Abstract
Lanthipeptides are a class of ribosomally-produced and post-translationally modified peptides (RiPPs) that possess a variety of biological activities, but typically act as antimicrobial agents (lantibiotics). Haloduracin is a lantibiotic that is composed of two post-translationally modified peptides, Halα and Halβ, which are biosynthesized from the precursor peptides HalA1 and HalA2 by their cognate lanthipeptide synthetases, HalM1 and HalM2, respectively. Co-expression studies of HalM1 and HalM2 with chimeric peptides consisting of the leader peptide of HalA1 and the core peptide of HalA2 (or vice versa) showed that the synthetases require both the cognate leader and core peptides for efficient processing. Investigation of the affinity in vitro showed that binding of the N-terminal leader peptide by HalM2 increases its affinity for the C-terminal core peptide. Thus, the two segments of the precursor peptide HalA2 synergistically bind to HalM2.
At present, lanthipeptides are the largest known family of ribosomally synthesized and post-translationally modified peptide natural products (RiPPs)1 that are produced by bacteria.2 Many lanthipeptides possess antimicrobial activity and are designated lantibiotics.3,4 The signature motifs of lanthipeptides are their lanthionine (Lan) and/or 3-methyl-lanthionine (MeLan) structures. These thioether bridged bisamino acids are generated by the dehydration of specific serine and threonine residues in the precursor peptide to give 2,3-dehydroalanine (Dha) and (Z)-2,3-dehydrobutyrine (Dhb), respectively.5 These unsaturated residues then serve as electrophiles for subsequent intramolecular cyclization, wherein cysteine residues within the peptide attack the Dha and/or Dhb residues in a regio- and stereoselective manner to form the characteristic (Me)Lan residues (Figure 1A). Following the post-translational modification of the core peptide, the leader peptide is proteolytically removed and the mature lanthipeptide is secreted. The general lanthipeptide maturation process is shown in Figure 1B for haloduracin, a class II lantibiotic for which the dehydration and cyclization reactions are catalyzed by a bifunctional synthetase (generically called LanM). For class I lanthipeptides, lanthionine formation is catalyzed by dedicated dehydratase (LanB) and cyclase enzymes (LanC).5
Figure 1.
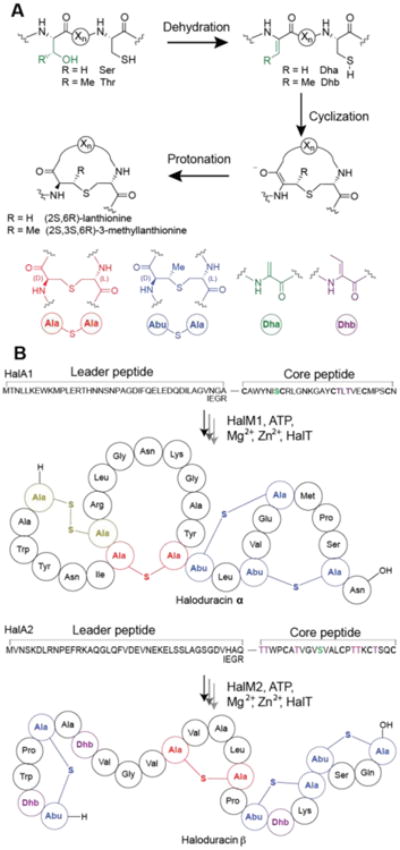
(A) The thioether bridges in lanthipeptides are formed by dehydration of Ser and Thr residues followed by a Michael-type addition of Cys residues to the dehydrated residues, Dha and/or Dhb. A shorthand notation is used, where lanthionine rings are red and methyl-lanthionine rings are blue. Dha is depicted in green and Dhb in purple. Abu, 2-aminobutyric acid; Dha, dehydroalanine; Dhb, dehydrobutyrine. (B) The general maturation process of Halα and Halβ. HalT is a conserved bifunctional lantibiotic transporter and protease that removes the leader peptide. In the HalM activity assays the last four amino acids in the leader peptide were replaced with the Factor Xa cleavage recognition site (IEGR) to allow leader peptide removal.
Haloduracin is a two-component lantibiotic produced by Bacillus halodurans C-125.6,7 In this organism, two lantibiotic synthetases, HalM1 and HalM2, process two different ribosomally-synthesized peptide substrates (HalA1 and HalA2, respectively) into the mature lanthipeptides (termed Halα and Halβ, respectively). Halα and Halβ function synergistically with 1:1 stoichiometry to exert antimicrobial activity.8 The two synthetases are very selective for their native substrate peptides in that HalM1 does not modify HalA2, and HalM2 only very poorly processes HalA1.6,9
As with all lanthipeptides, the full-length precursor peptide (generically termed LanA) can be divided into an N-terminal leader and C-terminal core peptide. Although the exact molecular mechanism is still not understood, the leader peptide has been proposed to play a role in assembling and/or activating the lanthipeptide biosynthetic enzymes. In addition, the leader peptide can act as a secretion signal and can reduce autotoxicity of the posttranslationally modified peptide to the producing organism.10-12 Recently, in another class of RiPPs, the ArgD leader peptide involved in the biosynthesis of the autoinducing peptide in Staphylococcus aureus was shown to have cytolytic and amyloidogenic properties after it was cleaved from the mature peptide, 13 suggesting that leader peptides may also have biological roles after RiPP biosynthesis.
The core peptide is the site of the post-translational modifications that give the lanthipeptides their unique structures and biological activities. To date, the molecular interactions between the leader and core peptide sequences and their cognate synthetases are poorly understood. Whereas site-directed mutagenesis studies have identified residues in the leader peptide that are important for dehydration and cyclization, 14-22 the manner in which the enzymes bind the leader and core peptides and the effects on catalysis are not known. Very recently, the first insights into leader peptide binding were obtained from a co-crystal structure of the class I lanthipeptide dehydratase NisB and its substrate NisA.23 Unfortunately, crystal structures of the bifunctional class II lanthipeptide synthetases are not available.
Among the models proposed for the effect of the leader peptide on LanM activity, we have favored the conformational selection model.11 In this model, the LanM enzyme exists predominantly in two conformations (active and inactive, Figure 2), with the inactive conformation being the dominant species in the absence of the precursor peptide. Binding of the LanA leader peptide to the active conformation of LanM would then shift the equilibrium towards the active state of LanM. In this leader peptide-docked Michaelis complex, the core peptide may gain access to the active site(s) of the lanthipeptide synthetase because of an increased effective concentration of the core peptide. Increased effective concentration is important because the shape of the core peptide changes considerably with each posttranslational modification, and hence the intrinsic affinity of the core peptide and its modified intermediates for the active sites is anticipated to be weak. Leader peptide binding may thus help the synthetase overcome weak core peptide binding affinity to allow a LanM synthetase to install multiple (Me) Lan rings in the same precursor peptide. Given the relaxed substrate specificity of many lanthipeptide synthetases, this conformational selection mechanism may also help to prevent non-specific post- translational processing of other cellular proteins.
Figure 2.
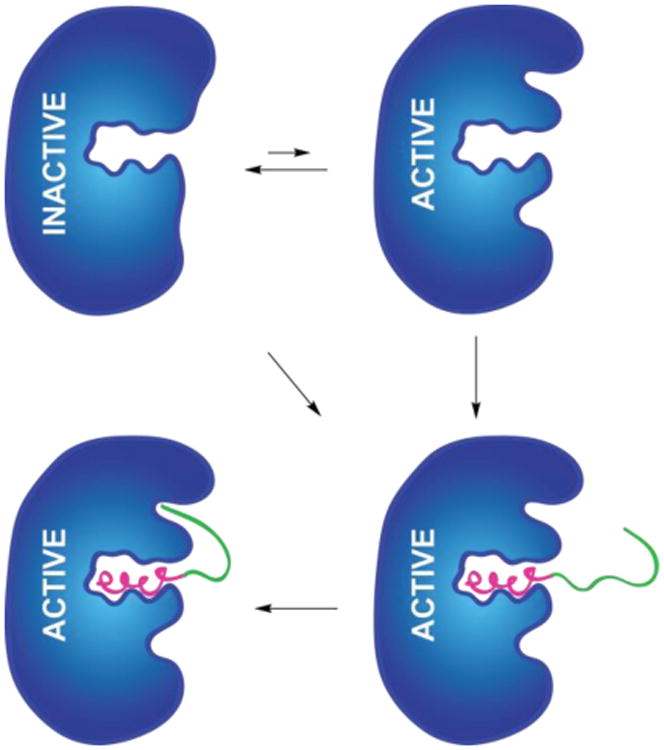
A putative model for the activation of a LanM lanthipeptide synthetase. Pink and green represent the leader and core peptide, respectively. Blue represents the lanthipeptide synthetase, with three possible binding pockets (dehydration active site at top, leader peptide binding in the center, and cyclization active site at bottom; the locations of the active sites are arbitrarily chosen). The diagonal arrow represents a model in which leader peptide binding is required for a conformational change that generates productive core peptide binding sites.
The model in Figure 2 predicts that two separate binding events (i.e. leader binding to a LanM docking site and core binding to the LanM active sites) should contribute to LanM catalytic efficiency. Substrate binding studies have not been reported previously for any class II lanthipeptide synthetases. For the class I cyclase NisC, substrate binding studies have shown that the leader peptide contributes nearly all of the substrate binding affinity as binding of the core peptide without the leader peptide attached was not observed.22 These findings are in agreement with a large number of studies that have shown that non-lanthipeptide sequences attached to leader peptides are processed by both class I and class II biosynthetic machinery, 24-28 suggesting that leader peptide binding is sufficient for activity, and by extrapolation for core peptide binding. However, recent studies in which the leader and core peptides were provided in trans led to partial activity on the core peptide, indicating that lanthipeptide biosynthetic proteins must possess some affinity for the core peptides of their LanA substrates.29,30 The model in Figure 2 provides a potential explanation if leader peptide binding increases the affinity of the protein for the core peptide. Thus far, the binding affinities of the leader and core peptides have not been systematically determined for any LanM enzyme, and the possibility of synergism in binding of the leader and core peptides has not been investigated for any lanthipeptide biosynthetic enzyme. We report here the substrate specificities of the haloduracin synthetases HalM1 and HalM2 using a variety of wild-type and chimeric HalA1 and HalA2 precursor peptides as well as the binding affinities of the HalA2 leader and core peptides, demonstrating synergism in the two binding events.
Results and Discussion
HalM1 and HalM2 assays with chimeric substrates
To probe the leader and core peptide specificity of the two synthetases we generated two chimeric substrates consisting of the HalA1 leader peptide and HalA2 core peptide (HalA1-A2) or the HalA2 leader peptide and HalA1 core peptide (HalA2-A1). These chimeras as well as the HalA1 and HalA2 peptides contained a Factor Xa cleavage site between the core and leader sequences to allow facile removal of the leader peptide. They were expressed in Escherichia coli and purified as N-terminally His6-tagged peptides using immobilized metal affinity chromatography (IMAC). N-terminally His6-tagged HalM1 and HalM2 were also expressed and purified. Since previous studies on LanM enzymes have shown that the substrates containing His6-tags do not affect catalysis, all experiments were performed with the tag attached, which greatly increases the solubility of the peptides. To verify that the His-tag did not affect the activity in this system, tagged and non-tagged HalA2 peptides were incubated with HalM2 and ATP, and the activity towards the two substrates was monitored over time by matrix-assisted laser-desorption time-of-flight (MALDI ToF) mass spectrometry (MS). The data showed, that indeed HalM2 has very similar activity with His6-tagged HalA2 and non-tagged HalA2 (Supporting Figure 1). From here onwards, all substrates discussed were His-tagged.
As expected, incubation of HalM1 with its cognate substrate HalA1 and ATP resulted in fully modified peptide as the major product as determined by MALDI ToF MS (three dehydrations, Supporting Figure 2). Similarly, co-expression of HalM1 with HalA1 in E. coli provided fully modified HalA1 (Supporting Figure 3). When HalM1 was supplied with HalA2 in vitro under the same conditions, dehydrations were not detected (Supporting Figure 2). Incubation of HalM1 with HalA1-A2 in the presence of ATP resulted in dehydration of five of the possible seven residues in the major product (Supporting Figure 2), whereas incubation of HalM1 with HalA2-A1 resulted in only one dehydration in the major product (Supporting Figure 2). These data support the expected activation of HalM1 by its cognate leader peptide, such that it can partially process a foreign core peptide. Furthermore, they also are consistent with weak recognition of the cognate core peptide by the enzyme, resulting in a low level of activity with HalA2-A1.
HalM1 was also co-expressed separately with the HalA1-A2 or HalA2-A1 chimeric substrates in E. coli, and the results correlated with the in vitro data, except for the HalA2-A1 substrate for which very little activity was detected (Supporting Figure 3). The low activity of the latter chimeric peptide may be caused by poor solubility, which was observed for all peptides containing the HalA1 core peptide, or because of lower concentrations of HalA2-A1 in E. coli compared to the in vitro experiments.
Next, we carried out a similar set of experiments with the lanthipeptide synthetase HalM2. As expected, incubation with its cognate substrate HalA2 and ATP resulted in predominantly seven dehydrations in vitro (Supporting Figure 4). More surprisingly, incubation with HalA1 resulted in predominantly one dehydration (Table 1, Supporting Figure 4). This finding suggests that HalM2 is more active or has a more relaxed substrate specificity than HalM1. Similarly, incubation of HalM2 with the chimeric substrate HalA1-A2 resulted in up to 6 dehydrations (Supporting Figure 4). This observation suggests that either HalM2 has considerably higher affinity for its cognate core peptide than HalA1, or that the HalA1 leader peptide can also bind to HalM2, or both (vide infra). Lastly, HalM2 was incubated with HalA2-A1 and the major product was dehydrated twice, with a peptide with three dehydrations observed as a minor product (Supporting Figure 4). Qualitatively similar results were obtained by co-expression of these substrates with HalM2 in E. coli (Supporting Figure 5). The dehydration assays of HalM1 and HalM2 with the two chimeric peptides suggest that either the enzymes have some affinity for the cognate core peptides, resulting in partial processing in the absence of the cognate leader peptide, or the enzymes have affinity for the non-cognate leader peptide. Regardless, it is clear that full dehydration activity is only observed when both cognate leader and core peptides are present, suggesting a synergistic mechanism of recognition.
Table 1. Dehydration of various substrates by HalM1 and HalM2 in vitro.
| Substrate | Possible number of dehydrations | HalM1 (major product) | HalM2 (major product) |
|---|---|---|---|
| HalA1 | 3 | 3 | 1 |
| HalA1-A2 | 7 | 5 | 6 |
| HalA2 | 7 | 0 | 7 |
| HalA2-A1 | 3 | 1 | 2 |
Cyclization cannot be detected directly by mass spectrometry since the Michael-type additions do not result in a change in mass. Therefore, N-ethylmaleimide (NEM) assays were carried out to detect free thiols, which indirectly reports on cyclization activity. The results of the NEM assays with the various co-expression products are shown in Supporting Figure 6 and summarized Supporting Tables 5 and 6. As anticipated,6 full cyclization was only observed when dehydration was complete (i.e. in the reactions of HalM1 with HalA1, and HalM2 with HalA2). HalM1 did not dehydrate HalA2 and as expected a peptide with 4 NEM alkylation adducts was observed. Similarly, the two chimeric peptides showed mostly fully alkylated (i.e. non-cyclized) peptide after co-expression with HalM1. Some minor dehydrated species showed partial alkylation, implying some cyclization had taken place. Although interpretation of these results is complicated by the low level of dehydration observed with some of the peptides (precluding cyclization), HalM1 did not appear to have robust cyclization with any of the peptides except its canonical substrate HalA1.
As observed with the dehydration assays, HalM2 appears to display stronger cyclization activity towards the various peptides than HalM1. Treatment of several of the partially dehydrated peptides with NEM resulted in less-than-maximal alkylations, implying that some of these peptides were partially cyclized. These findings are consistent with a recent kinetic study on HalM2 that demonstrated that cyclization commences before dehydration is completed.31
Given the higher level of activity observed for HalM2 and the higher solubility of the HalA2 peptide compared to HalA1, all further experiments were conducted with HalM2.
In trans activity of HalM2
Based on the model in Figure 2, a small population of the active conformation of the enzyme would be present in the absence of leader peptide. Therefore, HalM2 was incubated with the HalA2 core peptide and the products analyzed by MALDI-ToF MS. As expected, the majority of peptide was unmodified, but a small amount of mono-dehydrated product was also observed (Figure 3A). When the HalA2 leader and core peptides were incubated with HalM2 in trans up to six dehydrations were observed (Figure 3B). Although not conclusive, these findings are less easily explained by a conformational change induced by leader peptide binding that is required for core peptide recognition (diagonal arrow, Figure 2), and are more consistent with the conformational selection model in Figure 2, because leader peptide binding would increase the concentration of active HalM2.
Figure 3.
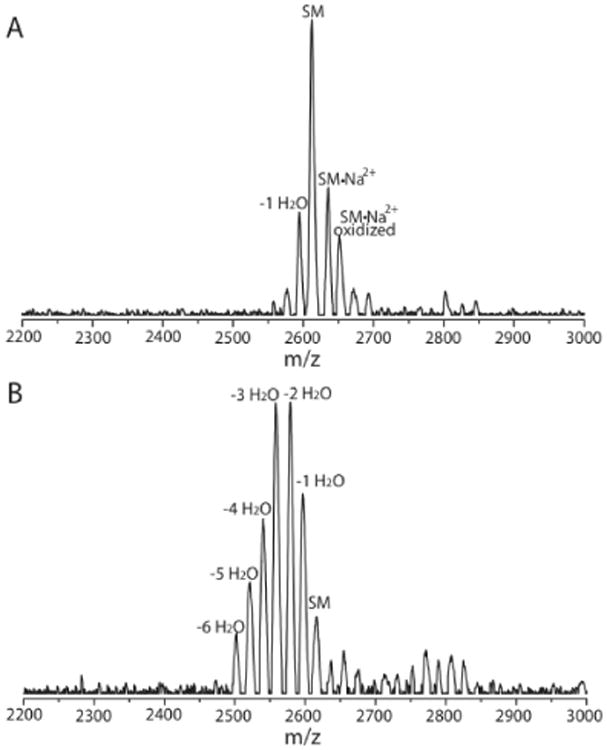
MALDI-ToF MS of the HalA2 core peptide after treatment with (A) HalM2 and (B) HalM2 in the presence of the HalA2 leader peptide. Expected and experimental m/z values are listed in Supporting Table 4. SM indicates starting material.
HalM2 fluorescence polarization binding studies
The activity assays report on overall activity but not on the actual binding events. Knowledge on substrate binding would be valuable, especially to distinguish between the three possibilities offered above for the activity observed for non-cognate leader and core peptides. Fluorescence polarization binding studies were therefore carried out to probe the affinities of various peptides for HalM2. A Cys residue was introduced by site-directed mutagenesis at the N-terminus of the His-tag of HalA2 and the HalA2 leader peptide (HalA2LP). After over-expression in E. coli and purification by IMAC, the N-terminal Cys was used to ligate these peptides by native chemical ligation (NCL)32 to a short synthetic peptide containing a 5-carboxyfluorescein group (5-FAM), a Strep tag sequence, and a C-terminal thioester (Supporting Figure 7). The NCL products were purified in two steps with a HisTrap column and Strep-Tactin Superflow Plus beads (Supporting Figures 8-10). The HalA2 core peptide containing a C-terminal 5-FAM was synthesized by peptide synthesis. This peptide did not contain a His6-tag.
The affinity of HalM2 for the fluorescently-labeled HalA2 and HalA2LP was similar (Kd = 1.9 ± 0.5 μM and 3.7 ± 1.3 μM, respectively, Figure 4A and B). Very weak affinity was observed for the HalA2 core peptide (Kd >500 μM; Figure 4C), indicating that the interactions between the HalA2 leader sequence and HalM2 are largely responsible for peptide binding under these conditions. Similar results were also reported for the class I cyclase NisC.22
Figure 4.
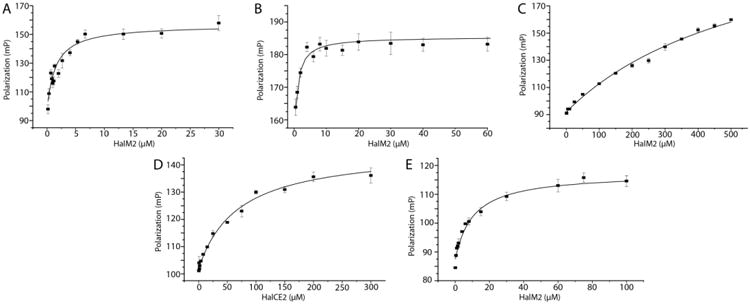
Fluorescence polarization was monitored to determine the dissociation constants of the leader and core peptides to the synthetase. (A) HalM2 was titrated into fluorescein-labeled HalA2, (B) HalM2 was titrated into fluorescein-labeled HalA2 leader peptide, (C) HalM2 was titrated into 5-FAM labeled HalA2 core peptide, (D) HalCE2 was titrated into 5-FAM labeled HalA2 core peptide, and (E) HalM2 was titrated into 5-FAM labeled modified HalA2 peptide. All experiments were performed in at least triplicate and fitted to a hyperbolic curve and a dose-response curve in order to extrapolate the Kd. The error bars represent the standard deviation from the average of the triplicate measurements.
The conformational selection model in Figure 2 in conjunction with the observed in trans activity predicts that binding of the leader peptide to the enzyme might increase the binding affinity for the core peptide. Attempts to test this hypothesis by measuring the binding of the core peptide to HalM2 in the presence of the leader peptide were unsuccessful because of problems with solubility. However, we were able to test the model with a variant of HalM2 in which the leader peptide of HalA2 was covalently attached to the protein. We have previously shown for the related class II lacticin 481 synthetase (LctM) that covalent attachment of the leader peptide results in a Constitutively active Fusion enzyme that we termed a ConFusion enzyme (LctCE) that displayed much higher activity towards the core peptide than when the leader peptide was provided in trans.33 Therefore, a HalM2 variant named HalCE2 was designed in which the HalA2 leader peptide was attached to the N-terminus of the HalM2 enzyme though a (GS)15 linker (the optimized linker length for LctM). This enzyme indeed was able to dehydrate the core peptide, but the activity was not as strong as for the LctCE protein (Supporting Figure 11). The difference may be related to suboptimal positioning of the leader peptide attached to the N-terminus of HalM2. Regardless, because activity was observed, the dissociation constant of the HalA2 core peptide with HalCE2 was measured using the C-terminally fluorescently labeled core peptide. As expected based on the model in Figure 2, the HalA2 core peptide bound considerably tighter to HalCE2 (Kd = 85 ± 12 μM) in comparison to wild-type HalM2 (Kd > 500 μM). This observation is the first experimental evidence for synergism in binding of the core and leader peptides and suggests that the conformation of the enzyme containing the bound leader peptide is different from that of the apo-enzyme.
The observed synergism also may provide a mechanism for product release. If leader peptide binding was the only driving factor in substrate recognition, product inhibition could be a problem. To test the affinity of the product for HalM2, the HalA2 variant containing an N-terminal Cys was co-expressed with HalM2 in E. coli to obtain fully modified peptide with 4 rings, as verified by MALDI-ToF MS, N-ethylmaleimide alkylation assay, and tandem electrospray ionization (ESI)MS (Supporting Figure 12). The purified product was conjugated to 5-FAM maleimide to obtain a fluorescently-labeled, fully-modified HalA2 peptide. The affinity of HalM2 for modified HalA2 was roughly 5-fold weaker (Kd = 10.4 ± 1.7 μM) than for unmodified HalA2 (Figure 4E). A similar decrease in affinity has also been reported for the class I dehydratase NisB for the fully cyclized NisA peptide,21 but a difference in affinity for unmodified and fully modified NisA peptide was not observed for the class I cyclase NisC.22
The class I NisA leader peptide contains a conserved FNLD motif that previous studies have shown is required for NisB and NisC binding.21-23 HalA2 does not contain the FNLD box, therefore, it is likely HalM2 recognizes the HalA2 leader peptide in a different manner. As a negative control, HalM2 was titrated into fluorescein-labeled NisA leader peptide and, as anticipated, binding was not observed (Supporting Figure 13).
HalM2 fluorescence polarization displacement studies
Because of the unexpected activity observed in the assays of HalM2 with HalA1 and HalA1-A2, we probed the affinity of HalM2 for the HalA1 peptide and its derivatives via displacement assays. To first validate the assay, increasing concentrations of unlabeled HalA2 peptide were added to the complex of HalM2 with fluorescein-labeled HalA2. The decrease in polarization signal demonstrated that unlabeled HalA2 peptide displaced the fluorescein-labeled HalA2 peptide from HalM2, providing an independent determination of the affinity that agrees well with that determined by direct titration of fluorescein labeled HalA2 (Figure 5A and Table 3).
Figure 5.
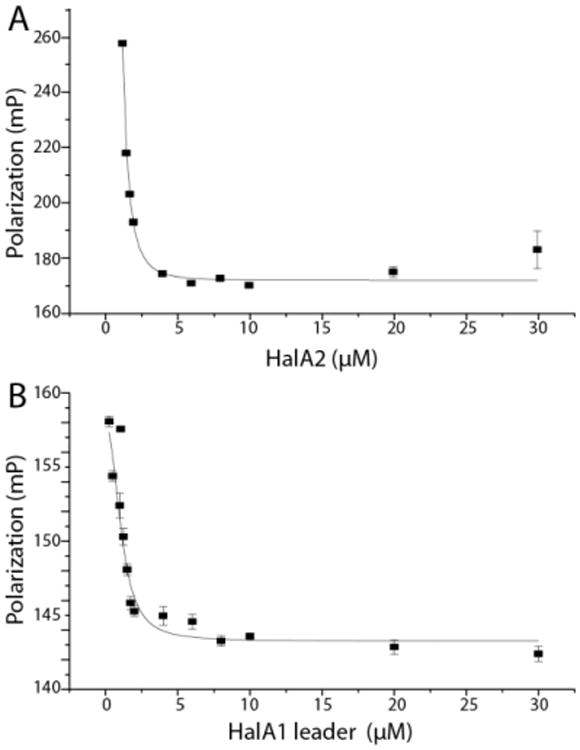
(A) Fluorescein-HalA2 displacement from HalM2 by unlabeled HalA2. (B) HalA1 leader peptide displaces the fluorescein-HalA2 leader peptide from HalM2. The curves were fit to a dose-response curve using Origin 9.0 software. The error bars correspond to the standard deviation of the average of the data that was recorded in triplicate.
Table 3.
Apparent binding affinities for HalM2 via displacement assays.
| Complex | Titrant | IC50 | Ki (μM) |
|---|---|---|---|
| HalM2:HalA2 | HalA2 | 1.2 ± 1.1 μM | 1.2 ± 1.1 μM |
| HalM2:HalA2LPa | HalA2LP | 5.6 ± 1.2 μM | 5.5 ± 1.2 μM |
| HalM2:HalA2LP | HalA1LP | 4.1 ± 1.1 μM | 4.1 ± 1.1 μM |
LP = leader peptide
As expected, unlabeled HalA2 leader peptide was also able to displace fluorescein-labeled HalA2 leader peptide from HalM2 (Supporting Figure 14). The affinity of the leader peptide by itself determined in this manner is close to, but slightly lower than, that determined by direct binding assays (Table 3). To test the hypothesis that the greater than anticipated activity of HalM2 towards HalA1 and HalA1-A2 was because of recognition of the HalA1 leader peptide, displacement of the HalA2 leader peptide with the HalA1 leader peptide was investigated (Figure 5B). Interestingly, the HalA1 leader peptide was indeed found to have a similar dissociation constant as the HalA2 leader peptide (Kd = 4.1 ± 1.1 μM). This observation explains why the HalA1-A2 and HalA1 peptides were substrates for HalM2, but not why both peptides were only partially modified by the enzyme.
The latter observation, as well as other findings discussed below, suggests that the enzyme has selectivity for the cognate core peptide. We also showed that the leader peptide of HalA2 has about the same affinity as the full length HalA2 peptide, and that the HalA2 core peptide by itself has very little affinity and is only dehydrated at most once in the absence of the leader peptide. Collectively, these findings reinforce the general notion that most of the binding affinity in lanthipeptide synthetases comes from the leader peptides. The observed increase of the affinity for the core peptide of HalCE2 compared to wild-type HalM2 supports a model in which leader peptide binding changes the conformation of the enzyme, and explains why in trans assays increase the activity over that seen with just the core peptide. Clearly, the observed binding constant of the HalCE2 enzyme for the HalA2 core peptide (85 μM) is still significantly weaker than that observed for the full length HalA2 peptide or the HalA2 leader peptide, once again indicating that the leader peptide contributes most to the affinity.
We attribute the partial activity observed with HalCE2 to suboptimal positioning of the leader peptide on the N-terminus of HalM2. In this regard, HalM2 differs from the lacticin 481 synthetase LctM, which can completely modify its cognate core peptide either in trans or with the leader peptide attached to its N-terminus. It is therefore possible that the observed Kd of 85 μM of the core peptide with HalCE2 underestimates the true affinity and that the affinity for the core peptide upon leader peptide binding is increased more substantially in the context of the wild type enzyme and substrate.
The importance of appropriate positioning of the substrate is also illustrated by other observations in this study. The incomplete modification of HalA1-A2 and HalA1 by HalM2 despite strong affinity for the HalA1 leader peptide shows that the cognate leader peptide may be important for precise placement of the core peptide. In other words, the HalA1 leader peptide may not be able to optimally position the HalA2 core peptide for catalysis by HalM2. Indeed, the HalA1 and HalA2 leader peptides have low sequence homology and different lengths (Figure 1).
One important unresolved aspect of the LanM enzymes that complicates the exact interpretation of binding and activity studies is that the proteins catalyze two reactions in two different active sites. It is currently not known if the dehydration and cyclization activities share one leader peptide binding site or if each has an independent binding site. Neither HalM1 nor HalM2 display sequence homology to known or anticipated leader peptide binding sites.23,34 Thus, binding of the HalA2 substrate could involve four different binding sites (two leader peptide and two core peptide binding sites). Indeed, for the class I nisin synthetase, both the dehydratase NisB and the cyclase NisC independently bind the leader peptide.21,22 Structural studies with and without leader peptide bound to LanM enzymes may provide insights into this question.
Conclusions
In summary, this study reports the first determination of dissociation constants of class II lanthipeptide synthetases and their substrates, as well as fragments thereof. We provide evidence that for the haloduracin synthetase HalM2, most of the affinity of the enzyme for its substrate comes from the leader peptide. We also demonstrated that the enzyme has weak affinity for the core peptide and that this affinity is increased by the presence of the leader peptide. The affinity for the core peptide is still lower than that for the leader peptide, such that the leader peptide determines the overall affinity for the substrate. Finally, precise positioning of the core peptide appears critical for HalM2 activity.
Materials and Methods
Cloning, Protein and Peptide Expression
Detailed procedures are described in the Electronic Supporting Information (ESI). Primer sequences for peptide mutants are included in Supporting Table 1. The fluorescently labeled HalA2 core peptide was obtained from United Biosystems.
HalM1 and HalM2 activity assays
HalM2 (4 μM) reactions were carried out at room temperature in 1 mL solutions containing substrate (40 μM) and 50 mM 4-2-hydroxyethyl-1-piperazineethanesulfonic acid (HEPES, pH 7.5) supplemented with 10 mM MgCl2, 5 mM ATP, and 1 mM TCEP. The reaction was quenched with TFA (final concentration 0.5%) after 12 h. The reaction mixtures were subjected to C4 solid phase extraction (SPE) columns (Grace Vydac) to remove HalM2. The C4 SPE eluant was lyophilized and re-suspended in 50 mM Tris, pH 8.0, then Factor Xa, trypsin or LysC was added dependning on the substrate peptide (see ESI), and the solution was incubated for an additional 6-12 h at room temperature. The modified cleaved substrates were zip-tipped and directly analyzed by MALDI-ToF MS. HalM1 reactions were completed in the same manner as described for HalM2 with the exception of buffer; HalM1 reactions were performed using 50 mM 4-morpholinepropanesulfonic acid (MOPS) rather than 50 mM HEPES.
HalM2 in trans assay
Purified HalA2 core peptide (40 μM) and HalA2 leader peptide (40 μM) were incubated with HalM2 (4 μM) overnight at room temperature in 50 mM HEPES, pH 7.5 supplemented with 10 mM MgCl2, 5 mM ATP, and 1 mM TCEP. The reaction was quenched with TFA (final concentration 0.5% v/v), partially purified by Zip-Tip, and directly analyzed by MALDI-ToF MS.
Fluorescence polarization (FP) studies
All FP experiments were performed on a Synergy H4 Hybrid plate reader (BioTek) and data recorded with Gen5 software. Increasing amounts of HalM2 were added to various fluorescein-labeled peptides (20 nM) in FP buffer (50 mM HEPES, 10 mM MgCl2, 1 mM TCEP, 0.25 mM AMP-PNP, pH 7.5). Each binding assay was repeated in triplicate in a 96 well black special optics polystyrene plate with a clear flat bottom (CoStar 3615). Data analysis was performed using Origin 9.0 and curves were fitted to a dose-dependent and a hyperbolic curve. Displacement assays were completed with HalM2 (1.5 μM) and a fluorescein-labeled peptide (20 nM) in FP buffer. Increasing amounts of unlabeled peptide were added to displace the labeled peptide.
Supplementary Material
Table 2.
HalM2 binding affinities for various substrates.
| Enzyme | Ligand | Kd (μM) |
|---|---|---|
| HalM2 | HalA2 | 1.9 ± 0.5 μM |
| HalM2 | HalA2 leader peptide only | 3.7 ± 1.3 μM |
| HalM2 | HalA2 core peptide only | > 500 μM |
| HalCE2 | HalA2 core peptide only | 85 ± 12 μM |
| HalM2 | modified HalA2 | 10.4 ± 1.7 μM |
Acknowledgments
This work was supported by the US National Institutes of Health (GM 58822 to W.A.V.) and the Howard Hughes Medical Institute (W.A.V.). Mass spectra were determined in part on an instrument purchased with grant S10RR027109-01 from the US National Institutes of Health. We thank C. Thibodeaux, M. Ortega, and M. Walker for supportive scientific discussions, and T. Oman for the HalCE2 clone.
Footnotes
Associated Content: Supporting Information. Description of all molecular biology procedures, protein purifications, and supporting figures. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RE, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Ribosomally Synthesized and Post-translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doroghazi JR, Albright JC, Goering AW, Ju KS, Haines RR, Tchalukov KA, Labeda DP, Kelleher NL, Metcalf WW. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol. 2014;10:963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnell N, Entian KD, Schneider U, Götz F, Zahner H, Kellner R, Jung G. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature. 1988;333:276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- 4.Cotter PD, Hill C, Ross RP. Bacterial lantibiotics: strategies to improve therapeutic potential. Curr Protein Pept Sci. 2005;6:61–75. doi: 10.2174/1389203053027584. [DOI] [PubMed] [Google Scholar]
- 5.Knerr PJ, van der Donk WA. Discovery, biosynthesis, and engineering of lantipeptides. Annu Rev Biochem. 2012;81:479–505. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- 6.McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA. Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc Natl Acad Sci U S A. 2006;103:17243–17248. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawton EM, Cotter PD, Hill C, Ross RP. Identification of a novel two-peptide lantibiotic, Haloduracin, produced by the alkaliphile Bacillus halodurans C-125. FEMS Microbiol Lett. 2007;267:64–71. doi: 10.1111/j.1574-6968.2006.00539.x. [DOI] [PubMed] [Google Scholar]
- 8.Oman TJ, Lupoli TJ, Wang TS, Kahne D, Walker S, van der Donk WA. Haloduracin alpha binds the peptidoglycan precursor lipid II with 2:1 stoichiometry. J Am Chem Soc. 2011;133:17544–17547. doi: 10.1021/ja206281k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper LE, McClerren AL, Chary A, van der Donk WA. Structure-activity relationship studies of the two-component lantibiotic haloduracin. Chem Biol. 2008;15:1035–1045. doi: 10.1016/j.chembiol.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oman TJ, van der Donk WA. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat Chem Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, van der Donk WA. Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products: New Insights into the Role of Leader and Core Peptides during Biosynthesis. Chem Eur J. 2013;19:7662–7677. doi: 10.1002/chem.201300401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plat A, Kuipers A, Rink R, Moll GN. Mechanistic Aspects of Lanthipeptide Leaders. Curr Protein Pept Sci. 2013;14:85. doi: 10.2174/1389203711314020001. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz K, Sekedat MD, Syed AK, O'Hara B, Payne DE, Lamb A, Boles BR. The AgrD N-terminal leader peptide of Staphylococcus aureus has cytolytic and amyloidogenic properties. Infect Immun. 2014;82:3837–3844. doi: 10.1128/IAI.02111-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Meer JR, Rollema HS, Siezen RJ, Beerthuyzen MM, Kuipers OP, de Vos WM. Influence of amino acid substitutions in the nisin leader peptide on biosynthesis and secretion of nisin by Lactococcus lactis. J Biol Chem. 1994;269:3555–3562. [PubMed] [Google Scholar]
- 15.Neis S, Bierbaum G, Josten M, Pag U, Kempter C, Jung G, Sahl HG. Effect of leader peptide mutations on biosynthesis of the lantibiotic Pep5. FEMS Microbiol Lett. 1997;149:249–255. doi: 10.1111/j.1574-6968.1997.tb10337.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen P, Qi FX, Novak J, Krull RE, Caufield PW. Effect of amino acid substitutions in conserved residues in the leader peptide on biosynthesis of the lantibiotic mutacin II. FEMS Microbiol Lett. 2001;195:139–144. doi: 10.1111/j.1574-6968.2001.tb10511.x. [DOI] [PubMed] [Google Scholar]
- 17.Patton GC, Paul M, Cooper LE, Chatterjee C, van der Donk WA. The importance of the leader sequence for directing lanthionine formation in lacticin 481. Biochemistry. 2008;47:7342–7351. doi: 10.1021/bi800277d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagao J, Morinaga Y, Islam MR, Asaduzzaman SM, Aso Y, Nakayama J, Sonomoto K. Mapping and identification of the region and secondary structure required for the maturation of the nukacin ISK-1 prepeptide. Peptides. 2009;30:1412–1420. doi: 10.1016/j.peptides.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 19.Khusainov R, Heils R, Lubelski J, Moll GN, Kuipers OP. Determining sites of interaction between prenisin and its modification enzymes NisB and NisC. Mol Microbiol. 2011;82:706–718. doi: 10.1111/j.1365-2958.2011.07846.x. [DOI] [PubMed] [Google Scholar]
- 20.Müller WM, Ensle P, Krawczyk B, Süssmuth RD. Leader peptide-directed processing of labyrinthopeptin A2 precursor peptide by the modifying enzyme LabKC. Biochemistry. 2011;50:8362–8373. doi: 10.1021/bi200526q. [DOI] [PubMed] [Google Scholar]
- 21.Mavaro A, Abts A, Bakkes PJ, Moll GN, Driessen AJ, Smits SH, Schmitt L. Substrate recognition and specificity of NisB, the lantibiotic dehydratase involved in nisin biosynthesis. J Biol Chem. 2011;286:30552–30560. doi: 10.1074/jbc.M111.263210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abts A, Montalban-Lopez M, Kuipers OP, Smits SH, Schmitt L. NisC binds the FxLx motif of the nisin leader peptide. Biochemistry. 2013;52:5387–5395. doi: 10.1021/bi4008116. [DOI] [PubMed] [Google Scholar]
- 23.Ortega MA, Hao Y, Zhang Q, Walker MC, van der Donk WA, Nair SK. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature. 2014 doi: 10.1038/nature13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kluskens LD, Kuipers A, Rink R, de Boef E, Fekken S, Driessen AJ, Kuipers OP, Moll GN. Post-translational Modification of Therapeutic Peptides By NisB, the Dehydratase of the Lantibiotic Nisin. Biochemistry. 2005;44:12827–12834. doi: 10.1021/bi050805p. [DOI] [PubMed] [Google Scholar]
- 25.Kuipers A, Meijer-Wierenga J, Rink R, Kluskens LD, Moll GN. Mechanistic dissection of the enzyme complexes involved in biosynthesis of lacticin 3147 and nisin. Appl Environ Microbiol. 2008;74:6591–6597. doi: 10.1128/AEM.01334-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levengood MR, van der Donk WA. Use of Lantibiotic Synthetases for the Preparation of Bioactive Constrained Peptides. Bioorg Med Chem Lett. 2008;18:3025–3028. doi: 10.1016/j.bmcl.2008.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rink R, Kluskens LD, Kuipers A, Driessen AJ, Kuipers OP, Moll GN. NisC, the Cyclase of the Lantibiotic Nisin, Can Catalyze Cyclization of Designed Nonlantibiotic Peptides. Biochemistry. 2007;46:13179–13189. doi: 10.1021/bi700106z. [DOI] [PubMed] [Google Scholar]
- 28.Rink R, Wierenga J, Kuipers A, Kluskens LD, Driessen AJM, Kuipers OP, Moll GN. Production of dehydroamino acid-containing peptides by Lactococcus lactis. Appl Environ Microbiol. 2007;73:1792–1796. doi: 10.1128/AEM.02350-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levengood MR, Patton GC, van der Donk WA. The leader peptide is not required for post-translational modification by lacticin 481 synthetase. J Am Chem Soc. 2007;129:10314–10315. doi: 10.1021/ja072967+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khusainov R, Kuipers OP. When the Leader Gets Loose: In Vivo Biosynthesis of a Leaderless Prenisin Is Stimulated by a trans-Acting Leader Peptide. ChemBioChem. 2012:2433–2438. doi: 10.1002/cbic.201200437. [DOI] [PubMed] [Google Scholar]
- 31.Thibodeaux CJ, Ha T, van der Donk WA. A Price To Pay for Relaxed Substrate Specificity: A Comparative Kinetic Analysis of the Class II Lanthipeptide Synthetases ProcM and HalM2. J Am Chem Soc. 2014;136:17513–17529. doi: 10.1021/ja5089452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 33.Oman TJ, Knerr PJ, Bindman NA, Velasquez JE, van der Donk WA. An engineered lantibiotic synthetase that does not require a leader peptide on its substrate. J Am Chem Soc. 2012;134:6952–6955. doi: 10.1021/ja3017297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koehnke J, Bent AF, Zollman D, Smith K, Houssen WE, Zhu X, Mann G, Lebl T, Scharff R, Shirran S, Botting CH, Jaspars M, Schwarz-Linek U, Naismith JH. The cyanobactin heterocyclase enzyme: a processive adenylase that operates with a defined order of reaction. Angew Chem Int Ed. 2013;52:13991–13996. doi: 10.1002/anie.201306302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


