Abstract
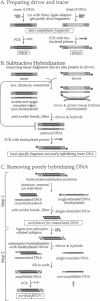
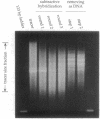
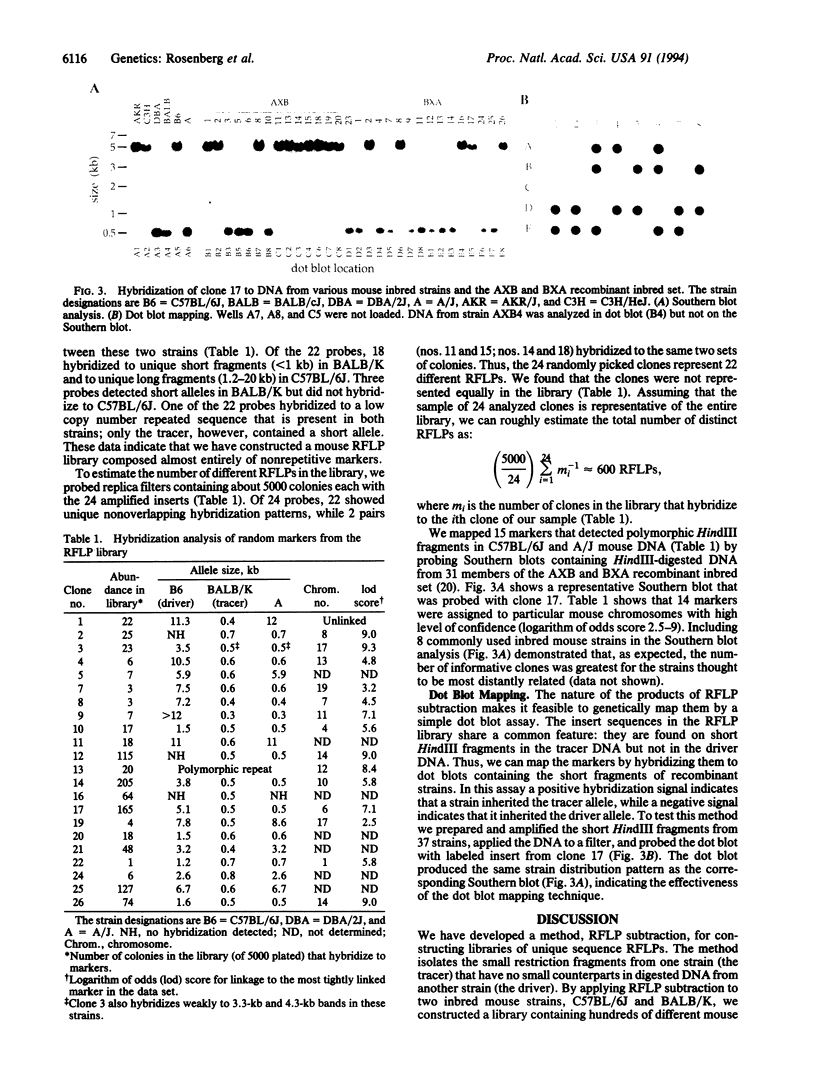
We developed a method, "RFLP subtraction," that isolates large numbers of unique sequence restriction fragment length polymorphisms (RFLPs) in a single experiment. The technique purifies small restriction fragments from one genome containing sequences that reside on large fragments in a related genome. We first isolate samples containing the small restriction fragments from two polymorphic strains. Subtractive hybridization then removes the fragments that are present in both samples. The remaining sequences are RFLPs: they occur on small fragments in one strain but not in the other. Here we use RFLP subtraction to make a library of hundreds of unique sequence RFLPs from two inbred mouse strains. We analyze and map a subset of the RFLPs and show that the genetic linkage of these markers can be rapidly determined by an efficient dot blot mapping technique. Several other potential applications of RFLP subtraction, including isolating region specific markers, are discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bautz E. K., Reilly E. Gene-specific messenger RNA: isolation by the deletion method. Science. 1966 Jan 21;151(3708):328–330. doi: 10.1126/science.151.3708.328. [DOI] [PubMed] [Google Scholar]
- Botstein D., White R. L., Skolnick M., Davis R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980 May;32(3):314–331. [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anollés G., Bassam B. J., Gresshoff P. M. DNA amplification fingerprinting using very short arbitrary oligonucleotide primers. Biotechnology (N Y) 1991 Jun;9(6):553–557. doi: 10.1038/nbt0691-553. [DOI] [PubMed] [Google Scholar]
- Dietrich W., Katz H., Lincoln S. E., Shin H. S., Friedman J., Dracopoli N. C., Lander E. S. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992 Jun;131(2):423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Hammer M. F., Silver L. M. Phylogenetic analysis of the alpha-globin pseudogene-4 (Hba-ps4) locus in the house mouse species complex reveals a stepwise evolution of t haplotypes. Mol Biol Evol. 1993 Sep;10(5):971–1001. doi: 10.1093/oxfordjournals.molbev.a040051. [DOI] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F. M. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993 Aug;4(2):403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Lamar E. E., Palmer E. Y-encoded, species-specific DNA in mice: evidence that the Y chromosome exists in two polymorphic forms in inbred strains. Cell. 1984 May;37(1):171–177. doi: 10.1016/0092-8674(84)90312-x. [DOI] [PubMed] [Google Scholar]
- Lisitsyn N. A., Rosenberg M. V., Launer G. A., Wagner L. L., Potapov V. K., Kolesnik T. B., Sverdlov E. D. A method for isolation of sequences missing in one of two related genomes. Mol Gen Mikrobiol Virusol. 1993 May-Jun;(3):26–29. [PubMed] [Google Scholar]
- Lisitsyn N., Lisitsyn N., Wigler M. Cloning the differences between two complex genomes. Science. 1993 Feb 12;259(5097):946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- Manly K. F., Elliott R. W. RI Manager, a microcomputer program for analysis of data from recombinant inbred strains. Mamm Genome. 1991;1(2):123–126. doi: 10.1007/BF02443789. [DOI] [PubMed] [Google Scholar]
- Marshall J. D., Mu J. L., Cheah Y. C., Nesbitt M. N., Frankel W. N., Paigen B. The AXB and BXA set of recombinant inbred mouse strains. Mamm Genome. 1992;3(12):669–680. doi: 10.1007/BF00444361. [DOI] [PubMed] [Google Scholar]
- Martin G. B., Williams J. G., Tanksley S. D. Rapid identification of markers linked to a Pseudomonas resistance gene in tomato by using random primers and near-isogenic lines. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2336–2340. doi: 10.1073/pnas.88.6.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R. W., Paran I., Kesseli R. V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J. L., Cheah Y. C., Paigen B. Strain distribution pattern of 25 simple sequence length polymorphisms in the AXB and BXA recombinant inbred strains. Mamm Genome. 1992;3(12):705–708. doi: 10.1007/BF00444366. [DOI] [PubMed] [Google Scholar]
- Myers R. M. The pluses of subtraction. Science. 1993 Feb 12;259(5097):942–943. doi: 10.1126/science.8094900. [DOI] [PubMed] [Google Scholar]
- Ostrander E. A., Jong P. M., Rine J., Duyk G. Construction of small-insert genomic DNA libraries highly enriched for microsatellite repeat sequences. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3419–3423. doi: 10.1073/pnas.89.8.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski J. A., Tingey S. V. Genetic diagnostics in plant breeding: RAPDs, microsatellites and machines. Trends Genet. 1993 Aug;9(8):275–280. doi: 10.1016/0168-9525(93)90013-8. [DOI] [PubMed] [Google Scholar]
- Shih T. Y., Martin M. A. A general method of gene isolation. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1697–1700. doi: 10.1073/pnas.70.6.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus D., Ausubel F. M. Genomic subtraction for cloning DNA corresponding to deletion mutations. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1889–1893. doi: 10.1073/pnas.87.5.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J. L., May P. E. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet. 1989 Mar;44(3):388–396. [PMC free article] [PubMed] [Google Scholar]
- Wieland I., Bolger G., Asouline G., Wigler M. A method for difference cloning: gene amplification following subtractive hybridization. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2720–2724. doi: 10.1073/pnas.87.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Kubelik A. R., Livak K. J., Rafalski J. A., Tingey S. V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990 Nov 25;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]