Abstract
DNA methylation is a key epigenetic modification involved in regulating gene expression and maintaining genomic integrity. Here we inactivated all three catalytically active DNA methyltransferases in human embryonic stem cells (ESCs) using CRISPR/Cas9 genome editing to further investigate their roles and genomic targets. Disruption of DNMT3A or DNMT3B individually, as well as of both enzymes in tandem, creates viable, pluripotent cell lines with distinct effects on their DNA methylation landscape as assessed by whole-genome bisulfite sequencing. Surprisingly, in contrast to mouse, deletion of DNMT1 resulted in rapid cell death in human ESCs. To overcome the immediate lethality, we generated a doxycycline (DOX) responsive tTA-DNMT1* rescue line and readily obtained homozygous DNMT1 mutant lines. However, DOX-mediated repression of the exogenous DNMT1* initiates rapid, global loss of DNA methylation, followed by extensive cell death. Our data provide a comprehensive characterization of DNMT mutant ESCs, including single base genome-wide maps of their targets.
Introduction
DNA methylation is a heritable and reversible enzyme-mediated modification to DNA that is implicated in a diverse range of biological processes1. In mammals, there are three catalytic active DNA methyltransferases (DNMTs): DNMT1, which is largely responsible for the maintenance of DNA methylation over replication, and DNMT3A and DNMT3B, which generally perform de novo methylation of either unmethylated DNA or hemimethylated DNA to assist in maintenance2. Deletion of these enzymes in mice results in embryonic (Dnmt1 and 3b) or postnatal (Dnmt3a) lethality, confirming their essential roles in development3,4. In line with knockout mouse models, mutations affecting the human DNA methyltransferases are linked to several diseases, including acute myeloid leukemia (AML) and Immunodeficiency, centromere instability and facial anomalies (ICF) syndrome5–8.
DNA methylation has been extensively mapped in somatic cells as well as ESCs in human and mouse9–14. Human ESC methylation patterns are most unique at hypomethylated regulatory elements that are enriched for binding of pluripotency-associated master regulators, such as OCT4, SOX2, and NANOG14. Additionally, both mouse and human ESCs display increased levels of non-CpG methylation that is not observed in most somatic lineages, consistent with the high activity of de novo methyltransferases in these cells11,15–17.
While deletion of Dnmt1 is lethal in all dividing somatic cells3,18–21, mouse ESCs are viable despite global loss of DNA methylation. In fact, all three Dnmts can be removed from these cells without any deleterious effects in the undifferentiated state22. As such, mouse ESCs have become a powerful tool to study the role and function of Dnmt enzymes, which clarified some of their specific targets and provided many general insights into the biology of DNA methylation23. No comparable efforts have been reported for human pluripotent stem cells and loss of function studies have been limited to the depletion of DNMT1 in the colon cancer cell line HCT116, which results in cell death24,25 and therefore indicates a similar requirement for maintenance of DNA methylation patterns in human cells. DNMT3B was reported to cooperate with DNMT1 to maintain methylation in HCT116 cells26,27 and its depletion results in altered timing of neuronal differentiation and maturation28. Recently, a human ESC model for ICF syndrome was reported by targeted disruption of DNMT3B29.
The limited number of human studies can likely be attributed to long-standing difficulties of genetic manipulation in the human ESC system, rather than lack of interest. However, new classes of genome-editing tools, transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems have been applied for genomic editing in mammalian cells30–34. These techniques have substantially enhanced targeting efficiency into human ESCs, allowing previously difficult studies to be undertaken33,35.
Here we report the inactivation of all three DNMTs in human ESCs and present a detailed analysis of the DNA methylation changes in DNMT1, DNMT3A, DNMT3B and tandem DNMT3A/3B double knockouts (DNMT3A−/−3B−/−). We further characterized the differentiation potential of the knockout lines using gene expression signatures, EB differentiation and teratoma formation. We also used directed differentiation towards the endoderm lineage to assess the role of DNMT3A in this transition. Most notably, we were unable to obtain DNMT1 homozygous deletions without implementing a doxycycline-repressible rescue line, demonstrating that loss of DNMT1 is lethal. Taken together, our results highlight several unique aspects of DNA methylation biology in the context of human ESCs and provide controlled, tractable systems to dissect the function of DNMTs in precise detail.
Results
Disrupting the catalytic domain of all three DNMTs
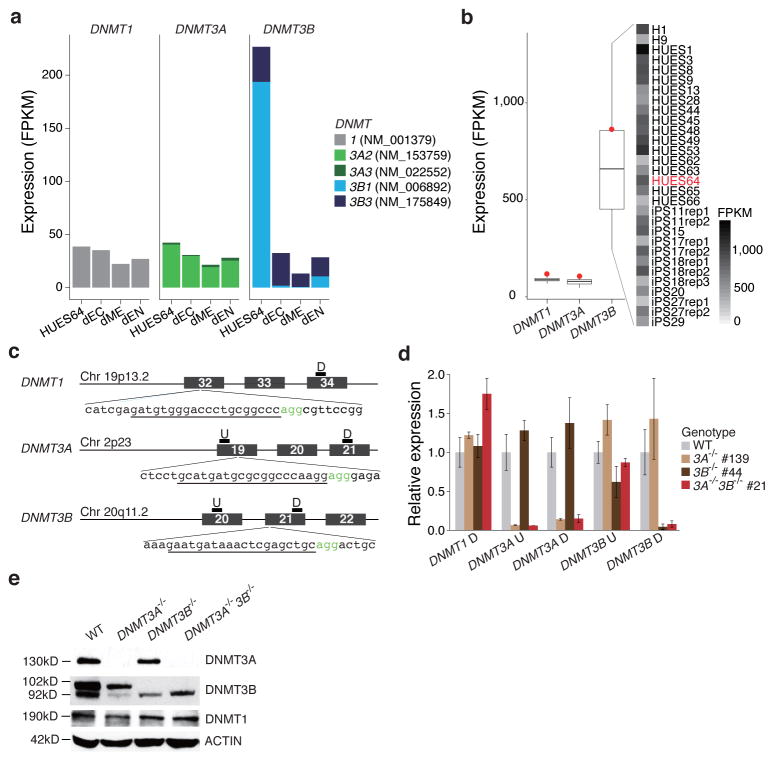
Of the three catalytically active DNMTs, DNMT3B shows the highest level of expression in undifferentiated ESCs (Fig. 1a), but also the most variation when examined across 25 pluripotent lines (Fig. 1b). As ESCs differentiate, DNMT1 and DNMT3A remain expressed at comparable levels, while DNMT3B is strongly downregulated and switches to predominant expression of a catalytically inactive isoform (isoform 3B1 NM_006892 to isoform 3B3 NM_175849; Fig. 1a, Supplementary Fig. 1a). While inactive, this isoform can likely still form complexes with catalytically competent DNMT3A and/or DNMT3B to contribute to DNA methylation activity36.
Figure 1. Targeted deletion of DNMT1, 3A and 3B in human ESCs.
(a) Expression levels of DNMT1, 3A and 3B in undifferentiated HUES64 ESCs and their derivatives, ectoderm (dEC), mesoderm (dME) and endoderm (dEN) in FPKM (Fragments Per Kilobase per Million fragments mapped) are shown. Only expression of the major isoforms is shown (see Supplementary Fig. 1a for all). (b) Left: Expression of DNMT1, 3A, 3B for 25 pluripotent (ESC and iPSC) lines (line: median, box: IQR, whiskers: furthest point within 1.5xIQR, red dot: HUES64). Right: Cell lines and DNMT3B expression. There is substantial variation of DNMT3B expression even among biological replicates (“rep”).(c) Overview schematic of the Cas9/gRNA-target sites. Genomic coordinates are shown on the right. The gRNA-targeting sequence is underlined, and the Protospacer-Adjacent Motif (PAM) sequence is labeled in green. Position of qPCR primers for RNA expression validation is shown on the top of the exons. P: primer pair; U: upstream; D: downstream.(d) RT-qPCR analysis for DNMT1, 3A and 3B in HUES64, DNMT3A−/− (#139), DNMT3B−/− (#44) and DNMT3A−/−3B−/− (#21). Primer details are in Fig. 1c. Error bars were generated from biological triplicates and represent one standard error.
(e) Western blot analysis for DNMT3A, 3B and DNMT1 in HUES64, DNMT3A−/− (#139), DNMT3B−/− (#44) and DNMT3A−/−3B−/− (#21). Arrow indicates the major DNMT3B isoform (NM_006892). The smaller band in the DNMT3B western blotting result is likely isoform 3 (NM_175849) as reported in Ref. 55, which was not targeted by our knockout strategy.
To better understand the role of the three DNMTs in human pluripotent stem cells and over cellular differentiation, as well as to study their specific target spectrum at high-resolution across the genome, we created targeted deletions for DNMT1, DNMT3A, DNMT3B and both DNMT3A and 3B in human ESCs. We selected the male line HUES64 for the following reasons: (i) it is on the NIH registry and generally available to researchers, (ii) it differentiates well into the three germ layers, (iii) it is karyotypically normal (Supplementary Fig. 1b) and grows well under standard culture conditions, and (iv) a substantial amount of publicly available transcriptional, epigenomic and transcription factor binding data have been generated for this line9,37.
In order to avoid hypomorphic effects24, we designed guide RNA G(N)19NGG sequences targeting the catalytic domains for DNMT1, 3A and 3B (Fig. 1c). After picking and expanding individual clones, we confirmed the targeted disruption of the catalytic domains by Sanger sequencing (Supplementary Fig. 1c, d). All experiments yielded high rates of mutations ranging from 53% to 66%, with homozygous deletion occurring in 3–6% of the clones for DNMT3A and B (Supplementary Fig. 1c). Double knockout cells were derived by targeting the already validated DNMT3A−/− clone (#139) with the DNMT3B gRNA and Cas9. To further validate the DNMT3 knockouts, we performed qPCR (Fig. 1d) and western blotting (Fig. 1e). Notably, while we obtained homozygous knockouts for DNMT3A and DNMT3B, both singly and in combination, we were unable to obtain any DNMT1−/− clones, despite high (>50%) efficiency when deriving heterozygous lines.
DNMT3 knockout ESCs remain pluripotent
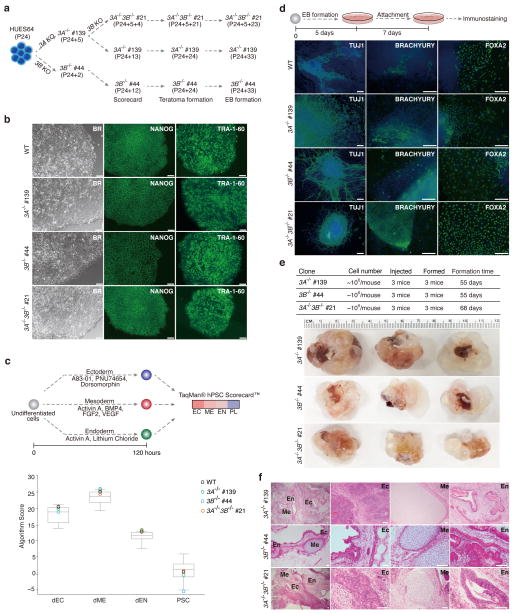
All of our DNMT3 knockout clones grew well and were morphologically normal. We selected representative clones for each knockout (DNMT3A−/−: #139; DNMT3B−/−: #44; DNMT3A−/−3B−/−: #21) and performed several assays to characterize these knockouts, both in terms of their molecular phenotype as well as of their differentiation potential (Fig. 2a). The clones were positive for the pluripotency-associated markers, NANOG and TRA1-60 (Fig. 2b) and showed tri-lineage differentiation potential based on the TaqMan® hPSC Scorecard™ (Fig. 2c; Supplementary Fig. 2)38. We differentiated these clones into embryoid bodies and the resulting cells were stained for markers of the three germ layers (Fig. 2d). Finally, we injected each clone into the kidney capsule of three independent NOD-SCID mice to test for teratoma formation (Fig. 2e) and confirmed the presence of cell types representing the three germ layers in several sections per knockout (Fig. 2f). These results show that deletion of the DNMT3 enzymes, either individually or in combination, has no obvious effect on morphology, viability and initial differentiation potential in human ESCs. This observation is consistent with prior studies of the respective knockouts in mouse ESCs, where both individual knockouts can differentiate, although the mouse double knockouts could only generate teratomas at early passage numbers4,39.
Figure 2. Assessing the differentiation potential of the DNMT3 knockouts.
(a) Overview of clones and passage numbers used for the various assays. HUES64 passage 24 (P24) was used to generate DNMT3A and 3B single knockouts. The number after “+” represents the passage number post targeting. The DNMT3A−/− clone #139 was passaged five times (P24+5) and then targeted using the DNMT3B gRNA. The DNMT3A−/−3B−/− passage numbers are then counted from there.
(b) Wild-type and DNMT3A−/−, 3B−/− and 3A−/−3B−/− cells show comparable morphology and expression of characteristic markers. Representative bright field (BR) and pluripotency-associated marker (NANOG, TRA-1-60) immunostaining images are shown. 10X magnification is shown. Scale bars, 100μm.
(c) TaqMan hPSC Scorecard analysis of direct differentiation potential for WT, DNMT3A−/−, 3B−/− and 3A−/−3B−/− cells. Top: Schematic overview of the conditions used for direct differentiation into ectoderm, mesoderm and endoderm. Bottom: Differentiation scores for WT, DNMT3A−/−, 3B−/− and 3A−/−3B−/− cells. Gray box represents the score range of 11 reference human PSC lines.
(d) Top: Schematic overview for the embryonic body (EB) formation. Bottom: Representative immunostaining images for TUJ1 (ectoderm), BRACHYURY (mesoderm) and FOXA2 (endoderm). 10X magnification is shown. Scale bars, 100μm.
(e) Top: Summary of injection and teratoma formation results. Bottom: Images of the nine teratomas collected. Ruler on top provides a size reference.
(f) Representative images of Hematoxylin and Eosin (H&E) staining of the teratoma from panel e. Neuroepithelium (ectoderm) (EC), cartilage with ossification center (mesoderm) (ME), and gut-like epithelium (EN). Scale bars, 50μm.
Passaging results in progressive loss of DNA methylation
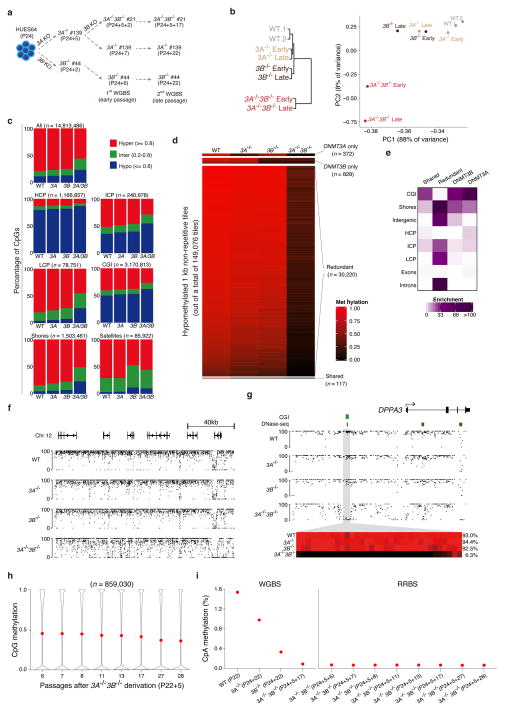
To characterize the effects of the DNMT3 knockouts on the human ESC methylome, we harvested genomic DNA from our mutant lines at an early (2–7) and late (17–22) passage and generated WGBS libraries (Fig. 3a). We utilized the methylation status of 6.9 million CpGs that were covered at 5x in all eight samples for the initial comparative analysis. Hierarchical clustering of the WGBS data shows that WT, DNMT3A−/− and DNMT3B−/− cells cluster closely together, but remain clearly segregated nonetheless (Fig. 3b, left). Despite being derived from the DNMT3A−/− clone, and therefore beginning in a DNMT3A-depleted background, the double knockout is clearly distinguishable, highlighting that complete de novo methyltransferase ablation dominates effects on global DNA methylation levels over those initiated by the original single knockout. To further assess the effects of the knockouts and of passage, we performed principal component analysis (Fig. 3b, right). The first two principal components explain 96% of the variation between the samples and separate them into two main groups: the first consists of the wild-type human ESCs, the DNMT3A and DNMT3B single knockouts, while the second includes the early and late passage double knockouts.
Figure 3. Global DNA methylation dynamics.
(a) Overview of the samples used for whole genome bisulfite sequencing (WGBS).
(b) Left: Hierarchical clustering using Euclidean distance based on mean DNA methylation levels of 1 kb tiles across the human genome. Right: Principal component analysis (PCA) based on mean CpG methylation levels for 1 kb tiles.
(c) Fraction of CpGs with high (≥ 0.8, red), intermediate (inter, >0.2 and <0.8, green) and low (≤ 0.2, blue) methylation values. Top left panel shows all CpGs while the other panels show CpG methylation distribution within different genomic features, including high CpG Promoters (HCP), intermediate CpG promoters (ICP), low CpG promoters (LCP), CpG islands (CGI), CpG island shores, and satellite repeats. The total number of CpGs associated with each feature is shown above each bar plot.
(d) Heatmap of non-repetitive differentially methylated 1kb tiles (q-value < 0.05 and methylation difference greater than 0.4).
(e) Enrichment [−log10(hypergeometric p-value)] of genomic features in the target classes identified in Fig. 3d.
(f) Genome browser tracks covering approximately 160kb in the different lines.
(g) Higher resolution view of the DPPA3 (STELLA) locus. The heat map below shows the DNA methylation values of individual CpGs within the grey region. The average DNA methylation value for the entire highlighted region is shown on the right.
(h) CpG methylation levels over several passages of the DNMT3A−/−3B−/− cells. Red dot indicates mean methylation.
(i) Global mean CpA methylation levels as assayed by WGBS and over several passages of the DNMT3A−/−3B−/− cells by RRBS.
We then analyzed the WGBS data from the later passage samples, which captured 14.8 million CpGs covered at 5x coverage in all four samples, to see if specific genomic features were particularly sensitive to the loss of de novo DNA methyltransferase activity. We divided all CpGs into three classes according to their methylation values—high (≥0.8), intermediate (>0.2 and <0.8) or low (≤ 0.2)—and examined the relative proportion falling into each category (Fig. 3c). The fraction of intermediate and low methylated regions in the knockouts gradually increases over passage, especially when comparing DNMT3A−/−3B−/− versus WT, which is similar to equivalent mouse knockout models (Supplementary Fig. 3a). Most genomic features show a mild decrease in DNA methylation in the single and a more substantial change in the double knockouts, with the exception of satellites, which appear particularly sensitive to the loss of DNMT3B (Fig. 3c, Supplementary Fig. 3b–d).
We next divided the genome into 1kb tiles and compared the distribution of CpG methylation values within each tile across the samples (Fig. 3d). We defined differentially methylated regions (DMRs) as those tiles that were significantly different (weighted t-test, q value < 0.05) between the wild-type and any of the knockouts and had an average methylation difference greater than 0.4, ignoring repetitive tiles. These DMRs can be divided into four classes: (i) DNMT3A specific targets, (ii) DNMT3B specific targets, (iii) shared targets that require both DNMT3A and DNMT3B, (iv) redundant targets that require either DNMT3A or 3B and are only lost on DNMT3A−/−3B−/− cells. As suggested by the CpG level analysis above, almost all DMRs (~96%) are redundantly targeted by DNMT3A and DNMT3B and only lose methylation when both are ablated. For each class, we calculated the enrichment of different genomic features. Redundant targets are notably broad in scope and are enriched for intergenic regions, CpG island shores, intermediate and low CpG-density promoters, and introns (Fig. 3e). In contrast, the other three classes are largely enriched for CpG islands (Supplementary Table 1). These enrichments suggest that DNMT3A and DNMT3B appear to act redundantly in regions of low CpG density, but may be more selectively targeted to regions of high CpG density.
The individual knockouts show a mild global reduction, an effect that becomes slightly more pronounced in the double knockouts (Fig. 3f). In contrast to this limited global decrease, several loci, including DPPA3 (STELLA), show complete loss of methylation in the DNMT3A−/−3B−/− cells (Fig. 3g), and form the set of redundant targets identified above. Another interesting pattern is seen at the NANOG promoter, which seems to be sporadically targeted by both de novo DNMTs, leading to low level methylation in the upstream region that is lost only in the DNMT3A−/−3B−/− cells (Supplementary Fig. 3c). NANOG is highly expressed, but the low level methylation in wild-type cells may reflect its known transcriptional heterogeneity within the ESC population.
We continued passaging of the DNMT3A−/−3B−/− cells for 22 passages, and profiled methylation over time using RRBS. In the absence of the de novo activity, CpG methylation is lost gradually due to the imperfect maintenance activity of DNMT1 (44.5% to 35.4%; Fig. 3h). Using an estimated doubling time of 28.8 hours, we estimate the fidelity of DNMT1 to be between 99.7% and 99.97%, with some noteworthy differences across genomic features (Supplementary Fig. 3b). In particular, features that are highly methylated and/or require continued repression by DNA methylation, such as transposable elements and satellite repeats40, seem to exhibit slightly superior maintenance than other genomic features.
Lastly, we investigated the global level of non-CpG methylation, which is known to be present in mouse15 and human11,17 ESCs. Consistent with our previous knockdown based experiments17, we find that DNMT3B depletion has a stronger effect than loss of DNMT3A (Fig. 3i, left). This could, however, simply be related to the comparatively high levels of DNMT3B in undifferentiated ESCs (Fig. 1a and b). In the DNMT3A−/−3B−/− cells, non-CpG methylation is rapidly reduced to background levels (Fig. 3i, right), confirming that continuous de novo DNA methyltransferase activity is required for non-CpG methylation in dividing cells, while CpG methylation can still be largely propagated by DNMT1, with a slow, gradual decline due to imperfect maintenance over successive cell divisions (Fig. 3h).
Shared and unique targets of DNMT3A and DNMT3B
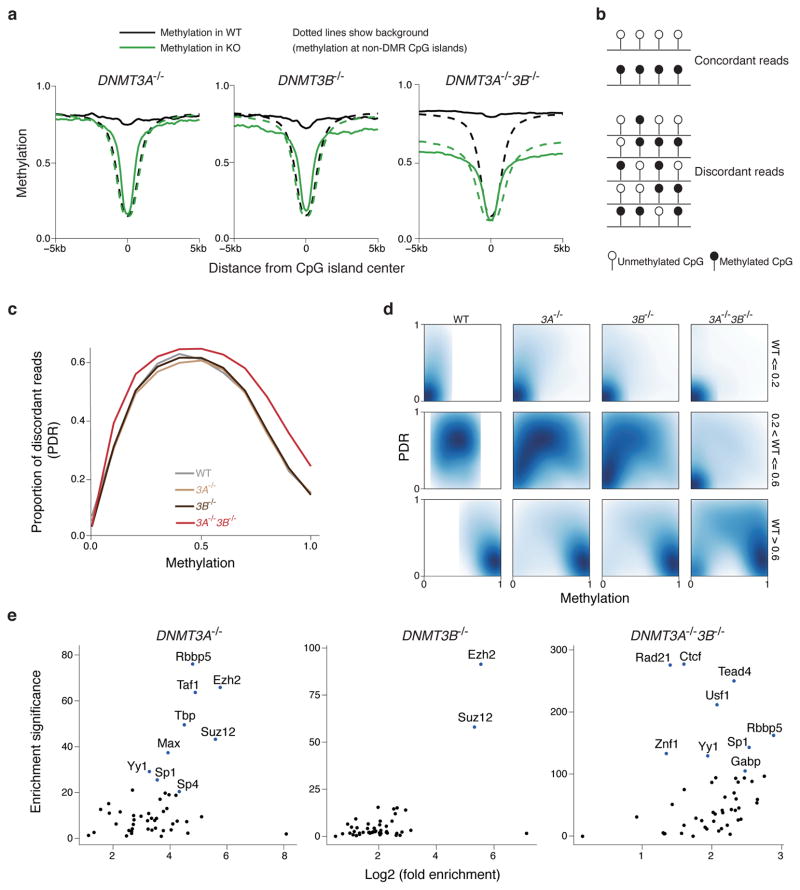
As noted above, we found a strong enrichment of CpG islands in the set of redundant DNMT3A/3B targets (Fig. 3e). To better understand this observation and gain a global perspective on the effect of DNA methylation on these targeted CpG islands and their neighboring regions (Supplementary Fig. 4a), we generated composite plots (Fig. 4a) and aggregate heatmaps (Supplementary Fig. 4b) for mean methylation levels across 25,489 CpG islands that were experimentally verified to be hypomethylated in somatic tissues41. We see a clear distinction in behavior between CpG islands that are hypomethylated in the knockouts, and the background set of CpG islands that are hypomethylated in both wild-type and knockout lines. These CpG islands are presumably specific targets of de novo methylation activity, since they become hypomethylated in the knockouts and then exhibit canonical CpG island methylation levels. In the DNMT3A−/−3B−/− cells, we see not only the focal loss of methylation seen at the center of the island, but also a general decrease in methylation at CpG island shores. These results are consistent with the enrichment results and suggest that DNMT3A and DNMT3B are specifically targeted to a subset of CpG islands.
Figure 4. Characterization of targets for DNMT3A and DNMT3B.
(a) Composite plot of methylation around CpG islands in WT and knockouts. The solid lines show the mean methylation of differentially methylated CpG islands, while the dotted lines show the mean methylation of all other CpG islands.
(b) Schematic for the definition of concordant and discordant reads.
(c) Proportion of discordant reads (PDR) in WT, DNMT3A−/−, 3B−/− and 3A−/−3B−/− cells.
(d) Two-dimensional density plots of PDR vs. methylation for CpGs with low (<= 0.2), medium(> 0.2 and <= 0.6) and high(> 0.6) methylation in WT cells.
(e) Enrichment of experimentally determined transcription factor binding sites in H1 ESCs generated by the ENCODE project for DMRs in DNMT3A−/−, 3B−/− and 3A−/−3B−/− cells compared to WT. Enrichment significance was defined as −log10(p-value) of the Fisher’s exact test, with the background being non-repetitive 1kb tiles that had at least 0.4 enrichment in WT.
Using our sequencing based data allows us to investigate the DNA methylation state of neighboring CpGs and classifying them into concordant or discordant patterns (Fig. 4b). We can then calculate the proportion of discordant reads (PDR) as the number of discordant reads divided by the total number of reads covering a CpG and plot the global distribution of this measure against CpG methylation42. In contrast to the single knockouts, the DNMT3A−/−3B−/− cells show a global increase in PDR (Fig. 4c), which suggests that DNMT3A and DNMT3B have similar mechanisms of action, and are likely to act redundantly to counteract global methylation loss due to imperfect DNMT1 fidelity. As expected, regions of low methylation in wild-type cells have low PDR, and these regions remain hypomethylated with low PDR in the knockouts (Fig. 4d, top panels). Regions of intermediate methylation show decreased methylation and lower PDR in knockout cells, with especially striking hypomethylation in the DNMT3A−/−3B−/− cells (Fig. 4d, middle panels). One exception are imprinting control regions that are known to be variable in ESCs but generally maintain stable, intermediate levels even in the DNMT3A−/−3B−/− cells (Supplementary Fig. 4c). Finally, regions of high methylation have low PDR in wild-type cells and the single KOs. These regions lose methylation specifically in the DNMT3A−/−3B−/− cells, with a corresponding increase in PDR (Fig. 4d, bottom panels).
To further assess the functional significance of the DMRs found above, we calculated the degree of overlap of the DMRs in the three knockout lines with published transcription factor binding sites43 and compared this overlap to that seen with a background set of all 1kb tiles with an average methylation of at least 0.4 in wild-type cells (Fig. 4e). Both DNMT3A−/− and DNMT3B−/− DMRs show enrichment for the PRC2 components EZH2 and SUZ12 (Fig. 4e). The DNMT3A−/−3B−/− DMRs shows strong enrichment for many sequence-specific transcription factors, including CTCF, which is known to be impacted with DNA methylation44. To see whether these targets are associated with any specific biological processes we took the set of 30,220 hypomethylated DMRs from the DNMT3A−/−3B−/− cells and used GREAT45 to associate them with genes and then calculate enrichments for functional ontologies. As expected, these regions are highly correlated with a large spectrum of developmental processes including patterning, regionalization, growth factor stimulation and corresponding pathways (Supplementary Fig. 4d and Supplementary Table 2). The enriched categories suggest that the DMRs we identify are generally hypermethylated in wild-type pluripotent cells by DNMT3A and DNMT3B, and normally lose methylation in a lineage and tissue specific manner as cells differentiate.
DNMT3A deletion prevents gain of DNA methylation
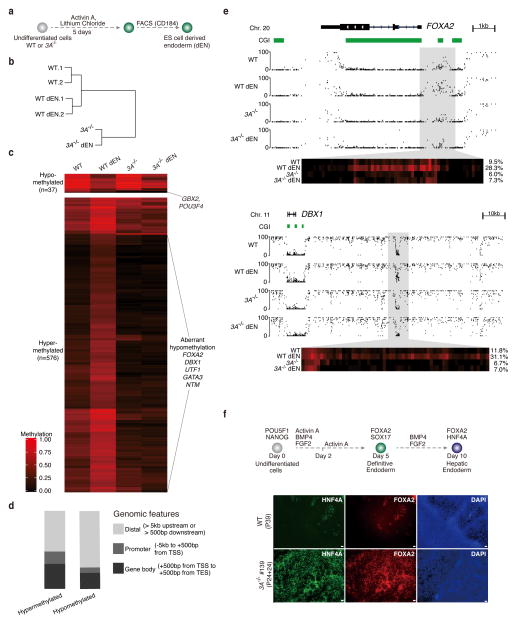
To further investigate the knockout effects during differentiation, we derived early endodermal cells from the DNMT3A−/− cells (Fig. 5a; Supplementary Fig. 5). We then generated WGBS data and performed hierarchical clustering analysis, which included undifferentiated and sorted endoderm cells (dEN) of both WT and DNMT3A−/− cells (Fig. 5b). These can be clearly segregated and undifferentiated cells in each case cluster more closely with their differentiated endoderm derivatives. We identified non-repetitive 1kb tiles that have significantly different DNA methylation levels between wild-type ESCs and dEN cells to determine their comparable DNA methylation state in undifferentiated and differentiated DNMT3A−/− cells (Fig. 5c). We identified 575 DMRs that gain DNA methylation in wildtype dEN and are enriched for promoters and gene bodies (Fig. 5d). Strikingly, most of these DMRs remain hypomethylated in the DNMT3A−/− derived dEN cells, suggesting that DNMT3A is required for their de novo methylation. For instance, we found that previously reported regions upstream of FOXA2 and downstream DBX1, respectively, were no longer methylated in the knockout cells (Fig. 5e). We also differentiated the cells further towards the hepatoblast stage and stained the population with antibodies for FOXA2 and HNF4A. The loss of DNMT3A had no immediately detectable negative impact on the downstream differentiation in this lineage (Fig. 5f). In summary, we detect a unique molecular role for DNMT3A during endoderm differentiation. Although we have not yet determined the functional relevance or downstream consequences of the targeted de novo methylation, our system provides a new, powerful platform to dissect these effects in this and other lineages.
Figure 5. Effect of DNMT3A deletion on endoderm differentiation.
(a) Schematic for the generation of CD184+ endodermal progenitor cells (dEN).
(b) Global analysis for the WGBS data of undifferentiated and dEN cells from WT (P23 and P26) and DNMT3A−/− (P24+13). Hierarchical clustering based on mean DNA methylation levels of 1 kb tiles across the human genome using Euclidean distance. 1 and 2 are two biological replicates. The passage number of the replicates is within 2 passages.
(c) Heat map of methylation levels (black, 0; red, 1) in WT, WT dEN, DNMT3A−/−, and DNMT3A−/− dEN of DMRs that change methylation significantly (q-value < 0.05 and methylation difference of at least 0.2) between WT and WT dEN.
(d) Classification of endoderm DMRs into promoters, gene bodies and distal regions as defined in the legend.
(e) Genome browser tracks of 1kb and 10kb regions highlight global and local methylation level for FOXA2 and DBX1. The heat map below shows the DNA methylation values of individual CpGs within the highlighted region. The average DNA methylation value for the entire highlighted region is shown on the right.
(f) Hepatoblast differentiation of WT (P39) and DNMT3A−/− (P24+24) cells. Top: Schematic of hepatoblast differentiation. Bottom: Representative images from two individual biological replicates are shown. 4X magnification of hepatoblast markers FOXA2, HNF4A immunostaining images for HUES64 (WT), DNMT3A−/− (#139) are shown. Scale bars, 100μm.
DNMT1 is essential for human ESCs
As described above, we were unable to obtain homozygous mutant lines for DNMT1, raising the possibility that human ESCs grown in standard FGF conditions require DNA methylation for viability. This hypothesis was further supported by the failure to create straightforward homozygous mutants using an alternative TALEN based strategy (Supplementary Fig. 6a) and our inability to obtain any viable lines with efficient DNMT1 knockdown despite having tested a total of nine different shRNAs (Supplementary Fig. 6b). Notably, the same TALEN based strategy readily generated DNMT3A and 3B homozygous lines, but no viable DNMT1 homozygous lines emerged, consistent with the CRISPR experiments.
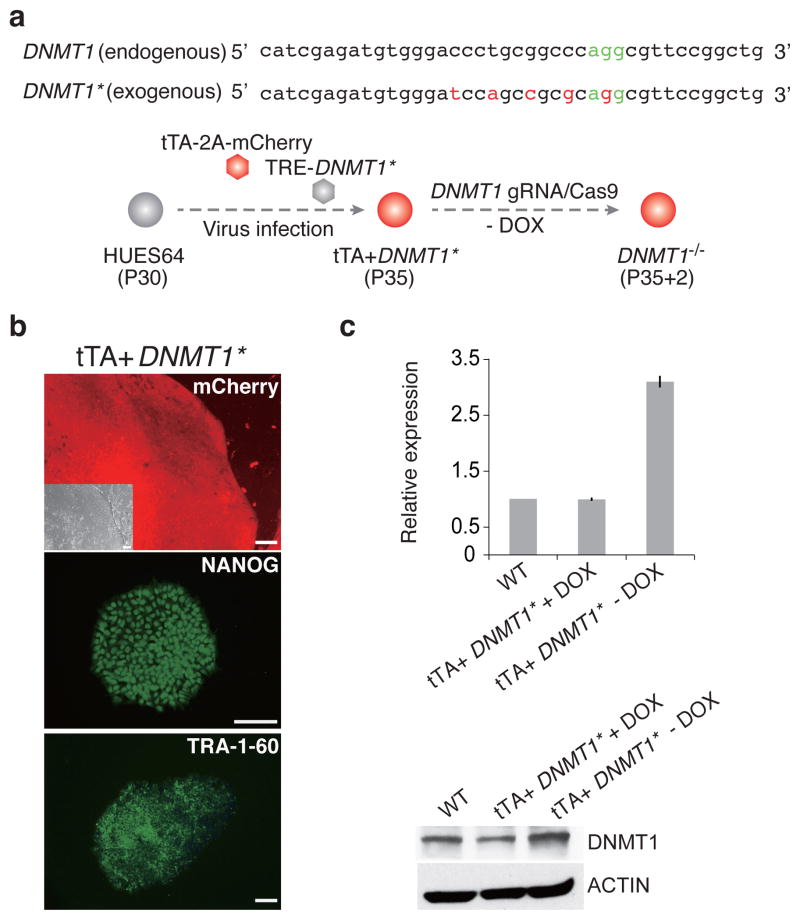
We therefore designed a rescue strategy that would enable the generation of homozygous mutant lines without the immediate lethality of DNMT1 loss (Fig. 6a). We first introduced two separate lentiviruses containing the tTA and TRE-DNMT1* into the wild-type HUES64 line, creating a doxycycline (DOX) responsive (TET/OFF) DNMT1* cell line that we could subsequently use for targeting (Fig. 6b and Supplementary Fig. 6c). We excluded the KOZAK sequence (GCCACC) upstream of the start codon to prevent overexpression to levels that might cause adverse effects prior to targeting and created five point mutations within the target sequence and the Protospacer Adjacent Motif (PAM) sequence that have no effect on the protein sequence (Fig. 6a and Supplementary Fig. 6d) to avoid the gRNA from disrupting the exogenous DNMT1*.
Figure 6. DNMT1 knockout strategy and targeting efficiency.
(a) Strategy to obtain DNMT1 homozygous clones. Top: We created mutations within the exogenous DNMT1 sequence that would prevent the targeting by our gRNA, but not alter the protein (Supplementary Fig. 6d). Asterisk (*) is used to distinguish the exogenous modified DNMT1* from endogenous DNMT1. Bottom: The DNMT1 rescue line was established by infecting HUES64 with two separate lenti virus, tTA-2a-mCherry and TRE-DNMT1* at passage 30. Then, DNMT1 was targeted as before into the tTA-DNMT1* line while the exogenous DNMT1* was expressed (no DOX).
(b) Representative bright field (BR), mCherry expression and pluripotent markers NANOG, TRA-1-60 immunostaining images for rescue line tTA+DNMT1*. Scale bars, 100μm.
(c) DNMT1 expression levels in our DNMT1* TET/OFF system. Left: RT-qPCR analysis for DNMT1. Primers amplify both endogenous and exogenous DNMT1. Error bars were generated from biological replicates and represent one standard error. Right: Western blotting analysis for DNMT1 under the control of DOX. The antibody can detect both endogenous and exogenous DNMT1.
Both qPCR and Western blot analysis confirm tight control of expression for the exogenous DNMT1*. In the absence of DOX, we observed an approximately threefold increase of overall DNMT1 levels that can be fully suppressed within 24 hours after supplementation with DOX (Fig. 6c). We next used the CRISPR/Cas9 approach to knock out the endogenous DNMT1 while maintaining expression of the exogenous DNMT1*. We could now readily obtain homozygous mutant lines in two independent rounds of targeting (Supplementary Fig. 6e, f), suggesting that our rescue strategy maintains a sufficient level of DNMT1 activity needed for survival. We selected two representative clones (#111, #122) for more detailed characterization and also confirmed by qPCR that DNMT1 targeting does not affect the expression of DNMT3A and DNMT3B (Supplementary Fig. 6g).
Loss of DNMT1 causes global demethylation and cell death
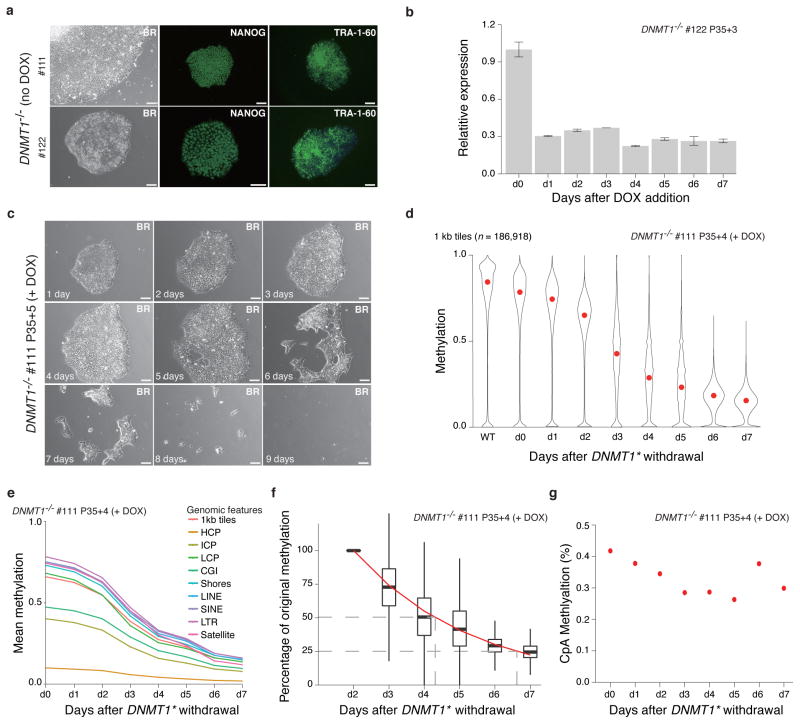
DNMT1 homozygous knockout ESCs proliferated normally and could be maintained over several passages as long as the exogenous DNMT1* was expressed. The cells displayed a normal morphology and stained positive for NANOG and TRA-1-60 (Fig. 7a). Addition of DOX caused rapid downregulation of DNMT1* (Fig. 7b) and cells began to die within a few days (Fig. 7c and Supplementary Fig. 7a). During this window we observed increased DNA damage and arrest in G1 (Supplementary Fig. 7b, c and Supplementary Table 3), consistent with previous studies that reported DNMT1 depletion in human epidermal progenitors and primary human fibroblasts (IMR90) results in G1 arrest20,46. Using a catalytic inactive version of DNMT1 (C1226W)47, which failed to provide a rescue, we confirmed that this effect is due to the catalytic activity of DNMT1 (Supplementary Fig. 7d–7g).
Figure 7. Loss of DNMT1 causes global demethylation and cell death.
(a) Representative bright field (BR) and NANOG/TRA-1-60 immunostaining for DNMT1−/− cells with the exogenous DNMT1* expressed (no DOX). 10X magnification. Scale bars, 100μm.
(b) RT-qPCR analysis after DNMT1* withdrawal relative to d0 level. Error bars are based on two replicates and represent one standard error.
(c) DNMT1* withdrawal causes massive cell death. Representative 10X magnification bright field (BR) images are shown. Scale bars: 100μm.
(d) Violin plots of the mean methylation (measured by RRBS) of 1 kb tiles across the human genome after DNMT1* withdrawal. Mean is indicated by red dot.
(e) Global reduction of DNA methylation in different genomic features. Overall CpGs (1kb tile) and selected features including CpG Islands (CGI), Shores, HCPs, ICPs, LCPs, LINEs, SINEs and satellite repeats are shown.
(f) Exponential model of methylation decay for 1kb tiles. Methylation at day 2 was normalized to 1 and methylation levels on subsequent days were calculated as a percentage of the day 2 value. An exponential model was fitted starting at day 2 and is shown by the red line. The dashed grey lines mark the timepoints when 50% and 25% of the day 2 methylation levels remain. Days 0 and 1 were excluded from this model to avoid noise from any remaining DNMT1 protein.
(g) Mean CpA methylation levels after DNMT1* withdrawal.
Despite G1 arrest and cell death, our inducible system enabled us to assess the effect on global DNA methylation after acute loss of DNMT1 activity, which highlighted a striking decrease over a one-week period (Supplementary Fig. 7h). We also confirmed that the catalytic inactive DNMT1C1226W could not rescue the cells and prevent the global loss of DNA methylation (Supplementary Fig. 7i). We then generated RRBS libraries to perform higher resolution analysis (Fig. 7d). Consistent with global, replication-dependent depletion, we find that all genomic features are affected similarly (Fig. 7e). We fitted an exponential model of the methylation decay of individual 1kb tiles and determined that it took 2.31 days for global DNA methylation to be reduced to half of the original levels (Fig. 7f). As expected, non-CpG methylation, which is the exclusive purview of de novo methyltransferases, was not affected by the acute loss of DNMT1 (Fig. 7g).
Discussion
DNA methylation is essential for normal development. Over the past three decades, major advances in understanding its role have included the deletion of the responsible enzymes in the mouse model as well as the more recent emergence of comprehensive mapping tools. Here, we have applied both to the human ESC model system to study their function, providing an unprecedented view of the respective genomic targets (shared and unique) of these enzymes.
All of the DNMT3 knockouts, including the DNMT3A−/−3B−/− cells, were able to differentiate into the three germ layers. Overall CpG methylation was only mildly affected in the single knockouts and even the DNMT3A−/−3B−/− cells retained around 65% methylation after 17 passages (wildtype HUES64 83%). This observation is consistent with previous mouse studies that suggested a dependence on the DNA methylation levels rather than the enzymes in the teratoma assay39. Our continued passing suggests an average fidelity for DNMT1 of around 99.8%, which is higher than that estimated for mouse ES cells2. Further experimentation is required to explore the underlying mechanism, however one can hypothesize that this simply reflects cell-type specific effects that have been noted for DNA methylation turnover rates48. We have recently observed increased DNA methylation discordance in chronic lymphocytic leukemia (CLL), which may be related to increased proliferation42. In this context, it is worth noting that mouse and human ESCs proliferate at very different rates (13.5±2.5 and 36±12 hours, respectively)49,50.
Our DNMT3 knockout human ESC lines provide a powerful model to study cell state-driven epigenetic effects during the differentiation into specific cell types. It is likely that phenotypes arise only post-differentiation, when de novo activity is required and/or compensation through DNMT3B is no longer available. Dnmt3a knockout mice develop to term, but die within a few weeks after birth4, suggesting some essential, though still poorly understood, developmental function.
It is well established that mouse and human ESC represent different pluripotent states, which may explain the sensitivity of the human ESCs to loss of DNA methylation. All somatic cells require DNA methylation and the more mature epiblast state modeled by current human ESCs may represent the first developmental period where maintenance becomes essential. In fact, although no data were shown, a similar effect has been suggested for mouse epiblast cells51. In the future, it will be interesting to test the effect of DNMT1 deletion under recently described human “naïve” conditions to dissect the transition to DNMT1-dependent pluripotency52,53,54.
Our study provides a comprehensive, high-resolution view of the largely conserved roles that the three catalytically active DNMTs play in human cells. The mutant lines represent powerful tools to study their function in various differentiation models and will therefore greatly accelerate our understanding of DNA methylation in human development and disease.
Online Methods
Cell culture
Human embryonic stem cells were expanded on murine embryonic fibroblasts CF1 (Global Stem) in KO-DMEM (Life Technologies) containing 20% Knockout Serum Replace (Life Technologies) and FGF2 (10 ng/ml) (Millipore). Cells were passaged by enzymatic dissociation using Collagenase IV (1 mg/ml) (Life Technologies). For feeder free culture condition, plates were coated with Geltrex (Life Technologies) and the culture medium was mTeSR1 (Stemcell Technologies). HUES64 was derived in the Melton/Eggan labs56 and maintained in our laboratory since then using the above conditions.
Electroporation
Human ESCs were harvested using 1mg/mL Collagenase Type IV (invitrogen) first and then dissociated into single cells using TrypLE™ (invitrogen). 1 × 107 cells resuspended in buffered saline (PBS) were electroporated with 15μg Cas9 and 15μg gRNA plasmids (Gene Pulser Xcell System, Bio-Rad: 250 V, 500μF, 0.4 cm cuvettes). Cas9 plasmid and gRNA backbone have been described previously35. Fluorescence-activated cell sorting (FACS) was carried out 48 hours posttransfection based on fluorescent-marker expression. Sorted cells were replated as single cells at very low density on irradiated CF1 feeders in hESC medium supplemented with Rho Kinase (ROCK)-inhibitor (Stemgent, Y-27632) for the first 24 hours. Individual colonies were picked and expanded on feeder free/mTeSR condition. Genomic DNA was then purified. Mutations were validated by PCR and DNA sequencing.
Constructs
GIPZ DNMT1 shRNA: Thermo Scientific, RHS4531-NM_001130823
TRIPZ DOX inducible DNMT1 shRNA: Thermo Scientific, RHS4740-EG1786
plenti-ef1a-tTA-2a-mCherry: tTA-2a-mCherry was inserted into lenti backbone (Addgene,17448). tTA was cloned from Addgene plasmid 2680357. mCherry was cloned from Addgene plasmid 2324358.
plenti-ef1a-puror-TRE-DNMT1*: DNMT1 was clone from Addgene plasmid 3693959. After creating mutations, DNMT1* was cloned into plenti-ef1a-puror-TRE backbone60 which was a gift from Dr Lei Xiao’s lab.
plenti-ef1α-DNMT1WT-IRES-nlsEGFP: DNMT1 was clone from Addgene plasmid 3693959.
plenti-ef1α-DNMT1C1226W-IRES-nlsEGFP: DNMT1 point mutation C1226W was created on the DNMT1WT.
Primers
DNMT3A U:
Forward: GACTCCATCACGGTGGGCATGG;
Reverse: TGTCCCTCTTGTCACTAACGCC.
DNMT3A D:
Forward: TCCACTGTGAATGATAAGCTG;
Reverse: GGAAACCAAATACCCTTTCCA.
DNMT3B U:
Forward: GAGTCCATTGCTGTTGGAACCG;
Reverse: ATGTCCCTCTTGTCGCCAACCT.
DNMT3B D:
Forward: GACTGCTTGGAATACAATAGGA;
Reverse: AAAGCCAAAGATCCTTTCGAG.
DNMT1 P:
Forward: GGTTTCCTTCCTCAGCTACTGCGA;
Reverse: CACTGATAGCCCATGCGGACCA.
ACTIN:
Forward: TTTGAGACCTTCAACACCCCAGCC;
Reverse: AATGTCACGCACGATTTCCCGC.
Sequence information for TALENs targeting strategy
For DNMT1 knockout:
tgtgggccatcgagatgtGGGACCCTGCGGCCCAggcgttccggctgaacaa
For DNMT3A knockout:
tcttctttgagttctaccGCCTCCTGCATGATGCgcggcccaaggagggaga
Capital letters are the spacer region. Lower case letters are the TALEN binding sites.
Virus production and infection on human ESCs
For virus production, virus was produced by co-transfecting lenti plasmid with delta 8.91 and VSVG on 293 cell by using FuGENE® HD (Promega). 24 hours after transfection, medium was changes to mTeSR1 for virus collection. Supernatant of virus was collected twice 48h and 72h after transfection separately. Virus was stored in 4°C. Before infection, virus was filtered through 0.45μm filter.
For virus infection on human ESCs, cells were plated as small clumps on geltrex pre-coated plate. 24 hour after plating, cells were infected with virus supernatant for 3 hours in incubator. Medium was changed back to mTeSR1. Repeat the 3 hours infection the next day. In this case, cells were maintained well in pluripotent state.
Isolation of genomic DNA from 96-well plate
Cells in each well were about 90% confluent. Washed twice with PBS and then lysed with 50μl/well of lysis buffer (10mM Tris PH7.5, 10mM EDTA, 10mM NaCl, 0.5% Sarcosyl or SDS, 1mg/ml Proteinase K) at 37°C overnight. Genomic DNA was precipitated by adding 100μl/well of cold NaCl/EtOH (1.5% vol 5M NaCl in 100% cold EtOH) at room temperature (RT) for 2–4 hrs. Genomic DNA precipitate was washed 3 times with 70% EtOH. Each time discarding the alcohol by inversion. Air-dry DNA for 15–20 min at RT. DNA was re-suspended in 30–100 μl of DNase-free, RNase-free ddH2O.
Antibodies
For immunostaining:
OCT4: BD Transduction Laboratories, 611202
NANOG: R&D, AF1997
TRA-1-60: Stemgent 090068
ANNEXIN V: Abcam, ab54775
FOXA2: R&D, AF2400
HNF4A: Abcam, Ab41898
DAPI: Hoechst 33342, Invitrogen, H3570.
γ-H2A.X (phospho S139): Abcam, ab2893.
For western blotting:
DNMT3A: Cell signaling, 2160S
DNMT3B: santa cruz biotechnology, sc-20704
DNMT1: Activemotif, 39204
ACTIN: Abcam, ab8226
Immunostaining
Human ESCs were passaged on Geltrex coated plates and cell were cultured in mTeSR1 medium. Then the cells were fixed in 4% paraformaldehyde (PFA) for 15 minutes at room temperature. Permeabilization and blocking was performed in 5% bovine serum albumin (BSA) and 1% Triton X-100 in PBS for 30minutes. Cells were stain with primary antibody in 4°C overnight. The secondary antibody was applied for 2 hours at room temperature. Nucleuses were stained with Hoechst 33342 (Life Technologies). Images were obtained using Olympus IX71 microscope and MetaMorph Advanced software.
RNA extraction
RNA was isolated from hESCs using TRIzol (Invitrogen, 15596-026), further purification was carried out with RNeasy columns (QIAGEN, 74104) and DNase treatment.
Embryoid body (EB) formation
Human ESCs were expanded in 10cm2 dishes on irradiated CF-1 MEFs (Global Stem). Cells were harvested using 1mg/ml Collagenase IV (Life Technologies) once they were confluent. After three times washes with 1 × phosphate-buffered saline (PBS), cell clumps were plated into 6-well low attachment dishes (Corning) in 20% Knockout Serum Replacement (KOSR) (Life Technologies), 200mM Glutamax (Life Technologies), MEM Non-Essental Amino Acid, in KO DMEM (Life Technologies). Medium was changed every 2–3 days.
Directed differentiation into the three germ layers
hESCs were plated as clumps on 6-well plates coated with Geltrex (Life Technologies) in mTeSR1 culture medium (Stem Cell Technologies). On day 3, directed differentiation was induced and medium was changed every day.
Ectoderm differentiation medium contains 2μM A83-01 TGFb inhibitor (Tocris), 2μM PNU-74654 WNT3A inhibitor (Tocris), and 2μM Dorsomorphin BMP inhibitor (Tocris) in 15% KOSR (Life Technologies), MEM Non-Essential Amino (Life Technologies), and 55μM 2-mercaptoethanol.
Mesoderm differentiation medium contains 100ng/ml Activin A (Life Technologies), 10 ng/ml bFGF (Millipore), 100ng/ml BMP4 (Life Technologies), 100ng/ml VEGF 100ng/ml (Life Technologies) in 0.5% FBS (Hyclone), MEM Non-Essential Amino Acid (Life Technologies), 55μM 2-mercaptoethanol and Glutamax (Life Technologies) in DMEM/F12 medium (Life Technologies) for the first 24 hrs of differentiation. For the rest 4 days, the medium contains 10 ng/ml bFGF (Millipore), 100ng/ml BMP4 (Life Technologies), 100ng/ml VEGF 100ng/ml (Life Technologies) in 0.5% FBS (Hyclone), MEM Non-Essental Amino Acid (Life Technologies), 55μM 2-mercaptoethanol and glutamax (Life Technologies) in DMEM/F12 medium (Life Technologies).
Endoderm differentiation medium contains 100ng/ml Activin A (Life Technologies) and 2μM/ml Lithium Chloride (Sigma) in 0.5% FBS (Hyclone), 200mM GlutaMax (Life Technologies), MEM Non-Essential Amino Acid (Life Technologies), 55μM 2-mercaptoethanol in RPMI medium (Life Technologies).
Teratoma formation
Teratoma formation assay was performed as a service by Harvard Genome Modification Facilities. Briefly, hESCs were injected into the kidney capsules of 3 immuno-suppressed mice as 1 million cells per animal to form teratomas. The formed teratomas were fixed in 4% paraformaldehyde (PFA) pH7.4, embedded in paraffin, sectioned 10–12μm thickness, and stained with Hematoxylin and Eosin (H&E) for examination. Histology examination was performed by histology core facility at Harvard Stem Cell Institute.
Hepatoblast Differentiation
Differentiation was performed as described previously61 with some modifications. Briefly, cells were dissociated with Accutase and seeded onto geltrex-coated dishes with ROCK-inhibitor at high confluence in mTESR1. Within twenty-four hours after plating, definitive endoderm differentiation was started and cells were cultured in RPMI/2% B27 (without Insulin), 100ng/ml Activin A, 10ng/ml BMP4, and 20ng/ml FGF2 for 48 hours. For days 3–5 of differentiation, cells were cultured in RPMI/2% B27 (without Insulin), 100ng/ml Activin A. For days 6–10 of differentiation, cells were cultured in RPMI/2% B27 (with Insulin), plus 20ng/ml BMP4 and 10ng/ml FGF2. Media was changed every twenty-four hours.
Cell cycle assay
Alive cells were incubated with Hoechst 33342 in the incubator for 15min. Then cells were trypsinized into single cells, washed with PBS once and analyzed by FACs directly.
Dot blot assay
Isolated genomic DNA was denatured in 0.4M NaCl and 10mM EDTA for 10 min at 99°C. The denatured DNA was spotted in a 2-fold serial dilution on a Zeta Membrane (BioRad) using a Bio-Dot apparatus (BioRad). The blotted membrane was rinsed in 2xSSC, air-dried and UV-crosslinked at 120,000 μJ cm−2. The membrane was blocked in 1xTBS containing 5% nonfat dry milk for 30min. Anti-5-methylcytosine monoclonal antibody (Diagenode, #33D3) was diluted 1:250 in TTBS, containing 0.3% Tween 20 and 5% nonfat dry milk, and was incubated overnight at 4°C. The membrane was washed in 3x 5min in TBS and was incubated for 1 hour at room temperature with HRP-conjugated goat anti-mouse IgG (Jackson Lab, #115-025-174). The membrane was washed 3x in TBS and visualized by chemiluminescence using the Gel Doc XR (BioRad). The same membrane was incubated 30min with SYBR Gold Nucleic Acid stain (Invitrogen, #S-11494) in 1xTAE buffer, total DNA loading was visualized by UV transillumination.
Scorecard Assay and Data Analysis
Experiment was carried out using TaqMan® hPSC Scorecard™Kit (Life Technologies, A15872). Data could be analyzed through TaqMan® hPSC Scorecard™ Analysis Software through Life Technologies website.
RRBS and WGBS Library Construction
RRBS library construction was described in Ref 62. WGBS library construction was well described in Ref 9.
RRBS and WGBS Data Processing and Analysis
WGBS raw sequencing reads were aligned using MAQ in bisulfite mode against human genome version hg19/GRCh37, discarding duplicate reads. DNA methylation calling was performed based on an extended custom software pipeline published previously for RRBS63. Briefly, the methylation level at a CpG or CpA site was the number of reads with that site methylated divided by the total number of reads covering the site.
To ensure comparability of region DNA methylation levels across all samples, only CpGs covered by ≥ 5x in all samples qualified for the computation of region DNA methylation levels. We defined the average methylation for a genomic region as the coverage-weighted mean of the methylation levels of the individual CpGs within the region. Subsequently, we averaged a region’s DNA methylation level over replicates. Differentially methylated regions were identified by using a two sample weighted t-test using the methylation values of the CpGs within each regions as individual sampling events (minimum number of CpGs in region for each sample is 2). We performed multiple testing correction using a method defined in Ref 64 using the R qvalue package.
We used two sets of CpG island annotations. The first uses a bioinformatics definition and designates a region as a CpG island if it has a GC content higher than 0.5 and a ratio greater than 0.6 of observed CpG dinucleotides to what would be expected based on the GC content over a length of 700 bp. The second is a set of experimentally verified CpG islands41.
We defined promoters as the regions 2000 bp upstream and 500 bp downstream of the Refseq TSS set. These were divided into high CpG promoters (HCP), intermediate CpG promoters (ICP), and low CpG promoters (LCP) as published previously65. Intergenic regions, exons and introns were defined on the basis of the RefSeq gene models downloaded from the UCSC browser.
We used GREAT (Genomic Region Enrichment of Annotation Tool)45 to associate DMRs with genes. We limited the associations to genes within 20kb of DMRs and used an FDR threshold of 10−6 and a fold enrichment threshold of 2. The complete set of enriched terms across all ontologies tested is in Supplementary Table 2.
Fidelity of DNMT1 was calculated by fitting an exponential model to the mean DNA methylation of different genomic features using the nls function in R. The exponential model has the form y=ax, where “a” is the fidelity and “x” is the estimated number of population doublings based on passage number, days per passage (8) and doubling time (28.8 hours).
The proportion of discordant reads (PDR) was calculated for each CpG by examining the individual reads covering that CpG and classifying them into concordant (all CpGs methylated or all CpGs unmethylated), filtering out reads with less than 3 CpGs covered.
We downloaded the conservative set of uniform peaks called by the ENCODE project43 and computed overlaps of these peaks with DMRs identified in DNMT3A−/−, DNMT3B−/− and DNMT3A−/−3B−/−. We calculated the significance of the enrichment using Fisher’s exact test, using the DMR set as the foreground and the set of non-repetitive 1kb tiles with a methylation level of at least 0.4 as the background.
Human imprinted control regions were taken from Ref. 66.
Mouse Dnmt3a−/−, 3b−/− and 3a−/−3b−/−, Dnmt1−/− and 3a−/−3b−/− with a Dnmt1 knockdown16 data were aligned to mm9 and otherwise processed similar to the human RRBS data.
Supplementary Material
Acknowledgments
We would like to thank all members of the Meissner lab, in particular Camille Sindhu for helpful discussion and Zachary D. Smith for critical feedback on the manuscript. We would like to thank Kiran Musunuru for providing the CRIPSR/Cas9 plasmids and thank Qiurong Ding from Musunuru lab for technical support. J.L. was supported by a postdoctoral fellowship from the Human Frontiers Science Program. J.K.J is supported by NIH Director’s Pioneer Award DP1 GM105378. A.M. is a New York Stem Cell Foundation Robertson Investigator. The work was funded by the US National Institutes of Health (NIGMS) grant (P01GM099117) and The New York Stem Cell Foundation.
Footnotes
Accession number
All data have been deposited in GEO under GSE63281.
Financial Disclosures
JKJ is a consultant for Horizon Discovery. JKJ has financial interests in Editas Medicine and Transposagen Biopharmaceuticals. JKJ’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Author Contributions
J.L. and A.M. designed and conceived the study, J.L. generated all the cell lines and performed the experiments, R.K. performed the analysis, H.G. generated the WGBS libraries, A.G and A.M supervised the DNA methylation experiments, M.J.Z. and K.C performed some analysis and assisted in the general data processing, A.M.T and V.A performed with the Scorecard assay, C.A.G. and J.D. assisted in the endoderm and hepatocyte differentiation, C.G performed the dot blot assay and R.P supported the RNA profiling and FACS, D.R, S.Q.T and J.K.J designed and generated the TALENs, W.M. and J.L.R performed expression analysis, J.L, R.K. and A.M. interpreted the data and wrote the paper.
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Jackson M, et al. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol. 2004;24:8862–71. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 4.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 5.Jin B, et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet. 2008;17:690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
- 6.Klein CJ, et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43:595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah MY, Licht JD. DNMT3A mutations in acute myeloid leukemia. Nat Genet. 2011;43:289–90. doi: 10.1038/ng0411-289. [DOI] [PubMed] [Google Scholar]
- 8.Winkelmann J, et al. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet. 2012;21:2205–10. doi: 10.1093/hmg/dds035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gifford CA, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–63. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Habibi E, et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–9. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith ZD, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–44. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–81. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E. De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene. 2002;289:41–8. doi: 10.1016/s0378-1119(02)00469-9. [DOI] [PubMed] [Google Scholar]
- 16.Meissner A, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–77. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziller MJ, et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011;7:e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan G, et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788–97. doi: 10.1523/JNEUROSCI.21-03-00788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson-Grusby L, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–9. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 20.Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463:563–7. doi: 10.1038/nature08683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5:442–9. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsumura A, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–14. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- 23.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–20. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 24.Chen T, et al. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat Genet. 2007;39:391–6. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 25.Egger G, et al. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc Natl Acad Sci U S A. 2006;103:14080–5. doi: 10.1073/pnas.0604602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee I, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 27.Rhee I, et al. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–7. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 28.Martins-Taylor K, Schroeder DI, LaSalle JM, Lalande M, Xu RH. Role of DNMT3B in the regulation of early neural and neural crest specifiers. Epigenetics. 2012;7:71–82. doi: 10.4161/epi.7.1.18750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horii T, Tamura D, Morita S, Kimura M, Hatada I. Generation of an ICF syndrome model by efficient genome editing of human induced pluripotent stem cells using the CRISPR system. Int J Mol Sci. 2013;14:19774–81. doi: 10.3390/ijms141019774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–4. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–8. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 35.Ding Q, et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393–4. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon CA, Hartono SR, Chedin F. Inactive DNMT3B splice variants modulate de novo DNA methylation. PLoS One. 2013;8:e69486. doi: 10.1371/journal.pone.0069486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsankov A, et al. Rewiring of transcription factor networks during human ES cell differentiation. Nature. doi: 10.1038/nature14233. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bock C, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–52. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith ZD, et al. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–5. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Illingworth R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landau DA, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26:813–25. doi: 10.1016/j.ccell.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–5. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 45.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barra V, Schillaci T, Lentini L, Costa G, Di Leonardo A. Bypass of cell cycle arrest induced by transient DNMT1 post-transcriptional silencing triggers aneuploidy in human cells. Cell Div. 2012;7:2. doi: 10.1186/1747-1028-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clements EG, et al. DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res. 2012;40:4334–46. doi: 10.1093/nar/gks031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shipony Z, et al. Dynamic and static maintenance of epigenetic memory in pluripotent and somatic cells. Nature. 2014;513:115–9. doi: 10.1038/nature13458. [DOI] [PubMed] [Google Scholar]
- 49.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–28. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 50.Li L, et al. Individual cell movement, asymmetric colony expansion, rho-associated kinase, and E-cadherin impact the clonogenicity of human embryonic stem cells. Biophys J. 2010;98:2442–51. doi: 10.1016/j.bpj.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hutnick LK, Huang X, Loo TC, Ma Z, Fan G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J Biol Chem. 2010;285:21082–91. doi: 10.1074/jbc.M110.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gafni O, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–6. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 53.Chan YS, et al. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–75. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 54.Theunissen TW, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–87. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Giaimo R, et al. The expression of de novo DNA methylase DNMT3b, of the methyl-CpG binding protein MBD2b and of 5-MCDG glycosylase shows two waves of induction during CaCO-2 cell differentiation. Gene. 2005;351:73–81. doi: 10.1016/j.gene.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 56.Chen AE, et al. Optimal timing of inner cell mass isolation increases the efficiency of human embryonic stem cell derivation and allows generation of sibling cell lines. Cell Stem Cell. 2009;4:103–6. doi: 10.1016/j.stem.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsiao EC, et al. Constitutive Gs activation using a single-construct tetracycline-inducible expression system in embryonic stem cells and mice. Stem Cell Res Ther. 2011;2:11. doi: 10.1186/scrt52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papapetrou EP, et al. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc Natl Acad Sci U S A. 2009;106:12759–64. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, et al. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem. 2006;281:19489–500. doi: 10.1074/jbc.M513249200. [DOI] [PubMed] [Google Scholar]
- 60.Rao L, et al. Highly efficient derivation of skeletal myotubes from human embryonic stem cells. Stem Cell Rev. 2012;8:1109–19. doi: 10.1007/s12015-012-9413-4. [DOI] [PubMed] [Google Scholar]
- 61.Si-Tayeb K, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boyle P, et al. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 2012;13:R92. doi: 10.1186/gb-2012-13-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bock C, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–14. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–7. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woodfine K, Huddleston JE, Murrell A. Quantitative analysis of DNA methylation at all human imprinted regions reveals preservation of epigenetic stability in adult somatic tissue. Epigenetics Chromatin. 2011;4:1. doi: 10.1186/1756-8935-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.