Abstract
Objective
We generated knock-in mice that express a tamoxifen-inducible Cre recombinase from the Prg4 locus (Prg4GFPCreERt2), and used these animals to fate-map the progeny of Prg4-positive articular cartilage cells at various ages.
Methods
We crossed Prg4GFPCreERt2 mice to Rosa26floxlacZ or Rosa26mTmG reporter strains, administered tamoxifen to the double heterozygous offspring at different ages, and assayed Cre-mediated recombination by histochemistry and/or fluorescence microscopy.
Results
In 1-month-old mice, the expression of the Prg4GFPCreERt2 allele mirrors expression of endogenous Prg4 and, when tamoxifen is given for 10 days, causes Cre-mediated recombination in ~70% of the superficial-most chondrocytes. Prg4GFPCreERt2 expressing cells are mostly confined to the top three cell layers of the articular cartilage in 1-month-old mice, but descendants of these cells are located in deeper regions of the articular cartilage in aged mice. At embryonic day 17.5, Prg4GFPCreERt2 expressing cells are largely restricted to the superficial-most cell layer of the forming joint, yet at approximately 1 year, progeny of these cells span the depth of the articular cartilage.
Conclusions
Our results indicate that Prg4-expressing cells located at the joint surface in the embryo serve as a progenitor population for all deeper layers of the mature articular cartilage. Also, our data reveal that Prg4GFPCreERt2 is expressed by superficial chondrocytes in young mice, but expands into deeper regions of the articular cartilage as the animals age. The Prg4GFPCreERt2 allele should be a useful tool for inducing efficient Cre-mediated recombination of floxed alleles at sites of Prg4 expression.
Keywords: Articular cartilage, superficial zone, chondrocytes, Lubricin, Prg4
Introduction
While much is known about the mechanisms that regulate the formation and maturation of growth plate cartilage (reviewed in (1, 2)), much less is known about the development of articular cartilage (reviewed in (3, 4)). In contrast to growth plate chondrocytes, which establish a transient cartilage template that is replaced by bone (i.e., endochondral ossification), articular chondrocytes establish a permanent cartilage tissue. Growth plate and articular chondrocytes arise from distinct progenitor populations, such that articular chondrocytes share a common origin with synovial cells that line the joint cavity (5, 6). In humans (and other large mammals), adult articular cartilage has been divided into four zones based upon histological features: The non-mineralized articular cartilage consists of a superficial zone, composed of flattened chondrocytes; a middle zone, composed of relatively small round chondrocytes; and a deep zone, that contains larger round chondrocytes arranged in a columnar fashion. Beneath these top three zones lies the mineralized articular cartilage (which is separated from the non-mineralized cartilage by the tidemark), and deeper still, the subchondral bone.
Both the identity of the cells that give rise to the articular cartilage and the mechanism by which the articular cartilage grows are incompletely understood. Two growth mechanisms seem plausible: interstitial and appositional. For interstitial growth a precursor cell population would give rise to distinct types of articular chondrocytes (i.e., superficial, middle, and deep) and these chondrocyte subtypes would populate their corresponding layers. For appositional growth, a precursor population would first give rise to cells in the superficial layer, which subsequently would undergo a maturation process to populate the other cartilage zones. Prior studies have attempted to delineate the mechanism of cartilage growth using metabolic labeling. Tritiated thymidine incorporation in the rabbit knee joint demonstrated two proliferative regions, one within the superficial zone of the articular cartilage and the other within the subchondral plate (7). Hunziker and colleagues noted that the superficial articular cartilage cells in the medial femoral condyle of 2-month-old New Zealand white rabbits proliferated more slowly than cells within the transitional and upper radial zones of this tissue (8). BrdU labeling in the opossum knee joint demonstrated that cells in both the superficial and middle layers incorporate BrdU (9). Pulse-chase experiments in mice have demonstrated that the superficial-most articular chondrocytes in newborn animals retain a pulse of BrdU/EdU for up to 6 weeks, suggesting that these cells may constitute a slowly cycling stem cell population (10, 11). Indeed, it has been proposed that the superficial cells of the articular cartilage may comprise a progenitor/stem cell population since only cells from this layer can give rise to colonies with high colony-forming efficiencies in a Notch-dependent manner, and show phenotypic plasticity when injected into the chick limb bud (12). However, none of these studies was able to trace the fate of superficial zone chondrocytes by lineage labeling in articular cartilage.
Lubricin, encoded by the Prg4 locus, is abundantly expressed by superficial zone chondrocytes and synoviocytes (13). Individuals with genetic deficiency of PRG4 have the camptodactyly-arthropathy-coxa vara-pericarditis syndrome (CACP) (13). Patients with CACP have normal appearing joints at birth, but with advancing age develop joint failure associated with noninflammatory synoviocyte hyperplasia and subintimal fibrosis of the synovial capsule (14). While Prg4−/− mice similarly display significant joint abnormalities, heterozygous Prg4 mutant (Prg4+/−) mice appear normal (15). Herein, we describe a mouse strain that has a chimeric GFP-tamoxifen-inducible Cre recombinase knocked into the endogenous Prg4 locus (Prg4GFPCreERt2). We demonstrate that GFPCreERt2 expression mirrors endogenous Prg4 expression in this strain and we use this strain to identify and lineage-trace descendants of Prg4GFPCreERt2 (and by extrapolation Prg4) expressing cells in articular cartilage. We report that young mice express Prg4GFPCreERt2 in cells located near the cartilage surface and that these cells serve as progenitors for cells located in both the superficial and deeper regions of the articular cartilage in older mice. We also find that Prg4GFPCreERt2 is expressed by superficial articular chondrocytes in young mice, but expands into deeper regions of the articular cartilage as the animals age
Materials and Methods
Mouse strains
Generating Prg4GFPCreERt2 mice
We designed a targeting vector (Figre 1A) that would insert a GFPCreERt2 and a PGKneo cassette (16) into the translation initiation codon site within exon 2 of the Prg4 locus. The targeting vector carried the GFPCreERt2 cassette followed by a PGKneo cassette flanked by FRT sites, which were bordered by approximately 2 kb of homologous Prg4 locus sequence on both ends. Prg4-containing BAC clone RP23-147F24 (BACPAC Resources) was used to PCR amplify the 2 kb 5′ homology and the 1.8 kb 3′ homology targeting arms. We flanked the PGKneo sequence with FRT sites in order to remove this cassette after correct targeting was achieved. PGK-DTA (encoding diphtheria toxin to select against non-homologous recombination) was inserted downstream of the 3′ homologous arm. We performed homologous recombination in the 129 ES cell line and chose a correctly targeted clone (Supplemental Figure 1A) to generate mice with the Prg4GFPCreERt2-PGKneo allele. Targeting in ES cells was assayed by PCR analysis, employing primers amplifying either 5′ or 3′ correctly targeted arms, followed by either EcoRI or SacI restriction digestion, respectively, of the PCR-generated fragments to ensure specificity of amplification. Correctly targeted ES cells were injected into mouse blastocysts to eventually generate a line of mice containing Prg4GFPCreERt2-PGKneo. We excised the PGKneo cassette using a FLP recombinase expressing mouse (ACT-FLPe) (17) to produce mice with the Prg4GFPCreERt2 allele alone. In subsequent crosses we distinguished the wild-type (Prg4+) and knock-in (Prg4GFPCreERt2) alleles using PCR (Supplemental Figure 1B). Primer pair F1/R1 produces a 337 bp amplimer from the Prg4GFPCreERt2 allele and primer pair F1/R2 produces a 258 bp amplimer from the Prg4+ allele (F1-TCAGGAATTCAAGCTGATTGC; R1-AACTTGTGGCCGTTTACGTC; R2- CCTTGAGATGAAACCTGTTGAATC). Prg4+/GFPCreERt2 mice have been maintained on a mixed genetic background (i.e., 129/Sv x C57BL/6) and donated to the Jackson Labs for distribution (Stock # 022757).
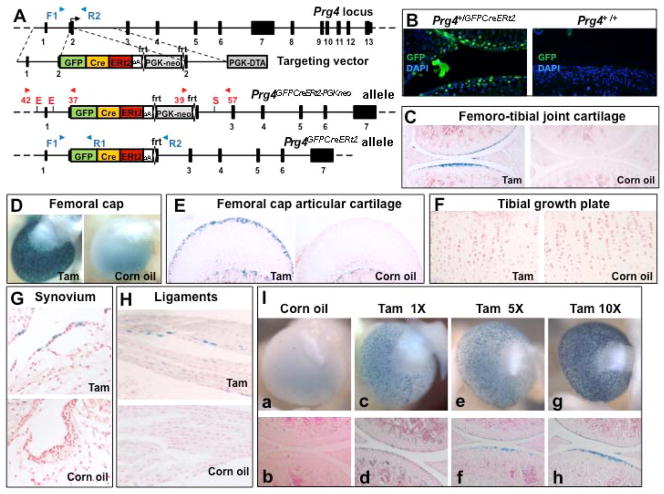
Figure 1. Prg4GFPCreERt2 drives robust recombination in superficial articular chondrocytes in 1-month-old mice.
(A) Schematic diagram of exon-intron structure of the wild-type Prg4 allele (not drawn to scale), the targeting vector, and the Prg4 knock-in allele prior to and after excision of the PGK-neo cassette. (B) Photomicrographs depicting immunofluorescence detection of GFPCreERt2 protein using a fluorescently-labeled anti-GFP antibody in the knee joints of 1-month-old Prg4+/GFPCreERt2 and Prg4+/+ mice. X-Gal stained (C) knee joints, (D) femoral heads, (E) femoral head sections, (F) tibial growth plates, (G) synovia, and (H) ligaments from P34 Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice that had been given daily IP injections of either tamoxifen (Tam) or vehicle (Corn oil) from P21 to P31. (I) Prg4+/GFPCreERt2; Rosa26+/floxlacZ were administered either corn oil (a,b) or a 1, 5, or 10 day course of tamoxifen (c–h). Animals were euthanized at P34 (3 days after the last injection), followed by whole mount X-Gal staining of their femoral heads and knee joints. Whole mounts of the femoral heads (a, c, e, g) and sections of the knee joints (b, d, f, h) are displayed.
The mouse reporter strains used
Rosa26floxlacZ (18); Rosa26mTmG ((19) (Jackson Labs Stock # 007576). Tg(Foxa3-cre); Rosa26+/floxlacZ mice were generated by crossing Tg(Foxa3-cre) animals (20) to homozygous Rosa26floxlacZ/floxlacZ animals. We induced Cre-recombinase activity in postnatal mice with the Prg4GFPCreERt2 allele by administering intraperitoneal (IP) injections of tamoxifen (100 mg/kg/dose) diluted in corn oil (10 mg/ml). Injection of corn oil alone served as a negative control. We induced Cre-recombinase activity in embryonic day 17.5 (E17.5) fetuses by giving a single IP injection of 4 mg tamoxifen in corn-oil to the dam. We studied a minimum of 3 animals per genotype, treatment and age group. Osx-Cre (21) and Col1-CreERt2-Cre alleles were previously described (22); Rosa26+/floxdtTomato (23) was employed as a recombination-reporter allele. 2 month-old Col1-CreERt2-Cre; Rosa26+/floxdtTomato mice were given a single injection of Tamoxifen and harvested 1 week after the injection. Osx-Cre; Rosa26+/floxdtTomato mice were harvested at 2 months-of-age, and processed for frozen sectioning. EdU (50mg/kg BW, Invitrogen) was administered either as a single IP injection to the dam for embryonic labeling, or daily (by IP injection) for 10 consecutive days to 1-month-old mice.
Immunofluorescence and fluorescence microscopy
Anti-GFP Rabbit IgG Antibody Fraction (Alexa Fluor 555 Conjugate; Invitrogen), anti-Aggrecan antibody (Millipore, AB1031), Anti-ASMA (Anti-Actin, α-Smooth Muscle; Sigma), Anti-CD31 (CD31/PECAM-1; Invitrogen), and Anti-Albumin (Abcam) were employed to detect their cognate proteins.
Results
Generation and characterization of Prg4+/GFPCreERt2 mice
We chose to knock-in a GFPCreERt2 cassette into the translation initiation site of the endogenous Prg4 locus (located in exon 2) to ensure that expression of this construct reflects the endogenous expression of the Prg4 gene (Figure 1A; Supplemental Figure 1A, B). Prg4+/GFPCreERt2 mice are viable, fertile, and lack the joint contractures, gait abnormalities, protein deposition on the articular cartilage, and synovial hyperplasia that are apparent in lubricin knock-out animals (15). Like Prg4−/− mice (15), the joints of 9-month old homozygous Prg4GFPCreERt2/GFPCreERt2 animals lacked the superficial-most layer of articular chondrocytes, exhibited a proteinaceous deposit on top of their articular cartilage (Supplemental Figure 1C) and displayed synoviocyte hyperplasia (data not shown). GFPCreERt2 expression in superficial zone chondrocytes of ~ 1-month-old Prg4+/GFPCreERt2 mice could be detected by employing an anti-GFP antibody (Figure 1B), but not by direct GFP fluorescence of the fusion protein (Supplemental Figure 2A), suggesting that GFPCreERt2 was expressed in superficial zone articular chondrocytes at a relatively low level. Consistent with this notion, RNA-seq analysis of articular cartilage revealed that GFPCreERt2 expression was 12- to 50-fold lower than endogenous Prg4 expression (Supplemental Table 1).
To assay recombination driven by the Prg4GFPCreERt2 allele, we crossed Prg4+/GFPCreERt2 animals with Rosa26floxlacZ/floxlacZ mice and analyzed β-galactosidase expression in the knee and hip joints of the double heterozygous offspring (i.e., Prg4+/GFPCreERt2; Rosa26+/floxlacZ) treated with tamoxifen. Expression of β-galactosidase was detected in femoro-tibial cartilage (Figure 1C), femoral cap cartilage (Figure 1D, E), synovial fibroblasts (Figure 1G), and the ligaments of the knee (Figure 1H) in Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice injected with tamoxifen. The observed pattern of expression reflects one previously reported for endogenous Prg4 expression using in situ hybridization (15), and also the expression pattern observed in mice containing a β-galactosidase-expressing Prg4 gene trap, Prg4+/lacZgenetrap (Figure 2W and unpublished observations). We further assessed the extent of Prg4GFPCreERt2–driven recombination in the articular cartilage by treating Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice with either a 1, 5, or 10 day course of tamoxifen, followed by euthanasia at P34, three days after the last injection. Whole mount X-Gal staining of femoral heads and knee joints in these mice demonstrated that after 10 days of tamoxifen injection, we observed high rates of Cre-mediated recombination in chondrocytes that were located in the superficial region of the articular cartilage (Figure 1I, compare c-f with g, h). In contrast, when we administered only vehicle (i.e., corn oil), very few cells (~ 5 cells per entire surface of the femoral cap) displayed evidence of Cre-mediated recombination (Figure 1Ia, b). Consistent with previous reports that Prg4/lubricin is expressed in articular but not in growth plate cartilage (15), we observed no evidence of Cre-mediated recombination in growth plate cartilage in tamoxifen-treated Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice (Figure 1F).
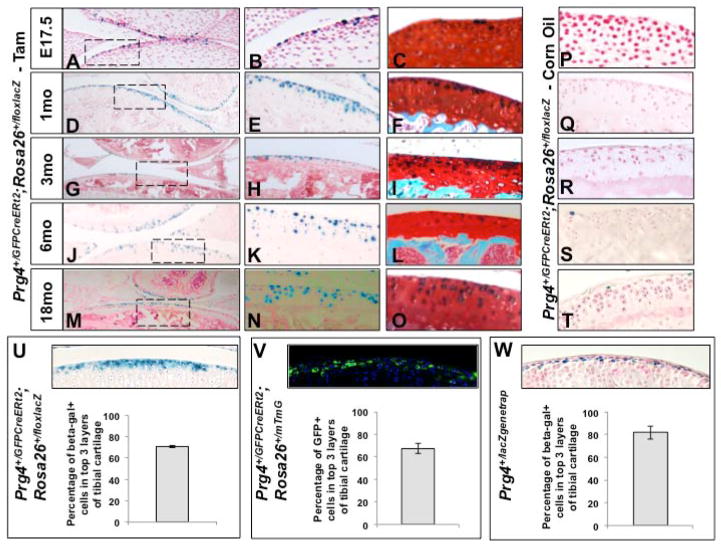
Figure 2. Prg4GFPCreERt2 is initially expressed by superficial zone chondrocytes and is additionally expressed by deeper zone chondrocytes as the mice age.
Photomicrographs of X-Gal stained knee joint cartilage from either P0 (A–C), 1-month-old (D–F), 3-month-old (G–I), 6-month-old (J–L), or 18-month-old (M–O) Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice that had been given either a single pulse (at E17.5) (A–C) or 10 consecutive days injection (D–O) with tamoxifen, 3 days prior to sacrifice. X-Gal/fast red staining (left panels). Higher magnification images of the boxed areas indicated in the corresponding left panel (center panels). High magnification images following X-Gal/safranin O/fast green staining (right panels). Knee joint tibial articular cartilage is displayed either on the bottom of the image or by itself. (P–T) Photomicrographs of X-Gal stained knee joint tibial articular cartilage from either P0, 1-month-old, 3-month-old, 6-month-old, or 18-month-old Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice treated with corn oil. (U, V) Either X-Gal staining (U) or Fluorescence microscopy (V) was performed on knee joint cartilage from either three P34 Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice (U) or three P34 Prg4+/GFPCreERt2; Rosa26+/mTmG mice (V) that were each given daily IP injections of tamoxifen from P21 to P31. Tibial cartilage of one such mouse of each genotype is displayed (top) and the average number (± 1SEM) of either X-Gal stained cells (U) or GFP fluorescing cells (V) within the 3 most superficial cell layers was determined (bottom). (W) X-Gal staining was performed on knee joint cartilage from a P34 Prg4+/lacZgenetrap mouse. The average number (± 1SEM) of β-galactosidase expressing cells within the 3 most superficial cell layers of tibial articular cartilage was determined.
Prg4GFPCreERt2 is expressed by superficial articular chondrocytes in young mice, but expands into deeper regions of the articular cartilage as the animals age
We evaluated the expression of Prg4 in the knee articular cartilage of mice at differing ages by pulsing either Prg4+/GFPCreERt2; Rosa26+/floxlacZ embryos with tamoxifen at embryonic day 17.5 (E17.5), or postnatal animals at 1, 3, 6, or 18 months-of-age. When given a single pulse of tamoxifen at E17.5 and harvested at postnatal day 0 (P0), β-galactosidase expressing cells were restricted to the superficial cell layers of the developing joint (Figure 2A–C). Mice given 10 days of tamoxifen, beginning at P21 and then sacrificed (at P34) 3 days after the last injection, displayed a majority of β-galactosidase-expressing cells in the 3 most superficial cell layers of the articular cartilage (Figure 2D–F). Interestingly, administration of tamoxifen to either 3-, 6- or 18-month-old Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice for 10 days (followed by sacrifice 3 days after the last injection of tamoxifen) resulted in β-galactosidase expression in deeper layers of the articular cartilage (Figure 2G–O). In contrast, in control Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice (that were injected with corn oil) very few cells displayed evidence of Cre-mediated recombination; with the oldest (18-month-old) mice showing the highest basal level of spontaneous recombination restricted to occasional superficial chondrocytes in their articular cartilage (Figure 2P–T). Consistent with these findings, we observed that a 15-month-old Prg4 gene-trap mouse (Prg4+/lacZgenetrap) similarly displayed β-galactosidase expression in both superficial and deeper regions of the femoral head articular cartilage, extending nearly to the subchondral bone (Supplemental Figure 3). Taken together, these findings indicate that expression of Prg4, which initially is restricted to the most superficial cells in the articular cartilage, is extended into deeper cells within this tissue as mice age.
Next, we determined the frequency of recombination driven by the Prg4GFPCreERt2 allele in the tibial articular cartilage of 1-month-old animals, using either Rosa26+/floxlacZ or Rosa26+/mTmG reporter alleles. We injected P21 mice with tamoxifen for 10 consecutive days, sacrificed the animals 3 days after the last injection at P34, and counted the number of recombined cells in the three most superficial cell layers of the tibial articular cartilage (Figure 2U, V). Recombination occurred in 70% and 67% of the three most superficial cell layers of the tibial articular cartilage in tamoxifen-treated Prg4+/GFPCreERt2; Rosa26floxlacZ and Prg4+/GFPCreERt2; Rosa26mTmG animals, respectively (Figure 2U, V). To estimate the percentage of cells that normally express Prg4 in these top three cell layers we used the β-galactosidase-expressing Prg4 gene-trap mouse strain (Prg4+/lacZgenetrap); this Prg4 gene-trap strain displayed detectable β-galactosidase expression in 82% of chondrocytes located in the top three cell layers of the articular cartilage (Figure 2W). Together, these data suggest that the effective recombination efficiency of the Prg4GFPCreERt2 allele (following 10 days of tamoxifen injection) is ~85% in Prg4 expressing articular chondrocytes.
Prg4GFPCreERt2 drives recombination outside of articular cartilage
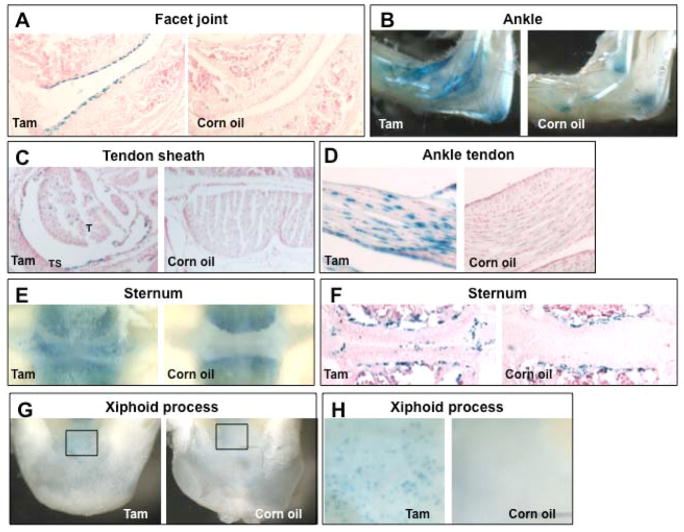
Extra-articular findings in patients with CACP syndrome, including pericarditis and intervertebral disc degeneration, suggested that PRG4 would be expressed at sites other than diarthrodial joints (14). This was confirmed by multi-tissue northern blot analysis (13) and other tissue-specific gene expression studies (e.g.,(25)). Therefore we looked for other sites of tamoxifen-induced Prg4GFPCreERt2-mediated recombination. In the skeletal system, we found recombination in tendons, tendon sheaths, vertebral facet joints, and sternal chondrocytes (Figure 3, Supplemental Figure 4). We also observed recombination in the heart and liver (Supplemental Figures 4, 5), consistent with prior findings that endogenous Prg4 is expressed in the liver (13). In this tissue, tamoxifen-induced Prg4GFPCreERt2 activity was detected in ~5% of the hepatocytes, but not in Kupffer, endothelial, or Ito cells (Supplemental Figure 5).
Figure 3. Prg4GFPCreERt2 is expressed in several cell types in 1-month-old mice.
Photomicrographs and photographs of X-Gal stained tissues that were harvested from P34 Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice that had been given daily IP injections of either tamoxifen (Tam) or vehicle (Corn oil) from P21 to P31. Sections displayed in A-F were counterstained with fast red. Note that Prg4+/GFPCreERt2 -mediated recombination is observed in vertebral facet joint chondrocytes (A), ankle tendons (B and D), tendon sheaths (C), sternal chondrocytes (E, F), and the Xiphoid process (G, H). T-tendon, TS-tendon sheath.
Prg4-expressing cells in both fetal and P21 mice are progenitors for chondrocytes in deeper zones of adult articular cartilage
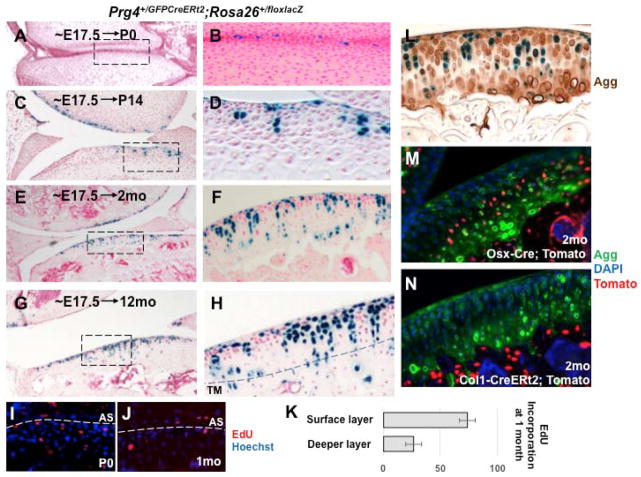
We performed lineage-tracing of Prg4-expressing cells in fetal (~E17.5) Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice by giving a single dose of tamoxifen to the dam. We then euthanized the dam’s offspring at either P0, P14, or at 2 months or 12 months-of-age; and determined the distribution of β-galactosidase-expressing chondrocytes in the femoro-tibial joint. We observed that β-galactosidase expression, which is specifically restricted to the superficial-most layer of the developing knee joint in P0 mice (Figure 2A–C; Figure 4A, B), expands into deeper layers of the knee joint in P14 animals (Figure 4C, D), extends throughout the depth of the articular cartilage in animals older than 2 months (Figure 4E, F), and beyond the tidemark in 12-month-old mice (Figure 4G, H). Of note, in all postnatal animals, the progeny of the recombined cells were present in both the superficial and deeper regions of the articular cartilage (Figure 4C–H). In addition, “columnar clones” of β-galactosidase-expressing chondrocytes that extended through the entire depth of the articular cartilage (extending from the unmineralized region into the mineralized matrix) were apparent in both 2- and 12-month-old animals (Figure 4E–H). In some cases, β-galactosidase expressing cells were also retained in the most superficial layer of such “columnar clones”, suggesting that the clonal progenitors gave rise to progeny in both the superficial cell layer and in deeper regions of the developing articular cartilage. These findings suggest that a radial expansion of Prg4-expressing cells in the articular cartilage takes place by 2 months of age to give rise to cells spanning the entire depth of this tissue. Indeed, a single pulse of EdU (at E17.5) was incorporated into proliferating cells in both the surface and deeper layers of the forming joint at P0 (Figures 4I). In contrast, postnatal EdU administration (by sequential injections of EdU from P21–P31) indicated that cellular proliferation in the articular cartilage was principally restricted to cells at or near the articular surface at P34 (Figure 4J, K), consistent with the notion that cells in the superficial region of the articular cartilage proliferate to give rise to cells that come to lie in deeper regions of this tissue. Importantly, β-galactosidase expressing chondrocytes in 2-month-old Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice (which had been administered tamoxifen at E17.5) were embedded throughout the aggrecan-positive articular cartilage matrix (Figure 4L), and extended well into the hypertrophic region of this tissue (marked by Osx-Cre and aggrecan expression; Figure 4M) but not into the underlying subchondral bone (marked by Col1-CreERt2 expression; Figure 4N). Taken together, these findings indicate that Prg4-expressing cells that are located in the superficial-most layer of the developing joints in the E17.5 mouse embryo give rise to deeper layers of articular chondrocytes, but not to subchondral osteocytes in the adult joint.
Figure 4. Prg4-expressing cells in the superficial-most cell layer of the forming joint in E17.5 embryos are progenitors for all zones of adult articular cartilage.
Pregnant dams were pulsed with tamoxifen at ~E17.5 and their Prg4+/GFPCreERt2; Rosa26+/floxlacZ progeny were harvested at either P0 (A, B), P14 (C, D), 2 months of age (E, F), or 12 months of age (G, H). Photomicrographs of X-Gal/fast red stained joints (left panels). Higher magnification images of the boxed areas indicated in the corresponding left panel (right panels). Knee joint tibial articular cartilage is displayed either on the bottom of the image or by itself. The tidemark (TM) was identified as a line of discontinuity in the intensity of fast red staining of the extracellular matrix, and is indicated by a dotted line. EdU incorporation into the knee joint cartilage in either P0 (I), or 1-month-old (J) animals. Location of the articular surface (AS) is delineated with the white dotted line. (K) Quantitation of the percentage of EdU-positive cells in either the superficial or deeper layers of the articular cartilage in 1-month-old animals injected for 10 days with EdU (as displayed in J; error bars indicate SD). (L) Immunodetection of aggrecan in 2-month-old Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice pulsed with tamoxifen at ~E17.5; lineage-traced cells are blue and remain within the articular cartilage as indicated by the aggrecan immunostaining. Expression of tdTomato and aggrecan in the knee joints of either 2-month-old Osx1-Cre; Rosa26+/floxdtTomato mice (M), or Col1-CreERt2-Cre; Rosa26+/floxdtTomato mice (that had been administered tamoxifen 7 days prior to sacrifice) (N).
We also performed lineage-tracing in P21 mice by treating them for 10 consecutive days with either tamoxifen or vehicle-alone and then analyzed their femoro-tibial joints at either P34 (i.e., 1 month), 6 months, or 18 months-of-age. Confirming what we previously had observed (Figure 2D–F), Prg4GFPCreERt2-mediated recombination occurred in cells closest to the articular surface in 1-month-old animals (Figure 5A, B). However, at 6 and 18 months of age, increasing numbers of recombined chondrocytes expressing β-galactosidase were present in both the superficial and deeper regions of the articular cartilage (Figure 5C–F), but rarely in the mineralized matrix (i.e., below the tidemark). The percentage of β-galactosidase expressing cells in the thickest region of the tibial articular cartilage to lie above the tidemark significantly increased between 1 and 18 months-of-age in Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice that had been injected with tamoxifen from P21-P30 (Figure 5G), suggesting that at least some of the Prg4GFPCreERt2-lineage labeled cells were proliferating during this time. Notably, the greatest depth of Prg4GFPCreERt2-lineage labeled cells in older animals was observed in the thickest region of the non-mineralized articular cartilage (usually near the middle of the condyle); the same region of the articular cartilage in which we previously observed the greatest amount of Prg4 induction in response to wheel running (27).
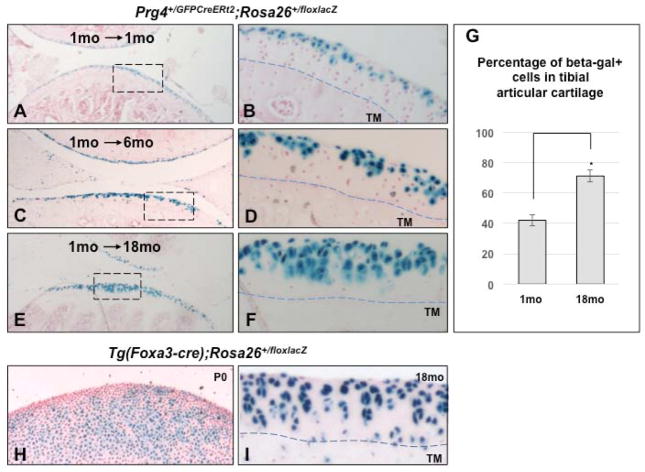
Figure 5. Prg4-expressing cells in 1-month-old mice are progenitors for both superficial and deeper regions of the adult articular cartilage.
Photomicrographs of knee joint cartilage from ~1 month (A, B), 6 months (C, D), and 18 months old (E, F) Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice that had been given daily IP injections of tamoxifen from P21 to P31. X-Gal/fast red staining (left panels). Higher magnification images of the boxed areas indicated in the corresponding left panel (right panels). (G) Quantitation of the percentage of cells expressing β-galactosidase located in the thickest region of the tibial articular cartilage (above the tidemark) in either 1- or 18-months-old Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice, that had been given daily IP injections of tamoxifen from P21 to P31 (error bars indicate SD; *p<0.05; Student’s test). P0 (H) or 18-month-old (I) Tg(Foxa3-cre); Rosa26+/floxlacZ knee articular cartilage stained with X-Gal/fast red. The tidemark (TM) was identified as a line of discontinuity in the intensity of fast red staining of the extracellular matrix, and is indicated by a dotted line.
A 1-day pulse of tamoxifen to fetal (~E17.5) Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice yielded “columnar clones” of β-galactosidase-expressing chondrocytes that extended the entire depth of the articular cartilage in 12-month-old animals, even extending below the tidemark in some cases (Figure 4G,H). In contrast, a 10-day pulse of tamoxifen to 1-month-old Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice resulted in β-galactosidase-expressing chondrocytes that extended to maximally two-thirds the depth of the articular cartilage in 18-month-old animals, and were not found below the tidemark (Figure 5E, F). Taken together, these findings imply that the deepest cells of the articular cartilage that lie adjacent to the tidemark are descendants of Prg4-expressing cells that were lineage- labeled at E17.5, but are not descendants of Prg4-expressing cells that were lineage- labeled at 1 month-of-age. In comparison, 18-month-old Tg(Foxa3-cre); Rosa26+/floxlacZ mice (20) display β-galactosidase-expressing chondrocytes throughout the full depth of their articular cartilage (Figure 5I). At P0, Tg(Foxa3-cre); Rosa26+/floxlacZ mice express β-galactosidase in all chondrocyte zones of the growth plate (26) and throughout the entire epiphyseal cartilage (Figure 5H). In summary, these results indicate that the earliest born cells (that are descendants of cells expressing Prg4 at E17.5 but not at P30) give rise to the deepest layer of the articular cartilage, and thus suggest that articular cartilage grows in an “appositional” fashion (much like the growth plate) with a progenitor population confined to the superficial region of the developing articular cartilage.
Discussion
We generated knock-in mice that express a tamoxifen-inducible Cre recombinase from the Prg4 locus (Prg4GFPCreERt2). In the knee joint, Prg4GFPCreERt2 is expressed by cells at the surface of the developing joint in E17.5 mice and by superficial articular chondrocytes in 1-month-old mice. Therefore, we employed the Prg4GFPCreERt2 allele along with the Cre-inducible reporter gene, Rosa26floxlacZ, to fate-map the descendants of Prg4-expressing cells in both fetal and 1-month-old animals. We found that Prg4-expressing cells, which were lineage-labeled at the surface of the developing joint in E17.5 mouse embryos, gave rise to columns of β-galactosidase expressing cells; that in some cases extended through the entire depth of the articular cartilage in 2-month-old animals, and to just below the tidemark in 12-month-old-animals. In some instances, these columns of β-galactosidase expressing cells (that had been lineage-labeled by Prg4GFPCreERt2 expression in E17.5 embryos) extended from only the middle of the non-mineralized matrix to the surface, or clusters of β-galactosidase expressing cells were restricted to either the deep or superficial regions of the articular cartilage. We interpret these findings to indicate that Prg4-expressing cells in the developing joint of the E17.5 embryo give rise to chondrocytes in all regions of the articular cartilage, and that the E17.5 lineage-labeled descendants of these Prg4-expressing cells display differing amounts of subsequent cell division. Prg4-expressing chondrocytes in 1-month-old mice also produced descendants in deeper regions of the articular cartilage. However, in this case the lineage-labeled descendants of Prg4-expressing cells (which were labeled at 1 month of age) extended from the articular surface to maximally two-thirds the depth of the articular cartilage in 18-month-old animals, and never reached the tidemark in these older animals. These findings indicate that the deepest cells of the articular cartilage that lie adjacent to the tidemark are descendants of Prg4-expressing cells that were lineage-labeled at E17.5 but are not descendants of Prg4-expressing cells that were lineage-labeled at 1 month-of-age. Thus our findings suggest that the deeper layers of the articular cartilage are “born” first. We interpret these observations to indicate that a significant component of articular cartilage growth is appositional, with Prg4-expressing progenitor cells first giving rise to cells that will eventually come to lie deep within this tissue, and subsequently giving rise to cells that will eventually come to lie in more superficial regions of the articular cartilage (outlined in Figure 6). Interestingly, we noted that Prg4-expressing cells that were lineage-labeled in 1-month-old Prg4+/GFPCreERt2; Rosa26+/floxlacZ mice gave rise to β-galactosidase expressing cells in the meniscus, synovium, and patella (Supplemental Figure 6), even though most β-galactosidase expressing superficial chondrocytes had been lost in these older animals. The presence of β-galactosidase expressing synoviocytes in 18-month-old mice suggests either that some Prg4GFPCreERt2 -expressing cells in 1-month-old-mice have “stem cell” like features that continue to generate synoviocytes or that some synoviocytes live for a very long time. Importantly, only a small fraction of synovial lining cells (~5%) were β-galactosidase-positive in these mice, implying that most synoviocyte stem cells were not expressing Prg4GFPCreERt2 at 1 month of age.
Figure 6. Model depicting the growth of articular cartilage based on Prg4GFPCreERt2 fate-mapping.
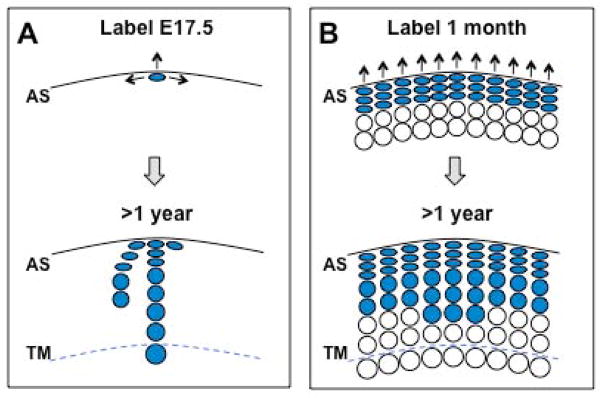
Growth occurs appositionally toward the articular surface (AS) and also laterally in the superficial zone. (A) The progeny of embryonic (E17.5) Prg4GFPCreERt2-expressing cells (colored blue) are present in all articular cartilage layers, and can in some cases extend as columnar clones from the superficial zone to the calcified cartilage beneath the tidemark (TM) in adult mice. (B) The progeny of Prg4GFPCreERt2-expressing cells in 1-month-old animals (colored blue) extend from the superficial zone to maximally two-thirds the depth of the articular cartilage. These findings indicate that the deepest cells of the articular cartilage that lie adjacent to the tidemark are descendants of Prg4GFPCreERt2 -expressing cells that were lineage labeled at E17.5 but are not descendants of Prg4GFPCreERt2 -expressing cells that were lineage-labeled at 1 month-of-age. Thus our findings suggest that the deeper layers of the articular cartilage are “born” first.
We noted that while there were only a relatively small number of recombined cells located at the surface of the developing joint in Prg4+/GFPCreERt2; Rosa26+/floxlacZ animals that had been injected with tamoxifen at E17.5 and sacrificed at P0, the number of recombined cells (located in both the superficial and deeper regions of the knee joint) in animals sacrificed at 12-months-of-age increased by several-fold. This increased number of recombined progeny cells in both the superficial and deeper layers of the articular cartilage in post-natal animals suggest that superficial progenitors in the embryo both self-renew (in the superficial zone) and give rise to cells located in deeper regions of the adult articular cartilage. In addition, because proliferation mostly occurs in relatively superficial articular cartilage cells in 1-month-old mice, it seems likely that superficial zone articular cartilage cells comprise a progenitor cell population for this tissue, in both prenatal and young mice. Indeed cells in the superficial zone of articular cartilage have previously been demonstrated to be label-retaining cells (10, 11), suggesting that these cells may constitute a slowly cycling stem cell population. Our findings do not rule out the possibility that the Prg4-expressing population (at either E17.5 or P30) is heterogeneous, with only a subset of these cells serving as progenitors for the expansion of the articular cartilage. Notably, these findings are consistent with prior observations, which have indicated that while transient epiphyseal chondrocytes (which are subsequently replaced by bone) are derived from matrilin-1 expressing progenitors, adult articular cartilage derives from progenitors that never have expressed matrilin-1 (6).
Interestingly, the greatest apparent expansion of Prg4-expressing progenitors in the knee joint (at either E17.5 or P30) occurred in the tibial articular cartilage, near the middle region of the condyles. This is the same region of the articular surface in which we previously observed the greatest amount of Prg4 induction in response to wheel running (27). In addition to inducing Prg4 expression, wheel running also promotes proliferation of Prg4-expressing cells in the articular cartilage (27). Thus, it seems plausible that the greatest amount of cell proliferation in Prg4-expressing articular cartilage progenitors occurs in the regions of this tissue that experience the highest levels of mechanical loading, and thus lie in the central domains of the condyles.
Inducing Prg4GFPCreERt2-mediated recombination in older mice (i.e., 3, 6, or 18 months-of-age) revealed Prg4GFPCreERt2 expression in both superficial and in deeper zone chondrocytes. This finding was confirmed in Prg4 gene-trap mice (Prg4+/lacZgenetrap), which similarly displayed β-galactosidase expression uniquely in the superficial zone of the articular cartilage at 1 month-of-age, but in both the superficial and deeper regions of femoral head articular cartilage at 15 months-of-age. While both Prg4GFPCreERt2 and Prg4lacZgenetrap transcripts lack exons 3 to 13 of native Prg4 (and thus may display differential mRNA stability compared to the native transcript) our findings suggest that expression of the Prg4 locus, which initially is restricted to the most superficial cells in the articular cartilage, is extended into deeper cells within this tissue as mice age. Thus, in contrast to either E17.5 or 1-month-old mice, in which Prg4GFPCreERt2 expressing cells are mostly restricted to the top layers of the articular cartilage, in older animals deeper zone chondrocytes (in addition to superficial zone chondrocytes) are capable of expressing Prg4. It seems unlikely that the age-dependent expansion of Prg4 expression into deeper regions of the articular cartilage in both Prg4+/GFPCreERt2 and Prg4+/lacZgenetrap mice is due to haploinsufficiency of Prg4 in these animals, as the articular cartilage of these heterozygous mice neither displays age-dependent fibrillation nor cartilage degradation. In addition, families carrying camptodactyly-arthropathy-coxa vara-pericarditis syndrome (CACP) mutations in PRG4 do not report an increased incidence of osteoarthritis among individuals who are heterozygous for mutations in this gene. Taken together, our findings indicate that chondrocytes that lie in deeper regions of the articular cartilage both originate from precursors that had initially expressed Prg4 within the superficial zone, and become capable of re-expressing Prg4 in response to aging.
As anticipated from the clinical consequences of genetic PRG4 deficiency, Prg4GFPCreERt2 expression was not limited to articular chondrocytes. We also observed expression in other tissues previously reported to express Prg4, including synovium, tendon, ligament, heart, and liver. In addition, we observed scattered Prg4GFPCreERt2 expressing cells in other sites, including sternal cartilage, the xiphoid process, and the proximal part of the aorta. Importantly, we never observed Prg4GFPCreERt2-mediated recombination in growth plate chondrocytes, which distinguishes Prg4GFPCreERt2 mice from both Col2CreERt2 and AggCreERt2 mice (28, 29) that express CreERt2 in both growth plate and articular cartilage. Ten consecutive days of tamoxifen treatment of 1-month-old Prg4GFPCreERt2/+ mice is capable of inducing recombination of either a Rosa26floxlacZ or a Rosa26mTmG allele in approximately 70% of the top 3 layers of the articular cartilage. We think that the relatively low level expression of the Prg4GFPCreERt2 allele (approximately 12 to 50-fold lower than endogenous Prg4) may account for the need for multiple tamoxifen injections to induce maximal levels of recombination in the articular cartilage. Unlike the GDF5Cre allele, which can drive recombination in articular cartilage progenitors but not in the growth plate in the embryo (5, 30), the Prg4+/GFPCreERt2 mouse strain uniquely drives recombination specifically in both adult articular cartilage and in synovial cells, and thus can be employed to determine the effect of altering expression of a floxed gene in these tissues in adult animals, without affecting that gene’s function in growth plate cartilage. On the other hand, because recombination driven by the Prg4GFPCreERt2 allele is only transiently restricted to the superficial zone of articular cartilage (in animals P30 or younger) and displays age-dependent expansion into deeper zones of the articular cartilage, the localization of genetically altered cells driven by this Cre driver changes as animals age.
Supplementary Material
Acknowledgments
This work was supported by grants from NIH/NIAMS (R21RAR055148A; R01-AR055552) to A.B.L., (R01-AR050180) to M.L.W., (P01 DK56246) to H. K, NIH/NIDDK (R01-DK094933) to A.K., and NIH/NIDCR (K99-DE022564) to N.O..
We thank “The Nikon Imaging Center at Harvard Medical School” for the use of their microscopes in this study.
References
- 1.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–6. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre V, Bhattaram P. Vertebrate skeletogenesis. Curr Top Dev Biol. 2010;90:291–317. doi: 10.1016/S0070-2153(10)90008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pacifici M, Koyama E, Iwamoto M. Mechanisms of synovial joint and articular cartilage formation: recent advances, but many lingering mysteries. Birth Defects Res C Embryo Today. 2005;75(3):237–48. doi: 10.1002/bdrc.20050. [DOI] [PubMed] [Google Scholar]
- 4.Onyekwelu I, Goldring MB, Hidaka C. Chondrogenesis, joint formation, and articular cartilage regeneration. J Cell Biochem. 2009;107(3):383–92. doi: 10.1002/jcb.22149. [DOI] [PubMed] [Google Scholar]
- 5.Koyama E, Shibukawa Y, Nagayama M, Sugito H, Young B, Yuasa T, et al. A distinct cohort of progenitor cells participates in synovial joint and articular cartilage formation during mouse limb skeletogenesis. Dev Biol. 2008;316(1):62–73. doi: 10.1016/j.ydbio.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hyde G, Dover S, Aszodi A, Wallis GA, Boot-Handford RP. Lineage tracing using matrilin-1 gene expression reveals that articular chondrocytes exist as the joint interzone forms. Dev Biol. 2007;304(2):825–33. doi: 10.1016/j.ydbio.2007.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mankin HJ. Mitosis in articular cartilage of immature rabbits. A histologic, stathmokinetic (colchicine) and autoradiographic study. Clin Orthop Relat Res. 1964;34:170–83. [PubMed] [Google Scholar]
- 8.Hunziker EB, Kapfinger E, Geiss J. The structural architecture of adult mammalian articular cartilage evolves by a synchronized process of tissue resorption and neoformation during postnatal development. Osteoarthritis Cartilage. 2007;15(4):403–13. doi: 10.1016/j.joca.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Hayes AJ, MacPherson S, Morrison H, Dowthwaite G, Archer CW. The development of articular cartilage: evidence for an appositional growth mechanism. Anat Embryol (Berl) 2001;203(6):469–79. doi: 10.1007/s004290100178. [DOI] [PubMed] [Google Scholar]
- 10.Yasuhara R, Ohta Y, Yuasa T, Kondo N, Hoang T, Addya S, et al. Roles of beta-catenin signaling in phenotypic expression and proliferation of articular cartilage superficial zone cells. Lab Invest. 2011;91(12):1739–52. doi: 10.1038/labinvest.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Candela ME, Cantley L, Yasuaha R, Iwamoto M, Pacifici M, Enomoto-Iwamoto M. Distribution of slow-cycling cells in epiphyseal cartilage and requirement of beta-catenin signaling for their maintenance in growth plate. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2014;32(5):661–8. doi: 10.1002/jor.22583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowthwaite GP, Bishop JC, Redman SN, Khan IM, Rooney P, Evans DJ, et al. The surface of articular cartilage contains a progenitor cell population. J Cell Sci. 2004;117(Pt 6):889–97. doi: 10.1242/jcs.00912. [DOI] [PubMed] [Google Scholar]
- 13.Marcelino J, Carpten JD, Suwairi WM, Gutierrez OM, Schwartz S, Robbins C, et al. CACP, encoding a secreted proteoglycan, is mutated in camptodactyly-arthropathy-coxa vara-pericarditis syndrome. Nat Genet. 1999;23(3):319–22. doi: 10.1038/15496. [DOI] [PubMed] [Google Scholar]
- 14.Bahabri SA, Suwairi WM, Laxer RM, Polinkovsky A, Dalaan AA, Warman ML. The camptodactyly-arthropathy-coxa vara-pericarditis syndrome: clinical features and genetic mapping to human chromosome 1. Arthritis Rheum. 1998;41(4):730–5. doi: 10.1002/1529-0131(199804)41:4<730::AID-ART22>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 15.Rhee DK, Marcelino J, Baker M, Gong Y, Smits P, Lefebvre V, et al. The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. J Clin Invest. 2005;115(3):622–31. doi: 10.1172/JCI200522263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mugford JW, Sipila P, McMahon JA, McMahon AP. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev Biol. 2008;324(1):88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25(2):139–40. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 18.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 19.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 20.Lee CS, Sund NJ, Behr R, Herrera PL, Kaestner KH. Foxa2 is required for the differentiation of pancreatic alpha-cells. Dev Biol. 2005;278(2):484–95. doi: 10.1016/j.ydbio.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133(16):3231–44. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 22.Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 2010;19(2):329–44. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13(1):133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Berger EJ, Zhao C, Jay GD, An KN, Amadio PC. Expression and mapping of lubricin in canine flexor tendon. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2006;24(9):1861–8. doi: 10.1002/jor.20239. [DOI] [PubMed] [Google Scholar]
- 26.Ionescu A, Kozhemyakina E, Nicolae C, Kaestner KH, Olsen BR, Lassar AB. FoxA family members are crucial regulators of the hypertrophic chondrocyte differentiation program. Dev Cell. 2012;22(5):927–39. doi: 10.1016/j.devcel.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogawa H, Kozhemyakina E, Hung HH, Grodzinsky AJ, Lassar AB. Mechanical motion promotes expression of Prg4 in articular cartilage via multiple CREB-dependent, fluid flow shear stress-induced signaling pathways. Genes Dev. 2014;28(2):127–39. doi: 10.1101/gad.231969.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128(24):5099–108. doi: 10.1242/dev.128.24.5099. [DOI] [PubMed] [Google Scholar]
- 29.Henry SP, Jang CW, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis. 2009;47(12):805–14. doi: 10.1002/dvg.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rountree RB, Schoor M, Chen H, Marks ME, Harley V, Mishina Y, et al. BMP receptor signaling is required for postnatal maintenance of articular cartilage. PLoS Biol. 2004;2(11):e355. doi: 10.1371/journal.pbio.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.