 2007;8:38. doi: 10.1186/1471-2350-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biesecker LG, Spinner NB. A genomic view of mosaicism and human disease. Nat Rev Genet. 2013;14:307–320. doi: 10.1038/nrg3424. [DOI] [PubMed] [Google Scholar]
METHODS REFERENCES
- 1.Logan M, et al. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 2.Engleka KA, et al. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev Biol. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Tang SH, Silva FJ, Tsark WM, Mann JR. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32:199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- 4.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 5.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 6.Vogan KJ, Epstein DJ, Trasler DG, Gros P. The splotch-delayed (Spd) mouse mutant carries a point mutation within the paired box of the Pax-3 gene. Genomics. 1993;17:364–369. doi: 10.1006/geno.1993.1333. [DOI] [PubMed] [Google Scholar]
- 7.Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathew SJ, et al. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development. 2011;138:371–384. doi: 10.1242/dev.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y, Otsuna H, Chien CB, Hansen C. An interactive visualization tool for multi-channel confocal microscopy data in neurobiology research. IEEE Trans Vis Comput Graph. 2009;15:1489–1496. doi: 10.1109/TVCG.2009.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maas SA, Ellis BJ, Ateshian GA, Weiss JA. FEBio: finite elements for biomechanics. J Biomech Eng. 2012;134:011005. doi: 10.1115/1.4005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veronda DR, Westmann RA. Mechanical characterization of skin-finite deformations. J Biomech. 1970;3:111–124. doi: 10.1016/0021-9290(70)90055-2. [DOI] [PubMed] [Google Scholar]
- 12.Strumpf RK, Humphrey JD, Yin FC. Biaxial mechanical properties of passive and tetanized canine diaphragm. Am J Physiol. 1993;265:H469–H475. doi: 10.1152/ajpheart.1993.265.2.H469. [DOI] [PubMed] [Google Scholar]
- 13.Mooney M. A theory of large elastic deformation. Journal of Applied Physics. 1940;11:582–592. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.