Abstract
Understanding of mammalian enhancer function is limited by the lack of a technology to rapidly and thoroughly test their cell type-specific function. Here, we use a nuclease-deficient (d)Cas9 histone demethylase fusion to functionally characterize previously described and novel enhancer elements for their roles in the embryonic stem cell state. Further, we distinguish the mechanism of action of dCas9-LSD1 at enhancers from previous dCas9-effectors.
Enhancers control development and cellular function, and intensive efforts are ongoing to elucidate cell fate-specific enhancer activity1,2 Indeed, a large number of genomic regions identified by genome-wide association studies (GWAS) of human disease fall within enhancer regions3,4. Thus, there is a pressing need for technologies to functionally annotate cell type-specific enhancer elements that control cellular function.
Engineered derivatives of CRISPR systems have enabled RNA-guided gene regulation through targeting of nuclease-deficient (d)Cas9-coupled transcriptional regulators to promoter regions5-10 and applicability of this approach in high-throughput contexts has been demonstrated11,12. Although a dCas9-KRAB repressor has previously been used to interfere with enhancer function, the study suggested steric rather than effector-mediated interference13. While targeted steric hindrance or nuclease-based disruption of an enhancer is possible, such approaches require in-depth knowledge of the enhancer, may be inefficient and could have potential side effects such as perturbation of chromatin folding or DNA damage stress responses.
The histone demethylase LSD1 has been previously implicated in repression of enhancers14,15 and TALE-LSD1 can target histone modifications that correlate with active enhancers16. Although changes in gene expression were detected16, the TALE-LSD1 approach only allows for low-throughput approaches and it remains unclear whether the expression changes were a result of decommissioning an enhancer, or due to an indirect effect on gene expression.
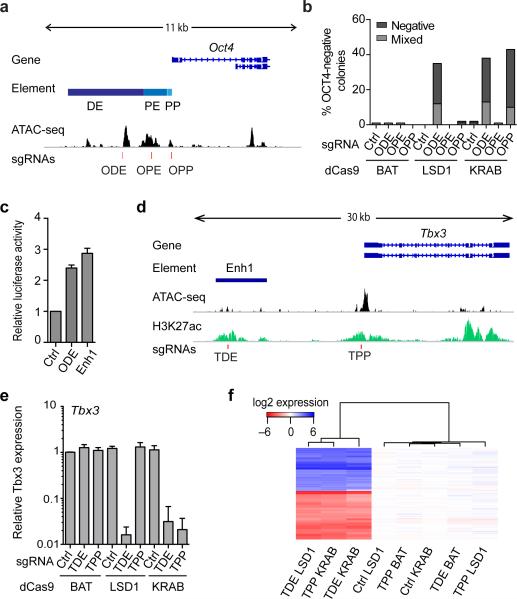
We generated mouse embryonic stem cells (mESCs) expressing versions of Neisseria meningitidis (Nm) dCas9 fused with LSD1, a non-effector BirA affinity tag (BAT), or a KRAB repressor (Supplementary Figs. 1-2) and used a viral delivery system for sgRNAs. We first targeted the well-characterized cis-regulatory region of Oct417 (Fig. 1a), a factor critical for the ESC state18. Oct4 expression is regulated by a proximal enhancer (OPE) active in epiblast cells, and a distal enhancer (ODE) active in mESCs and cells of the inner cell mass17,19. Targeting of LSD1 to the ODE resulted in loss of Oct4 expression and appearance of OCT4-negative colonies accompanied by phenotypic changes (Fig. 1b and Supplementary Fig. 3a,b) compared to control dCas9-BAT cells targeted to the same enhancer. No effects were observed when the OPE, the Oct4 proximal promoter (OPP) or a control locus was targeted in dCas9-LSD1 or dCas9-BAT cells. In contrast, targeting dCas9-KRAB to the OPP led to downregulation of Oct4, demonstrating that dCas9-KRAB is not enhancer specific (Fig. 1b and Supplementary Fig. 3a,b). dCas9-effector-dependent and sgRNA position-specific changes in ESC morphology correlate with loss of OCT4 and SOX2 pluripotency factor expression (Supplementary Fig. 3a) and genome-wide transcriptomic changes (Supplementary Fig. 3c and Supplementary Table 1), indicating a more profound change in the cellular state after interfering with ODE activity. Importantly, cluster analysis revealed high similarity between gene expression profiles from dCas9-KRAB OPP-sgRNA, dCas9-KRAB ODE-sgRNA and dCas9-LSD1 ODE-sgRNA cells compared to other dCas9-effector/sgRNA combinations. We conclude that the dCas9-LSD1 system can be used to delineate enhancers specifically, unlike the dCas9-KRAB system that more broadly affects the cellular state when targeted to promoters and enhancers.
Figure 1.
dCas9-effectors regulate cis-regulatory elements in an effector-dependent manner. (a) The genomic organization of the Oct4 locus: Distal enhancer: ODE; Proximal enhancer: OPE, Promoter: OP, accessible genome (ATACseq signal); red lines indicate the binding sites of the sgRNAs used in this study. (b) Percentage of colonies that do not contain OCT4-expressing cells (negative) or contain OCT4-negative cells among residual OCT4-expressing cells (mixed) following locus-specific sgRNA delivery. (c) Relative luciferase activity of reporter plasmids containing either a fragment of the ODE or of Enhancer 1 (Enh1). n=3 biological replicates +/− s.d. (d) The genomic organization of the Tbx3 locus (e) Quantitative PCR analysis for Tbx3 expression in Nm dCas9-effector mESCs treated with sgRNAs specific to an unrelated control genomic region (Ctrl) or the putative Tbx3 distal enhancer (TDE) or the Tbx3 promoter (TPP). n=3 biological replicates +/− s.d. (f) Heat map of gene expression microarray data from dCas9-effector mESCs with indicated sgRNAs displayed relative to the average expression levels of the dCas9-effector Ctrl-sgRNA samples. Unsupervised hierarchical clustering of 174 differentially expressed genes (listed in Supplementary Table 3) is displayed on the y-axis. Hierarchical clustering of samples based on similarities in gene expression profiles is displayed on the x-axis.
We next targeted eight candidate enhancers in dCas9-LSD1 ESCs (Supplementary Figs. 4-6 and Supplementary Table 2) and four showed both morphological changes and loss of ESC-associated alkaline phosphatase (AP) activity (Supplementary Fig. 7). These four enhancers are thus critical for the appropriate expression of genes required for the ESC state. Absence of differentiation phenotypes upon enhancer targeting could occur if the enhancer is dispensable for maintenance of the pluripotent state. Alternatively, some sgRNAs might be non-functional or chromatin state could counteract dCas9-LSD1 function. Importantly, no control-sgRNAs (FoxN1, N-Myc or Oct6) led to changes in morphology or AP activity, indicating sgRNA-specific changes in cellular state.
For follow-up, we focused on the putative enhancer with the highest differential score, Enh1. Test of Enh1 in a reporter assay confirmed its ability to enhance expression at comparable levels to an Oct4 DE sequence (Fig. 1c). This previously unannotated ESC-specific enhancer is positioned ~10kb upstream of the transcription factor Tbx3 (Fig. 1d), a gene previously implicated in the maintenance of pluripotency20. We therefore hypothesized that Enh1 may function in the ESC network by regulating Tbx3 expression. Indeed, we detected a relative reduction of Tbx3 mRNA and protein expression upon targeting Enh1 with dCas9-LSD1 (Fig. 1e and Supplementary Fig. 8a). This loss of Tbx3 could be similarly obtained when dCas9-KRAB was used to target Enh1 or the Tbx3 proximal promoter (TPP) but was not observed in control cells. Change in colony morphology and increase of differentiation-associated markers were detected in a dCas9-effector and sgRNA dependent manner (Fig. 1f, Supplementary Fig. 8 and Supplementary Table 3) concordant with the destabilization of the ESC state. Of note, none of the transcriptomic changes among any of the control conditions exceed 2-fold. While this finding does not rule out minor off-target effects, these data support a compelling level of specificity of our system.
Next we sought to dissect the mechanism by which targeting of Enh1, hereafter referred to as Tbx3 distal enhancer (TDE), results in Tbx3 downregulation by examining a time point prior to changes in cell morphology but when differential Tbx3 expression levels were detected (Supplementary Fig. 9a). Conducting chromosome conformation capture (3C) we observed a localized peak in interaction frequency between the TDE and the Tbx3 promoter in dCas9-BAT ESCs with a control-sgRNA supporting the presence of an enhancer-promoter loop (Supplementary Fig. 9b). Comparable interaction frequencies were observed for dCas9-LSD1 and dCas9-BAT with TDE-sgRNA, indicating that the looping interaction between the Tbx3 promoter and enhancer is not disrupted by dCas9-effector targeting per se.
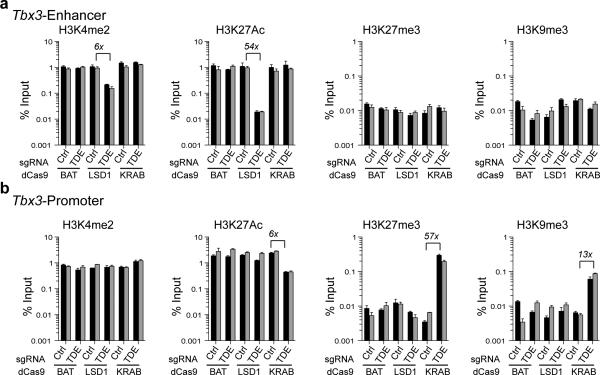
Investigating local histone modifications in the presence of either dCas9-KRAB or dCas9-LSD1, we observed relative loss of LSD1 substrate H3K4Me2 around the enhancer-sgRNA target site in dCas9-LSD1 cells (6 to 8 fold) (Fig. 2a and Supplementary Fig. 10). H3K27Ac, indicative of active enhancers2, was dramatically lost at the enhancer (33 to 54-fold) upon LSD1 targeting compared to other tested conditions consistent with prior studies of enhancer inactivation by LSD116 (Fig. 2a and Supplementary Fig. 10). Changes in H3 occupancy were not detected (Supplementary Fig. 10). Interestingly, although repressive marks have been previously associated with KRAB-mediated repression21, we could not detect concomitant increases in repressive marks, H3K27Me3 or H3K9Me3, at the enhancer after targeting of any of our constructs (Fig. 2a). To further understand the differences between the LSD1 and KRAB effects we surveyed chromatin state at the Tbx3 promoter. At the promoter we detected a loss of H3K27Ac, and an increase of H3K27Me3 (~57-fold) and H3K9Me3 (~13-fold) when the TDE was targeted with the KRAB effector but not with LSD1 (Fig. 2b). Taken together these data offer strong evidence that LSD1-induced enhancer deactivation results in Tbx3 downregulation while KRAB mediated Tbx3 downregulation is due to promoter silencing. We therefore caution that the dCas9-KRAB effector, besides having potential side effects such as heterochromatin spreading21, is silencing promoter activity rather than decommissioning the target enhancer.
Figure 2.
Enhancer targeting by dCas9-LSD1 or dCas9-KRAB. (a,b) ChIP qPCR analysis of H3K4Me2, H3K27Ac, H3K27Me3 and H3K9Me3 at the Tbx3 enhancer 345 bases upstream of the sgRNA target site (a) and the Tbx3 proximal promoter region (b) for dCas9-effector mESCs in the presence of control sgRNA (Ctrl) or a sgRNA specific to the putative Tbx3 DE (TDE). Locations of qPCR amplicons are indicated in Supplementary Fig. 4. The data are expressed as percent input values. Adjacent black and grey bars represent two independent experiments with mean +/− s.d. of three technical replicates.
Next, we showed that the LSD1-specific inhibitor trans-2-phenylcyclopropyl-amine hydrochloride (TCP), previously shown to overcome strong differentiation cues in mESCs such as loss of Oct4 expression14, inhibited the loss of Tbx3 mRNA and protein expression in dCas9-LSD1 cells but not in dCas9-KRAB cells after TDE targeting (Supplementary Fig. 11). We conclude that the loss of Tbx3 expression is directly dependent on the enzymatic activity of LSD1 and that KRAB can repress Tbx3 independently.
We conclude that the dCas9-LSD1 fusion protein allows for an effector dependent definition of functional, native enhancer elements that help to maintain a given cellular state. Accordingly, dCas9-LSD1 provides a rapid and powerful approach to understanding distal cis-regulatory regions such as enhancers without major disruption of the local genomic architecture. While we used the Nm dCas9, we expect that similar systems could be built on orthogonal CRISPR/Cas9 systems including the widely used Streptococcus pyogenes dCas9. Further, the use of chromatin modifiers with differential functionality and orthogonal dCas9 systems promises to enable even more complex epigenome engineering. This level of genome control has previously not been achieved with other RNA-guided regulatory technologies such as RNAi. Combined with rapid advances in cell type-specific annotation of regulatory elements and GWAS, this technology will be instrumental in dissecting the contribution of distal cis-regulatory elements to development and disease.
ONLINE METHODS
Effector and sgRNA plasmids
A human codon optimized, nuclease inactive version of Nm Cas9 was gene synthesized with two SV40 nuclear localization signals and a triple FLAG tag (Genscript Technologies) and cloned in frame with either a BirA affinity tag, a KRAB-repressor, or LSD1 (for amino acid sequences see Figure S1). A Rosa26 CAGS loxP stop loxP DEST vector was generated by inserting a gateway acceptor cassette into a Rosa26 CAGS loxP stop loxP targeting vector (a kind gift from A. McMahon). The various dCas9-effectors were inserted into the Rosa26 targeting vector by standard gateway cloning.
An sgRNA expression vector was generated by replacing the H1 shRNA and tetO IRES RFP-Puro expression cassettes in a pLV-H1TetO-RFP-Puro lentiviral backbone (Addgene plasmid number: 36297) with a U6 Nm sgRNA expression cassette harboring two BsmBI sites for gRNA cloning and EF1α Puro selection cassettes (see Supplementary Figure 1 for cloning sites). The Nm sgRNA acceptor and expression cassette was generated as a gene block containing a short form of the tracrRNA (8 and Supplementary Figure 1).
Note: Neisseria meningitidis (Nm) dCas9 is an orthogonal and substantially smaller Cas9 variant to the commonly used Cas9 from Streptococcus pyogenes (Sp) that is about 25% longer in its nucleotide sequence. While not tested in our study we expect an orthogonal Sp dCas9-LSD1 to perform similarly to the Nm dCas9-LSD1, and work on alternative PAM sequences. The ability to use either Sp Cas9 or Nm Cas9 broadens the spectrum of potential target sites in the genome8,22, although a thorough test in mammalian cells has to be conducted. Further, off-target effects are currently a major concern of Cas9 systems and appropriate software and multiple independent sgRNAs should be used to minimize the off-target effects that might influence interpretation of a given study.
sgRNA design
sgRNA target sites were obtained by using a custom script that searches for Nm Cas9 PAM sequences NNNNGATT, NNNNGCTT or NNNNGGTT within the enhancer or promoter regions of interest. Bowtie2 was used to map candidate targets to the mouse genome build GRCm38 with sensitive parameters (--local -f -k 10 --very-sensitive-local -L 9 -N 1) to detect potential off-target sites. Only sgRNAs were used that had two or more mismatches to other sites in the genome than the selected target site. All our sgRNAs had no other genomic matches at the alignment stringency used. Oligonucleotides for sgRNA cloning were obtained from Integrated DNA Technologies or Life Technologies and are listed in Supplementary Table 4.
ESC culture
V6.5 (a kind gift from K.Eggan) and Rosa26-targeted mouse ESC lines were maintained on mouse embryonic fibroblast feeder layers in ESC-Media (KnockOUT DMEM [GIBCO, 10829] supplemented with 15% Hyclone FBS [Thermo Scientific, SH30070.03], 1% Glutamax [GIBCO, 35050079], 1% NEAA [Cellgro, 25-025-CI], 100μM β-mercaptoethanol [Invitrogen, 21985023], and 1×103 U/mL LIF) according to standard methodology23. ESC lines were routinely tested to confirm the absence of mycoplasma.
Generation of stable Nm dCas9-effector mESCs
V6.5 mESCs were electroporated (230V, 500mF) with a Rosa26 targeting construct and selected from day 2-8 with 300μg/mL Geneticin (Life Technologies 10131035). Picked clones were screened by PCR analysis for integration (Supplementary Fig. 2; knockin primer fw: GCCGCCTAAAGAAGAGGCTGTGCTTTGG and reverse: TACCGTAAGTTATGTAACGCG). The following PCR conditions were used: 2-minute initial denaturation at 95°C, 30 cycles with 30-second denaturation at 95°C, annealing at 50°C for 30-seconds, extension at 72°C for 2-minutes, and a final extension at 72°C for 5-minutes. After transient transfection of a Cre recombinase expression plasmid (Addgene plasmid 13775), individual clones were picked, expanded and screened for removal of the loxP flanked stop cassette (Supplementary Fig. 2; stop removal primer fw: TGCTCCTGCCGAGAAAGTATCCATCATGGC and reverse: CGCCAAGCTCTTCAGCAATATCACGGGTAG). The following PCR conditions were used: 1-minute initial denaturation at 93°C and 30 cycles with 20-second denaturation at 95°C, annealing/extension at 68°C for 3-minutes.
Spontaneous differentiation capability of Nm dCas9-effector cells was assessed by injecting ~10E6 undifferentiated cells sub-cutaneous into NSG mice. Teratomas were retrieved and analyzed 3-4 weeks after injection. Hematoxylin and eosin images were acquired on a Nikon Eclipse 90i microscope using a 20x objective (Nikon Plan Fluor 20x/0.50 Ph1 DLL).
sgRNA delivery
Rosa26-targeted mESC lines were transduced with lentivirus containing an sgRNA specific to a genomic region of interest. HEK293T cells were used for lentiviral production as described9. ESCs were washed once with PBS and split with 0.25% trypsin (GIBCO, 25200114). Cells were MEF-depleted on 0.1% gelatinized plates for 30 minutes, then collected and counted. Cells were incubated with sgRNA lentivirus on low attachment plates for 3 hours in 2i media (ESC-media supplemented with 1μM PD0325901 [Tocris, 4192], and 3μM CHIR99021 [Tocris, 4423]), then plated onto 0.1% gelatin coated plates at 12.5×104 cells/cm2. After 48 hours, cells were split using 0.25% trypsin and plated in 2i media supplemented with 1μM puromycin to select for cells expressing the sgRNA. Transduced cells were maintained in 2i media supplemented with 1μM puromycin and split every 3 days. Cells were analyzed at the time of observed morphological changes (8 to 14 days after infection with sgRNA carrying lentivirus). To examine the effect of dCas9-LSD1 on local histone modifications, cells were analyzed 3 days prior to observed morphological changes. To assess the requirement for LSD1 enzymatic activity, experiments were carried out in 2i media supplemented with 500nM trans-2-phenylcyclopropyl-amine hydrochloride (TCP) (Tocris 3852). After 2 passages, TCP was withdrawn where indicated and cells were analyzed 14 days after TCP withdrawal.
Immunofluorescence analysis
Cells were fixed and processed as described9. Phase contrast images were acquired on a Nikon Eclipse TS100 microscope using a 20x objective (Nikon LWD 20x/0.40 Ph1 ADL). Fluorescent images were acquired on a Nikon Eclipse Ti microscope using a 20x objective (Nikon S Plan Fluor ELWD 20x/0.45). We used filter cubes ET CY3 (Chroma, 49004), ET FITC (Chroma, 49002), and ET DAPI (Chroma, 49000), to isolate fluorescence from Alexa-594, Alexa-488 and Hoechst respectively. Microscope and camera were controlled through NIS-Elements software (Nikon). Adobe Photoshop was used to apply linear contrast adjustments equally across all images. Further details are listed in Supplementary Note 1. Antibodies were diluted as indicated in Supplementary Table 5. OCT4 positive, negative, and mixed colonies were quantitated using NIS-Elements Analysis Software. Between 520-1300 colonies were identified for each condition by Hoechst staining of clusters of >5 cells. Colonies were scored as mixed if < 50% of cells in the colony expressed OCT4.
Quantitative PCR analysis
Cells were trypinsized, washed once with PBS, and total RNA was isolated with Trizol reagent (Invitrogen, 15596-018) following the manufacturer's instructions. 1 μg of total RNA was reverse transcribed with the Superscript III First Strand Synthesis System (Invitrogen, 18080-051). 15ng cDNA was utilized in quantitative PCR analyses with iTAQ Universal SYBR Green Supermix (Biorad, 172-5124) using specific primers listed in Supplementary Table 6. Relative gene expression was calculated using the ΔΔCT method. All genes were normalized to Gapdh.
RNASeq processing
RNASeq data for ESCs and Epiblast-like cells (EpiLCs) from a prior study24 were downloaded from the Short Read Archive (SRP040451). The raw fastq files were processed using the UMASS Medical School Bioinformatics Core Galaxy pipeline. Briefly, reads were aligned to the mouse ribosomal sequence and discarded from further analysis. Remaining reads were processed with RSEM25(version 1.2.7) using the RefSeq26 gene annotation set for the mouse build GRCm38. The inferred transcripts per million (TPMs) values were used for further analysis.
mESC-specific enhancer definition
Enhancers specific to ESC and EpiLC cellular states were identified on the basis of chromatin immunoprecipitation sequencing (ChIPSeq) for the H3K27Ac histone mark for each cell type24. Reads were aligned with Bowtie version 1.0.0 using -v 0 -a --strata --best -m 1 parameters, and peaks were identified with MACS version 1.4.2 using --tsize=35 --bw=300 -g 186550000 . Each peak called by MACS was initially considered a candidate enhancer. We then rescored the candidate peaks by way of a 300-nucleotide moving window, advancing 10 nucleotides for sequential windows as described27. An enrichment score, depth for H3K27Ac as compared to the null, was associated to each window and the maximum enrichment score across the windows was associated to the peaks. Any peaks with scores below a 2-fold enrichment relative to the null were filtered out. Low quality peaks, with scores below the 60% quantile, were also filtered out. Each remaining peak was then trimmed based on the read depth associated to 50-nucleotide moving windows. Ends were trimmed until a window score exceeding 20% of scale for the peak at hand was located. Each high quality peak for one stage, EpiLC or ESC, was then compared to the other by scoring the trimmed region for the alternate stage. After adding a pseudocount of 10 to the score for each stage, those peaks exhibiting at least two-fold enrichment were considered to be stage-specific.
For each cell type, any stage-specific peak with the nearest gene transcription start site more than 5000 bases away was identified as a potential distal enhancer. Peaks for which the nearest transcription start site was associated to a transcription factor were selected. To determine the strongest enhancer candidates for the ESC state, the list was further filtered to include only those peaks whose closest transcription factor exhibited at least a 4-fold enrichment in TPM for the ESC state relative to the EpiLC state, after adding a pseudocount of 5 TPM for each stage. The resulting list contained 24 candidates (Supplementary Table 2). We excluded the 13 intronic enhancers from further analysis.
We also excluded enhancer chr8:72326162-72328532 as it was within 5000 bases of a gene body and enhancer chr8:72332459-72335859 as it was closer to the gene body of a non-transcription factor than to the transcription factor, KLF2. We chose to test 2 of the 3 candidates near KLF5 and all other remaining candidates leaving eight putative distal enhancers that are listed in Supplementary Table 2a.
ATACseq
Assay for Transposase Accessible Chromatin (ATACseq) was performed on V6.5 mouse ESCs according to the protocol as described28. As starting material for the protocol, V6.5 mouse ESCs were grown on 0.1% gelatinized plates and maintained in 2i media. Cells were harvested with 0.25% trypsin for 5 minutes, collected, and counted. 50,000 cells were spun down at 500×g for 5 minutes at 4°C and washed once with 50μL of cold PBS. ATACseq library generation and analysis occurred according to standard methodology28. Seq reads were mapped to the genome using bowtie2 version 2.1.0 using the following parameters --phred33 -5 9 -3 2 . All ATACseq data are available through GEO under accession number GSE64059.
Microarray analysis
Total RNA was isolated with Trizol reagent (Invitrogen, 15596-018) following the manufacturer's instructions either directly (Oct4 experiments) or after trypsinization (Tbx3 experiments). 250ng of total RNA was amplified to generate labeled cRNA using the Illumina Total Prep RNA Amplification Kit (Ambion, AMIL1791). 750ng of each labeled cRNA sample was hybridized to MouseRef-8 v2 Expression BeadChip (Illumina, BD-202-0202) and the chip scanned on an Illumina BeadArray Reader. Raw probe level data were exported from Illumina Bead Studio for further processing in R (www.r-project.org). Raw probe level intensity values were adjusted using the R package limma29 to apply quantile normalization, background subtraction and to transform to log2 expression values. To identify differentially expressed genes, probes were ranked by highest difference in log2 expression value between any two populations. Genes with log fold changes greater than 3 were used for the generation of hierarchically clustered heatmaps. Data are displayed relative to the average log2 expression signal in dCas9-effector Ctrl-sgRNA samples. In instances where multiple probes mapped to the same gene, only expression values for the highest ranked probe were retained. All gene expression data are available through GEO under accession number GSE64059.
Luciferase Assay
Reporter plasmids containing a fragment of the Oct4 distal enhancer or putative Tbx3 distal enhancer were cloned into the pGL3-Promoter plasmid (Promega, E176A) upstream of an SV40 minimal promoter and firefly luciferase gene. For the luciferase assay, 5×104 V6.5 mouse ESCs were plated in 12-well plates and transfected with 900ng of the firefly reporter plasmid, and 100ng of pRL-SV0 renilla luciferase plasmid (Promega, E223A), using Lipofectamine 2000 (Invitrogen, 11668019). Cells were harvested 48 hours after transfection, and the luciferase activities were measured with the Dual-Luciferase Reporter Assay System (Promega, E1910) according to the manufacturer's protocol. The ratio of firefly luciferase to renilla luciferase was calculated and the data are expressed relative to an empty vector control.
Alkaline Phosphatase staining
Cells were harvested 14 days after infection with sgRNA carrying lentivirus. Cells were fixed with formalin solution (Sigma, HT5014) for 20 minutes at room temperature, and washed three times with 100mM Tris pH 8.5. Alkaline phosphatase (AP) expression was detected using the BCIP/NBT Alkaline Phosphatase Substrate Kit IV (Vector, SK-5400) according to the manufacturer's protocol. Overview images were acquired on a Nikon Eclipse SMZ1500 microscope with a 1x objective (Nikon HR Plan Apo 1x WD54).
Chromatin Immunoprecipitation
Nm dCas9-effector ESCs transduced with lentivirus containing an sgRNA were plated onto 0.1% gelatin coated plates at 5×104 cells/cm2 in 2i media. After 48 hours, cells were passaged and plated in 2i media supplemented with 1μM puromycin. Cells were maintained in 2i media with 1μM puromycin and harvested 9 days later. Cells were crosslinked with fixing solution ((11% formaldehyde, 100 mM NaCl, 1 mM EDTA, 50 mM HEPES-KOH [pH 7.6]) and incubated for 10 minutes at room temperature. The crosslinking reaction was stopped by the addition of glycine to 125mM. Cells were washed once with PBS, pelleted, frozen and stored at −80°C. Chromatin immunoprecipitation (ChIP) was performed as described30. 10×106 cells and 50μL of antibody-coupled protein A magnetic beads (NEB) were used for each ChIP. Protein A magnetic beads were coupled to either H3K4Me2 (Active Motif, 39141), H3 (Abcam, ab1791), H3K27Me3 (Millipore, 07-449), H3K9Me3 (Active Motif, 39161) or H3K27Ac (Abcam, ab4729) antibodies. ChIP and input samples were used as a template for qPCR analysis using SYBR FAST (KAPA Biosystems, KK4602) with the primers listed in Supplementary Table 7 or control primers (Active Motif, 71017 and Active Motif, 103727). Relative enrichment for each primer set was expressed as percent input.
Chromosome Conformation Capture (3C)
3C templates were generated as described previously31 with minor differences. Briefly, 30 × 106 cells were harvested and crosslinked per condition. Crosslinked cells were digested with HindIII (NEB, 400U) overnight at 37°C. Purified 3C templates were desalted and concentrated using Amicon Ultra Centrifugal Filter Units (Millipore, UFC503096). Control 3C template was generated from purified BAC DNA covering the Tbx3 locus (RP23-406M3, CHORI BACPAC). BAC DNA was purified using a QIAGEN Large-Construct Kit (12462) and 10 μg of purified BAC DNA was digested overnight with HindIII at 37°C. 3C primers (Supplementary Table 8) were designed around HindIII sites both upstream and downstream of the Tbx3 transcriptional start site using an online web module http://3DG.umassmed.edu32. PCR analysis was performed in triplicate with each primer paired with the anchor for each 3C template. Interaction frequencies were determined by quantifying PCR amplicons on a gel using Biorad Quantity One analysis software. Primer efficiencies were normalized by dividing the average value of each 3C template by the average value of the BAC 3C control template. 3C template generation efficiencies were normalized by calculating interaction frequencies through a control genomic region using gene desert primer pairs. 3C templates were normalized to each other by calculating the log ratio of the average interaction frequencies for each 3C template through the gene desert region. Final normalized interaction frequencies were calculated by multiplying the average BAC normalized interaction frequencies with the determined normalization factor for each 3C template.
Supplementary Material
Acknowledgements
We thank T. Fazzio, J. Dekker and S. Wolfe for helpful discussions and E. Sontheimer and M. Ziller for critical reading of the manuscript. We thank K. Eggan (Harvard University) and A. McMahon (University of Southern California) for providing the V6.5 ESCs and the Rosa26 targeting vector backbone respectively. We are grateful to S. Hainer, H. Belaghzal, K. Morrison and the DERC morphology core for technical help and discussions. R.M. is supported by The Leona M. and Harry B. Helmsley Charitable Trust (2015PG-T1D057 and 2015PG-T1D035), a Charles H. Hood Foundation Child Health Research Award, the Glass Family Charitable Foundation and the US National Institutes of Health (UC4 DK104218). M.G. is supported by a grant from the US National Institutes of Health (1R01HD080224-01A1).
Footnotes
Author Contributions
N.A.K., H.P. and R.M. were responsible for the conception, design and interpretation of experiments. M.G. conceived the bioinformatic approach for novel enhancer identification. N.A.K., H.P. and R.M.G. conducted experiments. B.T. and N.J.S. performed bioinformatics analyses. All authors wrote the manuscript.
Accession codes
ATAC-seq and microarray data are available through GEO at accession number GSE64059.
Reproducibility
No sample size estimates were performed to ensure adequate power to detect a pre-specified effect size. Experiments were not randomized and experimenters were not blinded to experimental conditions. No data were excluded from analysis.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parker SCJ, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proceedings of the National Academy of Sciences. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert LA, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Meth. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kearns NA, et al. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng AW, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014 doi: 10.1016/j.cell.2014.09.029. doi:10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konermann S, Brigham MD, Trevino AE, Joung J. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2014 doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao X, et al. Comparison of TALE designer transcription factors and the CRISPR/dCas9 in regulation of gene expression by targeting enhancers. Nucleic Acids Res. 2014;42:e155. doi: 10.1093/nar/gku836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whyte WA, et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray S, Levine M. Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes Dev. 1996;10:700–710. doi: 10.1101/gad.10.6.700. [DOI] [PubMed] [Google Scholar]
- 16.Mendenhall EM, et al. Locus-specific editing of histone modifications at endogenous enhancers. Nat. Biotechnol. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeom YI, et al. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- 18.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature Genetics. 2000 doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimizu T, et al. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev Growth Differ. 1999;41:675–684. doi: 10.1046/j.1440-169x.1999.00474.x. [DOI] [PubMed] [Google Scholar]
- 20.Ivanova N, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 21.Groner AC, et al. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kearns NA, et al. Generation of organized anterior foregut epithelia from pluripotent stem cells using small molecules. Stem Cell Res. 2013;11:1003–1012. doi: 10.1016/j.scr.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Buecker C, et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011 doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garber M, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Molecular Cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Meth. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth GK, Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S. Statistics for Biology and Health. Springer; New York: 2005. Bioinformatics and Computational Biology Solutions Using R and Bioconductor; pp. 397–420. [Google Scholar]
- 30.Chen PB, et al. Hdac6 regulates Tip60-p400 function in stem cells. eLife Sciences. 2013;2:e01557. doi: 10.7554/eLife.01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miele A, Gheldof N, Tabuchi TM, Dostie J, Dekker J. Mapping chromatin interactions by chromosome conformation capture. Curr Protoc Mol Biol. 2006 doi: 10.1002/0471142727.mb2111s74. Chapter 21, Unit 21.11. [DOI] [PubMed] [Google Scholar]
- 32.Lajoie BR, van Berkum NL, Sanyal A, Dekker J. My5C: web tools for chromosome conformation capture studies. Nat Meth. 2009;6:690–691. doi: 10.1038/nmeth1009-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.