Abstract
PFam Clan 0032, also known as the CDE superfamily, is a diverse group of at least 20 protein families sharing a common α, β-barrel domain. Of these, six different groups bind heme inside the barrel’s interior, using it alternately as a cofactor, substrate, or product. Focusing on these six, an integrated picture of structure, sequence, taxonomy, and mechanism is presented here, detailing how a single structural motif might be able to mediate such an array of functions with one of nature’s most important small molecules.
Introduction
When the human genome project began in 1990, sequencing one person’s genome may have seemed as overly ambitious as an earlier generation’s pledge to put a man on the moon. Yet, one decade and 3.4 gigabases later, the secret of the human blue print was unlocked.
Yet more inconceivable than those 3.4 billion base pairs, which fill the pages of 100 volumes on a bookshelf in the Wellcome Trust, is the size of their legacy. The European Bioinformatics Institute now maintains close to 80 million independent sequence entities in the TREMBL database, from organisms of all types. This number continues to rise exponentially with time. Understanding one’s own favorite sequence amid such a vast sequence space is akin to picking out a single star in a bright night sky. There, in the context of its neighbors, it may be possible to discern not only its properties but – continuing with the analogy – its membership in constellations and distance from other celestial landmarks. An early exercise in such sequence gazing led Maixner et al. to note that the gene encoding chlorite dismutase (Cld), an enzyme that catalyzes the detoxification of chlorite (ClO2−) to Cl− and O2, was found in nearly all Bacterial and many Archaeal phyla.1 This is in spite of the fact that very few microbes were known to require such a reaction, which acts as the terminal step in the respiration of perchlorate (ClO4−) or chlorate (ClO3−).2
This extraordinary observation, based in sequence, led our group to more closely examine the Cld structure in the context of known structural motifs.3 When the first Clds were crystallographically characterized, the heme-binding domain of Cld was considered unusual for its mixed α-helical/β-sheet composition, since well-studied heme proteins were mainly α-helical.4–7 However, we noted that the domain structure had been seen in heme-related contexts before,8 and appeared to be widespread enough to be the basis of a protein superfamily. In keeping with convention, we gave it the common name “CDE”, an acronym for three of its then partially-characterized families, all of which bound heme: Clds, dye-decolorizing peroxidases (DyPs), and EfeBs (an elision of “Escherichia coli” and “Fe”).
With the aid of improving analytical tools and a growing sequence database, it is now clear that the CDE superfamily is much larger and more diverse than initially expected. This article aims at highlighting what is currently known about the content and scope of the superfamily, with a particular emphasis on its heme-binding members. The latter proteins alone fall into at least six distinct families, which use a histidine-ligated heme b or a closely related tetrapyrrole as either a cofactor, a substrate, or reaction product. These families include the previously mentioned Clds, DyPs, and EfeBs, as well as the aldoxime dehydratase (OxdA), IsdG, and HemQ families (Table 1; each family to be described individually below). Within the Cld, DyP, and OxdA protein structures, heme is a tightly bound cofactor and the site of a catalytic process in which its iron directly binds and activates chlorite (ClO2−), peroxide (H2O2), or an aliphatic aldoxime (R-CH=NOH), respectively. Within the EfeBs, which belong to a subgroup of the large and diverse DyP family, 9–13 heme plays a role in the assimilation of iron, though its precise catalytic function has been the subject of some debate (discussed below). In two more families, the IsdGs and HemQs, heme serves respectively as a substrate and a product. In the IsdGs, heme is a non-innocent, O2-activating substrate in an oxygenase reaction that results in release of the ring-opened tetrapyrrole and iron. Finally, the HemQs are currently grouped as a Cld subfamily (see below), though they are clearly functionally distinct. It was recently shown that HemQs convert coproheme to heme b via two oxidative decarboxylations of the substrate’s propionate side chains (see below and elsewhere in this issue).14
Table 1.
Families constituting the CDE superfamily (SCOPe 54909, Pfam clade CL0032) and their properties
| Descriptive name | SCOPe | Pfam | Description | Taxonomic distribution |
|---|---|---|---|---|
| Families interacting with heme | ||||
| 1. Chlorite dismutase, Cld | 110965 | PF06778 | Catalyze the disproportionation of chlorite into chloride and oxygen using heme as a cofactor; characterized by a conserved distal arginine | Diverse Bacteria and Archaea |
| 2. DyP-type-peroxidase, DyP | 143265 | PF04261 | Heme peroxidases with wide substrate specificity, distinct active site and domain structure, and the ability to function well under lower pH conditions relative to other peroxidases | Diverse Bacteria and Fungi; Platyhelminthes, Nematoda, Chordata |
| 3. EfeB† | 143265 | PF04261 | DyPs with heme peroxidase activity, a twin-arginine motif targeting them for extracytoplasmic transport in their heme-bound form, and a role in iron assimilation | Gram-negative Bacteria; potentially others |
| 4. Aldoxime dehydratase, OxdA, OxdRE | -- | PF13816 | Catalyze the dehydration of aldoximes to their corresponding nitriles using heme as a cofactor | Fungi, Proteobacteria, Actinobacteria, Firmicutes |
| 5. HemQ† | 110965 | PF06778 | Cld-family proteins that catalyze the oxidative decarboxylation of coproheme to form heme b; characterized by a neutral distal residue in place of Arg | Gram-positive Bacteria and some (aerobic) Archaea |
| 6. IsdG family† | 82666, 110970 | PF03992 | Cofactorless monooxygenases involved in the oxidative degradation of heme; part of the ABM family | Gram-positive Bacteria; potentially others |
| Functionally characterized; no direct association with heme | ||||
| 7. Antibiotic monooxygenase, ABM; also called PG130 | 82666, 110970 | PF03992 | Small (~120 amino acid) cofactorless monooxygenases involved in the oxidation or oxygenation of organic molecules, particularly in the biosynthesis of antibiotics; frequently found as homodimers or fusions with a variety of other domains | Representatives in phylogenetically broad Bacteria, and Archaea; Fungi and Chordata |
| 8. EthD | 143272 | PF07110 | Involved in the degradation of ethyl tert-butyl ether | Diverse Bacteria; Fungi; Viridiplantae; Euryarcheota |
| 9. Lpr/AsnC transcriptional regulators | 69733 | PF1037 | Transcriptional regulators of the lysine-responsive protein type, frequently fused to helix-turn-helix-containing domains | Diverse Gram-positive and –negative Bacteria; Archaea; Nematoda |
| 10. Methylmucono-lactone methyl-isomerase, Mml-I | 160292 | PF09448 | Involved in the catabolism of methyl-substituted aromatics via a modified oxo-adipate pathway in bacteria | Proteobacteria |
| 11. Mucolactone delta-isomerase, MIase | 54910 | PF02426 | Catalyze the third step of catechol catabolism to succinate- and acetyl-CoA | Diverse Bacteria; Crenarchaeota |
| 12. Sulfur oxygenase reductase, SOR | 143278 | PF07682 | A non-heme-iron-containing 24-mer that catalyses the reaction: 4S + O2 + 4H2O → 2HSO3− + 2H2S + 2 H+ | Aquificae, Proteobacteria, Firmicutes; Crenarchaeota, Euryarchaeota |
| 13. NapD | -- | PF03927 | Involved in the formation of periplasmic nitrate reductase | Gram-negative Bacteria |
| 14. Polyketide synthesis cyclase | 110959 | PF04673 | Cyclase involved in polyketide synthesis and sharing structural similarity with polyketide monooxygenase | Actinobateria |
| Functions not well characterized | ||||
| 15. Yci-I | 102965 | PF03795 | Unknown function; contain a highly conserved Asp and His, suggesting an enzymatic function | Diverse Bacteria and Fungi |
| 16. DGPF domain | 110962 | PF03795 | Unknown function but thought to have a role in transcription initiation; specified by Pfam as a Yci-I subfamily, but listed as a separate family by SCOPe | Same as Yci-I |
| 17. Dabb-stress responsive protein | 89927 | PF07876 | Unknown function | Diverse Bacteria, Protists, and single-celled Eukaryotes; Fungi; Viridiplantae; Euryarchaeota |
| 18. YdhR | 117940 | PF08803 | Unknown function; homodimeric structure | Diverse Bacteria, Protists, and single-celled Eukaryotes; Fungi; Viridiplantae; Euryarchaeota |
| 19. NIPSNAP | 117943 | PF07978 | Many hypothetical proteins along with members of the NIPSNAP family which have roles in vesicular transport | Diverse Bacteria, Protists, and single-celled Eukaryotes; Fungi; Viridiplantae; Euryarchaeota |
| 20. Atu0297 (DUF‡1330) | 143275 | PF07045 | Unknown function; hypothetical proteins of ~90 residues in length | Diverse Bacteria, Protists, and single-celled Eukaryotes; Fungi; Viridiplantae; Euryarchaeota |
| 21. YbaA (DUF‡1428) | 160289 | PF07237 | Unknown function | Diverse Bacteria; Crenarchaeota |
| 22. YiiL (DUF718) | 160298 | PF05336 | Unknown function | Diverse Bacteria; Fungi; Chlorophyta; Euryarchaeota; Chordata |
| 23. Unnamed DUFs‡: 3291, 4188, 881 | -- | PF11695, PF13826, PF05949 | Unknown function; listed by PFam but not SCOPe | -- |
| 24. Marine metagenome family DABB1/2/3 | 160301 160295 160306 |
-- | Unknown function; listed by SCOPe but not Pfam | -- |
Proteins designated as part of other families by Pfam and/or SCOPe but having significant functional distinctions from the larger family cohort;
DUF = domain of unknown function
Based on what is currently known from experimental data, we propose below how the same basic protein architecture may accommodate such a diversity of functions with heme. Our treatment of the subject cannot be comprehensive in a short-format article and consequently focuses on salient ideas derived from an analysis of informatics, structure, and mechanism. We wish to acknowledge the many authors whose work cannot be fully addressed here, and encourage the interested reader to view the other outstanding articles in this special issue, which focus on individual proteins and families in greater depth.
Content and shape of the superfamily
Protein superfamilies are in concept about as old as the sequencing boom. In 1990, analyzing all 350 of the then-available protein structures, Farber and Petsko noted that 17 – close to 10% of all of the structurally characterized enzymes – shared the same α/β TIM barrel structure. This suggested that the TIM barrel could act as an exceptionally good scaffold for a variety of reactions, one that nature could in principle have arrived at independently, several times in evolutionary history. They instead argued that all TIM-barrel proteins descended from a single common ancestor, and detailed how this group could have diversified into (at least) four families with distinct enzymatic functions.15
The structures in the Protein Data Bank now number more than 100,000, but the early concepts of superfamily and family established by these and other authors remain roughly the same. The superfamily, of which there are ~2,000 in total, is the largest grouping (or clade) of proteins for which common ancestry can be inferred. This inference is based on structural alignment even if little or no sequence similarity is apparent.16 Families are defined as groups of proteins that, in addition to common ancestry and structure, share related functions and more overtly similar sequences. Notably, because the definition of family depends on function, distinguishing new families from old requires the experimental input of biologists and biochemists. Hence, new family or “subfamily” designations continue to emerge as more of sequence space is empirically explored. (See below.)
A number of efforts at organizing protein structures into superfamilies have been made, resulting for example in the Structural Classification of Proteins (MRL and Berkeley National Lab) or SCOPe17,18 and Pfam databases (European Bioinformatics Institute, the Wellcome Trust, and the Howard Hughes Medical Institute). These overlapping efforts have led to some redundancy in nomenclature. Hence, the CDE superfamily in the SCOPe database is known rather colorlessly as “dimeric alpha+beta barrel containing,” or via its numerical designation, 54909. The same superfamily is designated as clade CL0032 by the Pfam database, which lists 18 constituent families while SCOPe names 24. In spite of the differences in numbers, the family organization proposed by the two databases is nonetheless largely similar. The names and several salient properties for all of the CDE families, many of which are uncharacterized, are summarized in Table 1.
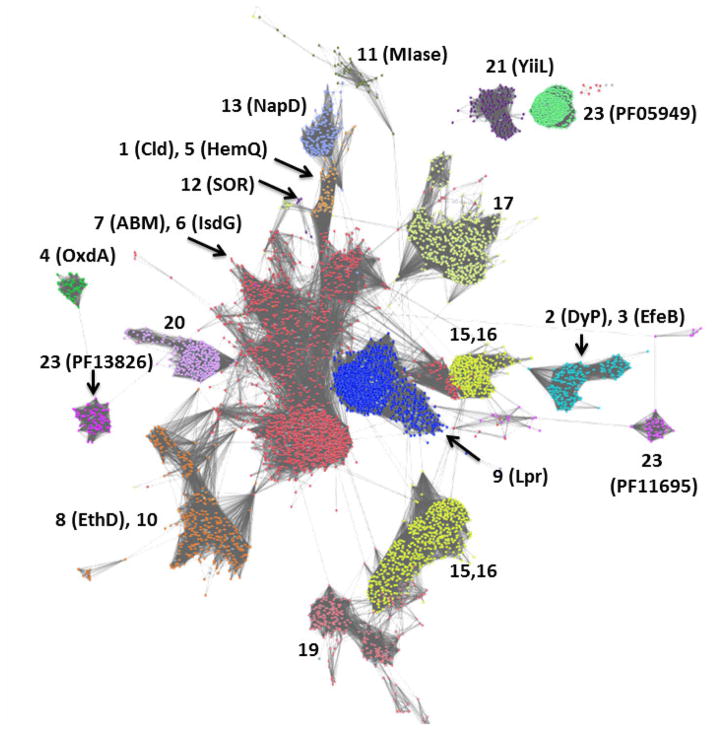
We have previously used phylogenetic trees to visually subdivide the superfamily and its constituent families;3 however, its sheer size now necessitates a more sequence-inclusive approach, such as that offered by sequence similarity network analysis (Cytoscape).19,20 This “all-by-all” method of sequence comparison is conceptually similar to phylogenetic tree drawing but capable of organizing thousands of sequences at a turn. Sets of closely related sequences are represented as dots or nodes, and sequences related to one another at the given stringency (similar to the BLAST e-value) are clustered or connected by lines. All of the CDE sequences (currently >100,000) grouped into their Pfam-designated families at the stringency shown (Figure 1). From this analysis, it is clear that some families within the superfamily are more closely related to one another than others; those which are connected by multiple lines between the nodes, for example, are more closely related to one another than those joined by few or none. Moreover, several of the Pfam-designated families actually encompass multiple functions, and could justifiably be subdivided further. Higher stringency analysis of the Cld and DyP families, for example, has been used here to break them into subfamilies, as discussed further below.
Figure 1. Network analysis of CDE superfamily (SCOP #54909, InterPro CL0032) showing its subdivision into families.
Groups of protein sequences with ≥50% pairwise identity are represented by individual dots colored according to Pfam family designation. Each family is labeled according to the number assigned to it in Table 1. Dots are shown overlapping or with lines connecting them if the least significant pairwise sequence-similarity score between the representative sequences of each is better than the threshold (BLAST E-value ≤1×10−7). The superfamily resolves into ≥18 families at this relatively low level of stringency, with at least one (group 7, the ABM family) beginning to resolve into subgroups. The network was generated from >100,000 sequences downloaded from Pfam and using the Enzyme Function Initiative Enzyme Similarity Tool (http://efi.igb.illinois.edu/efi-est/) and visualized using Cytoscape (www.cytoscape.org).
Domain architectures and oligomerization states
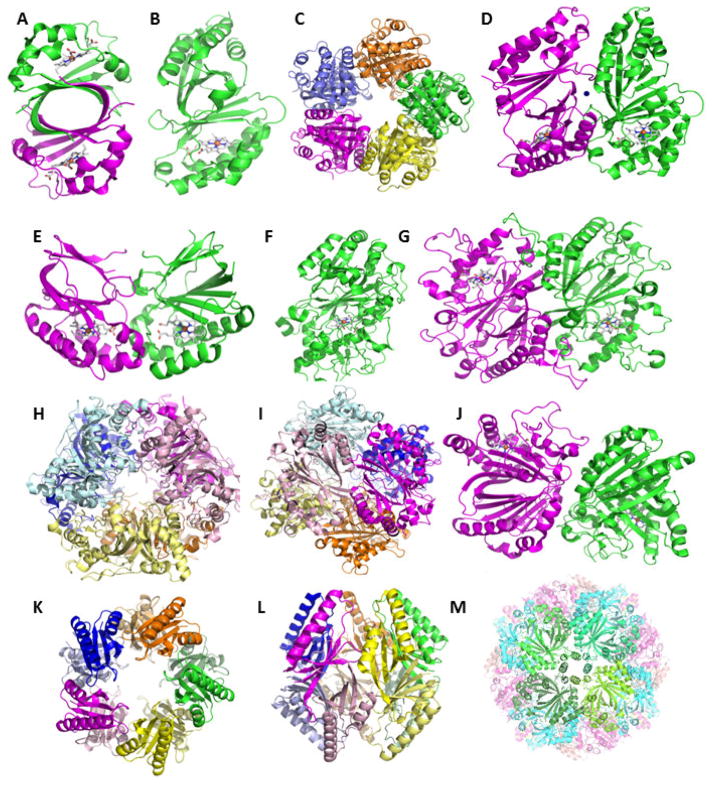
CDE proteins share a common domain structure consisting of ≥110 amino acids arranged in a series of alpha helices on one side of a barrel and beta sheets on the other. This domain is remarkably versatile with respect to the architectures and oligomerization states in which it is acts as a building block. Figure 2 shows examples of all of the architectures and oligomerization states described below. Table 2 provides a list of representative protein structures from the families of interest, with their protein data bank identification (PDB ID) codes and references.
Figure 2. CDE domain structures and oligomerization states, emphasizing the heme-binding proteins.
All structures are rendered as cartoons. Individual polypeptides are shown in different colors. Bound hemes are drawn as sticks with carbon colored grey. A. IsdGs have a single-domain monomer (PDB ID 3LGN). Two IsdG monomers form a homodimer that resembles the bidomain monomer found in almost all of the structurally characterized proteins from the superfamily. B. The bidomain monomer from a representative Cld (Dechloromonas aromatica) is shown, with heme bound in the lower C-terminal domain (left, 3Q08). C. These assemble into a homopentamer, shown looking down the 5-fold axis with the heme-binding domains oriented away from the viewer. D. The interface between two monomers from the pentamers is shown close up, illustrating the alignment between the heme binding domains. A Ca2+ ion is shown as a blue sphere. E. A subfamily of Clds have a single-domain monomer with extended sequence at the C-terminus (3QPI). These form homodimers in which the pathway for accessing the heme clearly differs from the structure shown in (B). F. DyPs from different sources have crystallized as monomers, homodimers, and homohexamers. The monomer structure (from Bjerkandera adusta Dec 1, 2D3Q) is similar to that in the Clds but longer and elaborated with several loops. G. Two DyP monomers oligomerize in a head-to-tail orientation, in contrast with the structure in (D). A representative dimer, the EfeB from E. coli (2Y4F) is shown. H. A hexameric DyP (from Rhodococcus jostii, 3QNS) is presented looking down the 3-fold axis and (I) at the interface between two monomers, illustrating the “trimer-of-dimers” arrangement of the homohexamer. Members of the pseudodimer are shown in same-color light/dark shades. J. The aldoxime dehydratases form a head-to-tail dimer that resembles the DyPs but with a wider solvated interface between the monomers (3W08). K. Homodecameric, “pentamer-of-dimers structure” assumed by proteins from the muconolactone isomerase family. The structure of chloromuconolactone dehalogenase (3ZNU) is shown looking down the 5-fold axis (left) and from the side (right). H. Hollow 24-mer formed by sulfur oxygenase reductase (2YAV), looking down a 4-fold axis flanked by 4 bidomain monomers (colored different shades of green). Figures generated using PyMol: www.pymol.org.
Table 2.
Representative available X-ray crystal structures for heme-binding CDE proteins
| Family | PDB IDs | Source | Description | Subfamily | Ref. |
|---|---|---|---|---|---|
| DyP | 2IIZ, 2HAG | Shewanella oneidensis: facultative aerobe, Proteobacterium | Dimeric; Heme-bound and heme-free forms available. Also called prephenate dehydrogenase (TyrA). | Group 2 (Fig. 6) | 22,29 |
| 2GVK | Bacteroides thetaiotaomicron: Gram-negative anaerobe; dominant species in human gut | Hexameric; Heme-free structure. | Group 2 (Fig. 6) | 29 | |
| 3QNS,3QNR, 3VEC, 3VED,3VEE, 3VEF, 3VEG, 4HOV | Rhodococcus jostii: soil-dwelling member of phylum Actinobacterium with robust biodegradative capabilities | Hexameric; homologous to YfeX (E. coli). | Group 2 (Fig. 6) | 24, 25 | |
| 2D3Q, 3MM1, 3MM2, 3MM3, 3AFV,3VXI,3VXJ | Bjerkandera adusta (formerly Thanatephorus cucumeris): fungus from phylum Basidomycota; causative agent of white rot | Monomeric; structures of mutants and different heme- ligand complexes available. | Group 3 (Fig. 6) | 30 31, 76, | |
| 4AU9, 4W7J-O | Auricularia auricula-judae: saprophytic fungus from phylum Basidomycota; | Monomeric | Group 3 (Fig. 6) | 26, 40 | |
| EfeB | 2Y4D,2Y4E, 2Y4F,3O72 | Escherichia coli: phyum Proteobacteria | Dimeric; Heme-free, protoporphyrin IX- and heme-bound | Group 1 (Fig. 6) | 11, 32 |
| 4GS1 | Thermobifida cellulosilytica: lignocellulose-decomposing member of phylum Actinobacterium | Dimeric | Group 1 (Fig. 6) | 33 | |
| 4GRC, 4GT2, 4GU7 | Streptomyces coelicolor: member of phylum Actinobacterium with robust biodegradative capabilities | Dimeric | Group 1 (Fig. 6) | 33 | |
| Cld | 2VXH | Azospira oryzae: Perchlorate respiring member of phylum Proteobacteria | Pentameric in solution, hexameric in solid state | Respiratory Clds (Fig. 7) | 4 |
| 3Q08, 3Q09 | Dechloromonas aromatica: Perchlorate respiring member of phylum | Pentameric | Respiratory Clds (Fig. 7) | 5 | |
| 3NN1-4, 4M05-4M09 | Candidatus Nitrospira defluvii: non-perchlorate respiring member of phylum Nitrospira. The cld gene is suggested to have been laterally transferred from a respirer. | Pentameric | Respiratory Clds (Fig. 7) | 6, 38 | |
| 3QPI | Nitrobacter winogradskyii: non-perchlorate respiring member of phylum Proteobacteria. | Dimeric | Non-resp. Clds (Fig. 7) | 7 | |
| HemQ† | 3DTZ | Thermoplasma acidophilum: phylum Crenarchaeota | Pentameric; ligand-free | Group 1 HemQ (Fig. 7) | 35 |
| 1T0T | Geobacillus stearothermophilus: phylum Firmicutes | Pentameric; ligand-free | Group 2 HemQ (Fig. 7) | 36 | |
| 1VDH | Thermus thermophilus: phylum Deinococcus-Thermus | Pentameric; ligand-free | Group 2 HemQ (Fig. 7) | 37, 39 | |
| 4WWS | Listeria monocytogenes, phylum | Pentameric; heme-bound | Group 2 HemQ (Fig. 7) | ||
| OxdA | 3W08 | Pseudomonas chlororaphis: phylum Proteobacteria | Homodimer | -- | 27 |
| 3A15, 3A16, 3A17, 3A18 | Rhodoccus erythropolis: phylum Actinobacteria | Homodimer | -- | 27 | |
| IsdG | 2ZDO, 1XBW, 1SQE, 3LGN, 3LGM | Staphylococcus aureus: phylum Firmicutes | Homodimer | -- | 42, 43, 45 |
| 4NL5, 3HX9 | Mycobacterium tuberculosis: phylum Actinobacteria | Homodimer | -- | 41, 44 |
Listed as to be published by the PDB.
Tentative functional assignment based on unpublished data (DuBois et al.) and informatics (Figure 7).
In the Cld, HemQ, DyP, EfeB, and OxdA families, with at least one notable exception,7 the domain occurs twice in tandem on the same polypeptide. In the structurally characterized proteins from these families (see references for several examples),4–6,8,11,20–40 heme is found only in the C-terminal domains. This is in spite of the presence of both a cavity and in some cases potential iron ligands inside the empty domains, which tend to be more compact than their C-terminal counterparts.3 The bidomain subunit can exist as a lone monomer, as it does in some DyPs.22,40 Alternatively, it forms a “head-to-tail” homodimer in other DyPs as well as EfeBs and OxdAs,26,27 in which the N-terminal domain of one monomer is adjacent to the C-terminal, heme-binding domain of the next. In some DyPs, the dimer further assembles into a trimers-of-dimers or homohexamer.24 By contrast, in the oxochlorate-respiration-associated Clds and in the HemQs, all of the C-terminal (heme-binding) portions mutually align on one face of a homopentamer.4–6,35–39 Exceptions to this rule are the non-respiratory Clds, which are monodomain proteins that form dimers in which the two C-termini align.7
In contrast, the IsdGs are all single-domain proteins. These assemble into dimers that resemble the figure-8-shaped, bidomain subunit of the families described above.41–45 The larger antibiotic biosynthesis monooxygenase (ABM) family, of which the IsdGs form a relatively small part, is structurally very diverse. It contains proteins with one or two ABM domains on the same polypeptide. These domains are also found appended to other structurally distinct proteins with a wide variety of functions. Notably, the substrates and heme-binding properties of many ABM proteins have not been determined, though the family as a whole appears to be functionally very versatile. The diversity of the family is reflected in its sprawling representation, relative to all of the other families, in Figure 1.
As illustrated in Figure 2, the different oligomerization states and subunit interactions observed in these structures clearly influence the solvent and substrate accessibility of the heme-binding pocket. Coupled with their distinct sets of conserved active site residues (discussed below), these structural differences likely give rise to the spectrum of catalytic properties observed for these families.
Finally, in some of the CDE families outside the heme-binders highlighted here, several impressively large homo-oligomers have been structurally characterized. Members of the muconolactone delta-isomerase (MIase) family (Table 1), for example, form homodecamers which resemble the Cld/HemQ homopentamers (Figure 2).46,47 The sulfur-oxidoreductases (SORs) are non-heme-iron-binding 24-mers with a large open cavity in the center (Figure 2).48 These structures again illustrate the tremendous flexibility of the domain as a building module.
One pocket, many binding modes
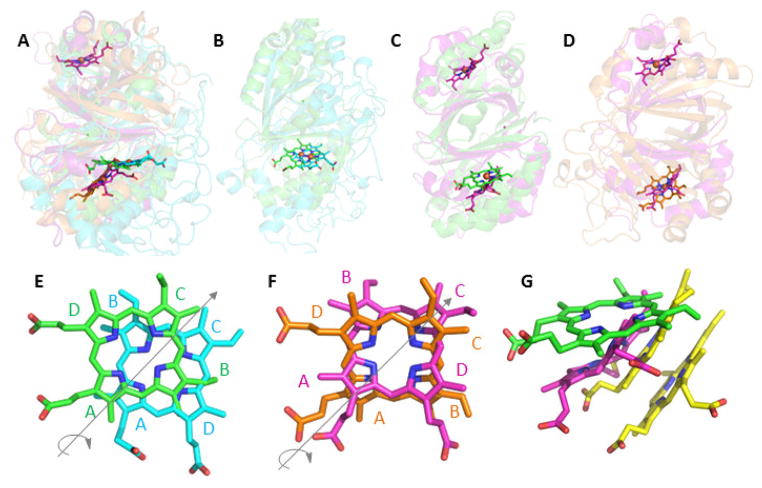
Since several CDE families bind heme b or closely related molecules, one might reasonably predict that the heme binding mode to the domain would be relatively static. However, structural biology has shown otherwise. Heme in fact binds in two distinct, non-overlapping regions of the domain pocket. In each case, it assumes multiple orientations with respect to the positions of its methyl, vinyl, and propionate side chains. The overlaid structures of representative heme-binding domains illustrate this point best (Figure 3A–D). The Cld (green) and DyP (cyan) hemes occupy the same plane and roughly the same position in the cavity (Figure 3B). However, their orientations differ by an approximate 180° (C2) rotation around an axis passing through N22 and N24 (IUPAC numbering) and bisecting the A and C rings of the porphyrin (Figure 3E). This rotation swaps the positions of the B and D rings, switching a vinyl with a propionate ring substituent. (Note that the hemes in available EfeB structures superimpose with those in their sequence cousins, the DyPs, and are consequently not discussed separately here.11,32)
Figure 3. Superimposition of representative CDE protein structures illustrating their diverse heme binding modes.
In each panel, the proteins are shown as cartoons in roughly similar orientations in order to emphasize similarities and differences in the positions of their hemes. The bound hemes are rendered as sticks: Cld (carbon-green, PDB ID 3Q08), DyP (carbon-cyan, 2D3Q), OxdA (carbon-orange, 3W08), IsdG (carbon-magenta, 3LGN), and MhuD (carbon-yellow, 3HX9). A. Superimposition of Cld, DyP, OxdA, and IsdG shows the two distinct planes occupied by the hemes in Cld/DyP and IsdG/OxdA. Note that IsdG is a dimer in which each monomer binds heme, while the other proteins are bidomain monomers where heme binds only in the C-terminal domain. B. Cld/DyP monomer superimposition illustrating the approximate C2 rotation through the heme rings A and C that relates the heme in one of these proteins to the heme in the other. C. Superimposition of the Cld monomer/IsdG dimer. In the lower domain, the side chains on the IsdG heme are oriented similarly to those in DyP, but the heme occupies a different plane. D. Superimposition of the OxdA monomer/IsdG dimer. In the lower domain, the hemes assume roughly the same plane but, once again, are distinguished from one another by a C2 rotation. E–G. Close-up views of the superimposed hemes generated from overlaid CDE protein structures. The heme rings are labeled and the iron atoms have been removed for clarity: E. Cld (green)/DyP (cyan); F. IsdG (magenta)/OxdA (orange); G. Cld (green)/IsdG (magenta)/MhuD (yellow). The side chains of the lower heme in MhuD, an IsdG-family protein from Mycobacterium tuberculosis, are oriented similarly to the heme in IsdG. The upper heme is oriented roughly like the heme in Cld, though in a different plane. Figures generated using PyMol: www.pymol.org.
The OxdA and IsdG hemes, by contrast, occupy a position close to one another but far lower in the pocket and on an entirely different plane from Cld/DyP (Figure 3B). Their heme orientations are likewise related to one another by an approximate C2 rotation through the A–C ring axis (Figure 3F). The flexibility in this region of the pocket extends to accommodating two bound hemes in the IsdG-family protein from Mycobacterium tuberculosis (MhuD) (Figure 3G).41 One heme occupies a position similar to that of single-heme-binding homologs from Staphylococcus aureus. The second stacks beneath the first, and is related to it by the same C2 rotation observed elsewhere in this superfamily. This arrangement appears to minimize steric clashes between methyl and vinyl side chains on the adjacent macrocycles. It also brings the lower ring into more or less the same orientation, with respect to the heme side chains, as the single heme in the other crystallographically characterized proteins from the IsdG family.
Finally, the structures of four probable HemQ-family proteins are available, from Geobacillus stearothermophilus, Thermoplasma acidophilum, Thermus thermophilus, and most recently, Listeria monocytogenes; however, as heme is a product in the HemQ-catalyzed reaction, it appears to bind characterized members of the family with relatively weak affinity.33,34,38 Hence, the available structures have either an empty cavity or a solvent molecule above the conserved histidine where the coproheme substrate is expected to bind. Notably, the HemQs and Clds are very closely related in sequence (Figure 1) and are consequently members of the same Pfam family (Table 1); yet, the Clds bind heme avidly as a cofactor. The superimposed structures of a solvent-bound HemQ and heme-bound Cld suggest several reasons for their distinctly different interactions with heme. First, the probable tetrapyrrole binding site in HemQ has more room for accommodating the two extra propionate groups that distinguish coproheme from heme (Figure 4). In particular, a leucine residue present in Clds but not HemQs occludes a potential mooring place for a coproheme propionate. This region is occupied by a molecule of polyethylene glycol in the HemQ structure from G. stearothermophilus (Figure 4A). Moreover, the Cld/HemQ monomer secondary structures largely cohere, with the exception of a large helix-loop segment (residues ~110–140) where they diverge widely. The lifting of this segment on all four HemQ structures relative to the Clds creates an obvious opening where the coproheme substrate and heme product could enter and leave the protein (Figures 4B and 4C). The appearance of a partially conserved, positively charged KRR or RRR motif in the helical portion of the segment, in HemQs but not Clds, is further suggestive of a mechanism by which this “gate” could close around the substrate or interact with another protein. These hypotheses are currently being tested by our research group.
Figure 4. Superimposition of Cld and HemQ structures illustrating features that may be important for their different interactions with heme.
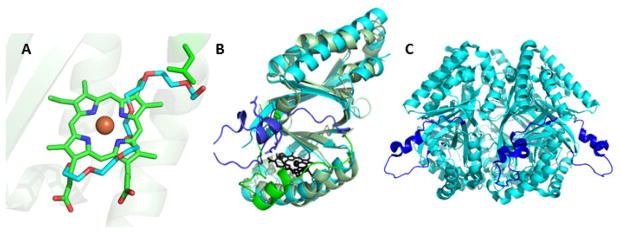
A. Superimposition of Cld (3Q08) and HemQ (1T0T) structures showing the bound heme in the Cld (carbon-green) and polyethylene glycol (carbon-cyan) in the HemQ. The polyethylene glycol extends into a region of the pocket which is blocked in the Cld by a leucine side chain (L178, D. aromatica Cld numbering). B. The overlaid monomer structures are shown as cartoons in light green (Cld) and cyan (HemQ), and their heme/polyethylene glycol ligands in black. Residues 110–140, where the two structures sharply diverge, have been highlighted in darker shades of green/blue. A stretch of three arginines on the helical portion of this region in the HemQ are rendered as sticks. C. HemQ pentamer rendered as cyan cartoon, shown from the side. Residues 110–140 are shown in a darker shade of blue. These form a mixed loop/helical region on the protein’s surface and hence could be the site of substrate entry/product egress. Figures generated using PyMol: www.pymol.org.
Understanding function: Heme as a cofactor
The superfamily’s diversity of heme binding modes, oligomerization states, and conserved active site residues (discussed below) supports a variety of reactions within the same structural scaffold. These include the catalysis of three distinct reactions with heme as the cofactor: O2 generation (Clds), peroxidase-style one electron oxidations (DyPs, EfeBs), and dehydration of aldoximes to yield nitriles (OxdAs). What is known about how these reactions work and how the chemical environment of the heme promotes them? Briefly:
Chlorite dismutases
The chlorite dismutases detoxify ClO2− by catalyzing its rapid conversion into Cl− and O2. This unusual reaction constitutes the terminal step in the respiration of perchlorate (ClO4−), a metabolic pathway which was considered unusual when it was discovered in the late 1990s because the substrate was thought to be almost entirely unnatural.2 Subsequent investigation, however, has identified photochemical reactions in the stratosphere as a robust and potentially ancient source of ClO4−,49,50 on Earth as well as Mars. Microbial removal is now believed to account for the anion’s scarcity in most terrestrial environments. Though the real diversity of perchlorate-respiring microbes may not be known, many Proteobacterial species with this metabolism have been identified.51,52 In each case, the organism possesses either a dedicated ClO4− or chlorate (ClO3−) reductase in addition to a chlorite dismutase, whose major biological function is to detoxify ClO2− by producing oxygen gas.2,53
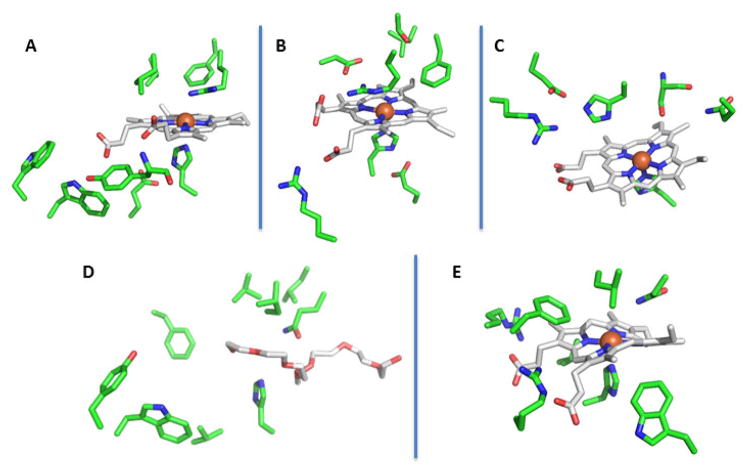
Clds from these respirers share several common active site features (Figure 5A). They have a proximal histidine ligand which, in spite of its hydrogen bond to a conserved glutamic acid, has vibrational properties consistent with a more neutral imidazole ring than in heme peroxidases, akin instead to the proximal ligand in myoglobin.54,55 On the heme periphery are a pair of conserved tryptophan residues (W155 and 156 in the Cld from Dechloromonas aromatica) very near to the monomer’s surface. Mutation of these residues to phenylalanine appears to destabilize the oligomerization state of at least the D. aromatica Cld, suggesting that they are important for maintaining the protein’s subunit-subunit interface.56 The heme propionic acids are stabilized by hydrogen bonds to backbone amide nitrogens and a conserved tyrosine (Figure 5A).5
Figure 5. Active site structure comparisons for representative heme-binding CDE protein families, illustrating the conserved amino acids in each.
The hemes (carbon-gray, oxygen-red, nitrogen-blue) are shown in similar orientations in order to facilitate comparison of the environments of the bound hemes. Conserved and/or functionally significant amino acid side chains are shown as sticks in carbon-green, oxygen-red, and nitrogen-blue. Potential hydrogen-bonding interactions are shown as dashed lines: A. Cld (3Q08); B. DyP (2D3Q); C. OxdA (3W08); D. HemQ (1T0T, polyethylene glycol solvent molecule in the likely heme-binding site); and E. IsdG-family protein (IsdI, 3LGN). Figures generated using PyMol: www.pymol.org.
The distal pocket acts as a sterically confined “roof” over the heme’s open coordination position. A strictly conserved arginine (R183 in the D. aromatica Cld) is a conspicuous exception in this otherwise hydrophobic distal pocket. The arginine is absolutely critical for efficient oxygen formation and a signature residue distinguishing O2-generating from non-generating members of this family (see below). It has been shown to have roles in recruiting and stabilizing bound anionic ligands, and has been proposed to stabilize the anionic, acetate leaving group in the oxygen-atom transfer reaction between ferric Cld and peracetate (CH3C(O)COO−).57 An analogous role seems highly plausible for the reaction between Cld and chlorite. The hypochlorite leaving group has a near-neutral pKa but would be stabilized in deprotonated form by the positively charged arginine. This would render it more nucleophilic and hence better poised to react with the electron-deficient oxygen of a Cld ferryl porphyrin cation radical species (Compound I).
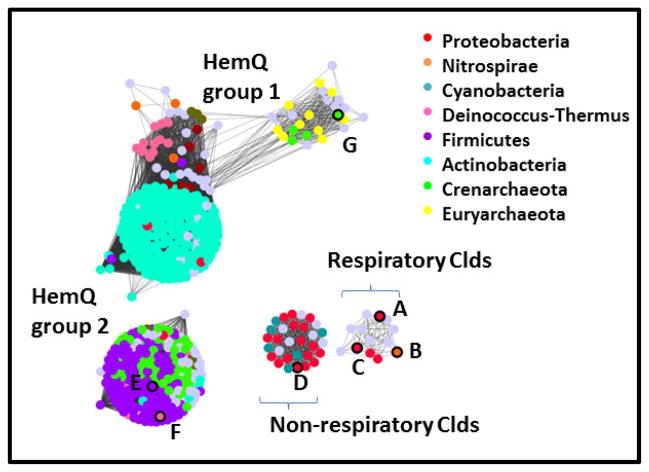
There is additionally a second group of Clds which, as discussed above, form homodimers instead of pentamers and which have a single domain on each polypeptide instead of a pair.7,58 These Clds come from non-perchlorate or –chlorate respirers, and have been identified primarily in diverse Proteobacterial species.58 They form a distinct group in phylogenetic7 or network analyses of the Cld protein family (discussed in Figure 7 below). Unlike the respiratory Clds, which are preceded by a SecB peptide earmarking them for the periplasmic space, these Clds appear to be cytoplasmic. They moreover possess the critical distal arginine and indeed have been shown to produce O2 from chlorite.7,58 What is the selective advantage for having a chlorite detoxification system in so many bacteria, especially pathogenic species? It has been suggested that toxic chlorate, which many bacteria can incorporate via nitrate uptake systems, might be transformed by respiratory nitrate reductases to water and chlorite.59 A functional Cld would then afford some protection to the organism from ClO2− produced endogenously in the cytoplasm. Using a Δcld strain of Klebsiella pneumoniae, we indeed observed a substantial growth defect due to chlorate specifically under nitrate-respiring conditions.60 This suggests that environmental chlorate, which like perchlorate has until recently flown under the geochemical radar,61 might be pervasive enough in some environments to promote the genomic retention and transfer of cld genes. The chemistry of these “non-respiratory” Clds, as well as the Cld-like HemQs, lies at the frontier of this field. The latter proteins are further described below.
Figure 7. Network analysis of the Cld family (PF06778) showing its division into subfamilies.
Groups of protein sequences with ≥85% pairwise identity are represented by dots colored according to phylum domain of life of their host organism, as shown in the legends. Gray dots indicate that no assignment is presented. Proteins with crystal structures in the the Protein Data Bank are outlined in black on the left panel, and their PDB IDs given here: A. 2VXH; B. 3NN1; C. 3Q08; D. 3QPI; E. 1T0T; F. 1VDH; G. 3DTZ. See Table 2 for further information on these structures. Dots are shown overlapping or with lines connecting them if the least significant pairwise sequence-similarity score between the representative sequences of each is better than the threshold (BLAST E-value ≤1×10−25). The Cld family forms 4 subgroups at this relatively high level of stringency, which are labeled here and discussed further in the text. The network was generated from 1117 sequences downloaded from Pfam and using the Enzyme Function Initiative Enzyme Similarity Tool (http://efi.igb.illinois.edu/efi-est/) and visualized using Cytoscape (www.cytoscape.org).
Dye-decolorizing peroxidases and EfeBs
The DyPs are a diverse group of enzymes, predominantly from Bacteria and Fungi (and a few higher Eukaryotes), first explored by Shoda, Kim, Sugano, et al. in the late 1990s-early 2000s.62 This group showed that DyPs catalyze peroxidase-type reactions at a ferric heme. The peroxidase reaction is canonically described as a three-step process:
Peroxide activation: FeIII(Porphyrin) + H2O2 + H+ → H2O + FeIV=O(Porphyrin▪+) (Compound I)
Oxidation 1: FeIV=O(Porphyrin▪+) + substrate-H → FeIV-OH(Porphyrin) (Compound II) + substrate▪
Oxidation 2: FeIV=O(Porphyrin) + substrate-H → FeIII(Porphyrin) + H2O + substrate▪
The initially characterized members of the family had the unique ability to degrade dyes sharing the 3-ring aromatic core structure of 9,10-anthraquinone; hence, they were called “dye-decolorizing.63,64” Having the capacity to oxidize these chemically intransigent, biotechnologically important substrates is the first hallmark of this group. Currently known DyP substrates are diverse, and include reduced transition metal cations such as Fe(II) and Mn(II),65,66 aromatic species,63,64,67 porphyrins,13 and very large polymers including lignin.40,68,69 DyPs are also structurally distinct from the many plant, fungal, and mammalian peroxidases studied before their discovery, not only in the overall heme scaffold but also at the active site (Figure 5B), where they possess a conserved His-Asp-Arg motif. These structural and consequent mechanistic distinctions from the well-studied peroxidases are a second major hallmark of the DyPs
The diversity of this family has become better appreciated as more sequences have been deposited and characterized. Peroxibase (http://peroxibase.isbsib.ch/), an online repository for information about peroxidases, lists subfamilies A–D of DyPs based on phylogenetic analysis. Members of the A subfamily are also known as the EfeBs. EfeBs are found in diverse bacteria as part of an iron-regulated, three-gene efeUOB operon (orthologs in some species are known as fepABC).9 EfeU is a transmembrane protein that is homologous to the high-affinity iron permease, Ftr1p, of Saccharomyces cerevisiae. EfeO is a periplasmic protein with a cupredoxin N-terminal domain and possible metal transporting capabilities.70 EfeB is also periplasmic, brought there in its heme-bound form via the twin-arginine transporter (Tat) system.71 The exact role of EfeB in iron assimilation is not known. It has been described as an Fe(II)-oxidizing peroxidase, producing Fe(III) for delivery to inner membrane ABC transporters.9 Supporting this description, Fe-55 transport and in vitro catalytic assays have demonstrated the protein’s involvement in Fe(II) import and its chemical capabilities as a peroxidase.9,21 Heme itself has also been proposed as a substrate;10,72 however, the proposed reaction, involving liberation of iron from the tetrapyrrole, has not been directly demonstrated.
At least one EfeB (DyP-A), from the Actinomycete Rhodococcus jostii RHA1, has been studied biochemically.24 Two paralogous enzymes from this organism, designated DyP-A and DyP-B, due to their membership in the Peroxibase DyP-subfamilies A and B, were compared in order to assess their possible utility in lignin degradation. Using genetic knockouts and a lignin-degrading assay, RjDyP-B was shown to be capable of manganese-dependent lignin degradation.24,65 The role of manganese may be as an electron conduit: Mn(III) is enzymatically reduced to Mn(II), which in turn is released from the cell where it acts extracellularly as an oxidant toward complex organic substrates. Biochemically, RjDyP-B behaves like a peroxidase. Upon reaction with one equivalent of H2O2, RjDyP-B formed a spectrum characteristic of a typical peroxidase Compound I intermediate. Its second-order rate constant for Compound I formation and steady-state kcat/Km(H2O2) and kcat/Km(dye) were moreover indicative of a respectable level of peroxidase activity. By contrast, RjDyP-A did not degrade lignin in the same assay. When mixed with an equivalent of H2O2, a ferryl (Fe(IV)=O) spectrum with a Soret peak at 419 nm and α and β bands at 557 and 528 nm formed, rather than Fe(IV)=O(Porphyrin▪+). Compared with RjDyP-B, RjDyP-A reacted much more slowly with H2O2 and had a ~100-fold lower kcat/KM(dye), suggesting that it is not as clearly peroxidase-like under the conditions tested. In short, phylogenetic distance and differences in operonic structures, regulatory motifs, subcellular localization, and catalytic properties all suggest that the EfeBs are a distinct DyP subtype, with a likely role in Fe uptake.
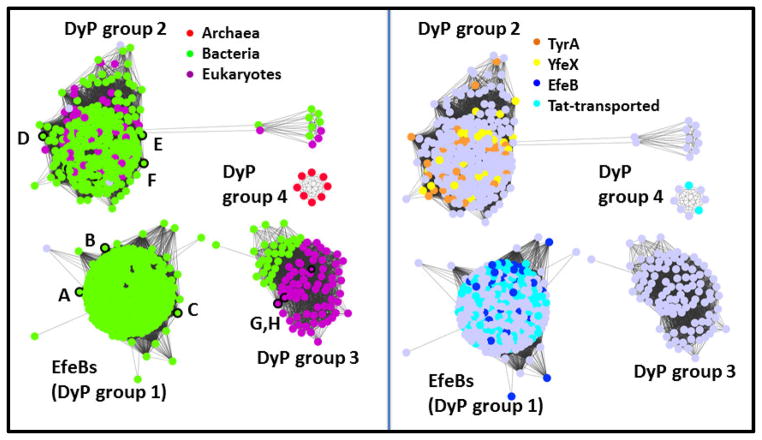
Generally speaking, the characterized DyPs outside the EfeB group appear to catalyze peroxidase reactions with a diverse range of preferred substrates, kinetic properties, biological roles, and sources. How diverse are they? Network analysis of the DyP family indicates that, at a moderate level of stringency, the family groups into four clusters (Figure 6). Proteins in the EfeB cluster (group 1) are almost entirely from bacteria. Many of these proteins are annotated as having a Tat-transporter peptide (Figure 6, right panel), and many appear to share an efeOUB-like operonic context. Members of a second cluster include a mixture of Bacterial and Fungal proteins, where the peroxidase reactions they catalyze serve a variety of proposed cellular housekeeping functions, including melanin or tyrosine biosynthesis (TyrA) and cytochrome maturation (YfeX). This group is dominated by members of the Peroxibase DyP-Bs. A third group contains characterized DyP-C and -D proteins, many from species known to have a biological need for peroxidase-mediated biodegradation of lignin or other plant products. For example, Bjerkandera adusta, the source of one of the first studied DyPs, is a fungal pathogen of plants and a causative agent of white rot. Auricularia auricula-judae, another source of fungal DyPs from the third group is a saprophyte, gaining nutrition from dead or dying elder trees. The need to tackle such large or structurally diverse substrates could distinguish the proteins from this cluster from the peroxidases in cluster 2. The DyP from A. auricula-judae, for example, has been shown to accommodate large substrates at its surface, which is connected to the heme site via a proposed conduit of amino acids.26, Notably, the proteins in the third group neatly partition along taxonomic lines, into Bacterial and Fungal subgroups (Figure 6, left panel). Finally, a small cluster of DyP proteins deriving predominantly from halophilic Archaea forms a fourth group. The functions of these are not known, though some of them are predicted to have an N-terminal Tat sequence (Figure 6, right panel).
Figure 6. Network analysis of the DyP family (PF04261) showing its division into subfamilies.
Groups of protein sequences with ≥70% pairwise identity are represented by dots colored according to (left panel) the domain of life of their host organism or (right panel) their Uniprot-assigned annotations, as shown in the legends. Gray dots indicate that no assignment is presented here. Proteins with crystal structures in the the Protein Data Bank are outlined in black on the left panel, and their PDB IDs given here: A. 2Y4D; B. 4GS1; C. 4GRC; D. 2IIZ; E. 2GVK; F. 3QNS; G. 2D3Q; H. 4AU9. See Table 2 for further information on these structures. Dots are shown overlapping or with lines connecting them if the least significant pairwise sequence-similarity score between the representative sequences of each is better than the threshold (BLAST E-value ≤1×10−25). The DyP family forms 4 subgroups at this relatively high level of stringency, which are labeled here and discussed further in the text. The network was generated from 2768 sequences downloaded from Pfam and using the Enzyme Function Initiative Enzyme Similarity Tool (http://efi.igb.illinois.edu/efi-est/) and visualized using Cytoscape (www.cytoscape.org).
Structurally and mechanistically, the DyPs are distinct from the better-known peroxidases. 73–75 In the latter, a distal pocket histidine (pKa = 7) is stabilized in its neutral/deprotonated state by a nearby arginine, allowing it to act as a base toward H2O2 (pKa = 12).76 Removal of the peroxide proton facilitates binding of the HOO− anion to the ferric iron. Subsequent heterolytic cleavage of the Fe(III)O-OH bond is 5-orders of magnitude faster in wild type cytochrome c peroxidase, relative to a mutant in which the distal histidine is substituted by leucine.77 In DyPs, there is no critical distal histidine; however, there is a conserved arginine-aspartic acid pair (Figure 5B). The latter was shown to act, in lieu of the histidine, as an active site base in the DyP from the fungus Bjerkandera adusta.78 However, a mutagenesis study of the DyP from the bacterium Rhodococcus jostii demonstrated that mutants at this position had very little effect on catalysis, while the companion arginine residue was absolutely essential and proposed to itself act as the active site base.25 The discrepancy has not been fully explained. However, these two DyPs come from very different organisms, could act on very different preferred substrate types, and represent distinct branches of the DyP family tree (Figure 6). It is possible that they could use distinct catalytic strategies.
The DyPs not only have a distinct complement of active site residues but also, as shown in Figures 3B and 3F, a unique heme binding mode. The heme is rotated around the A–C ring axis relative to the heme in Clds. This places the propionate on ring A within hydrogen bonding distance of the distal arginine (Figure 5B). This residue forms bridging hydrogen bonds between the propionate and the conserved distal aspartic acid, placing all 3 moieties near the center of the action, catalytically speaking. This is in contrast to the Clds, where the distal aspartate is absent and the propionate is not in a position to form a hydrogen bond to the distal arginine (Figure 5A). The Cld arginine therefore either hydrogen bonds to the heme ligand or adopts an open conformation, pointing away from the heme iron, which enables access to the active site.5
Aldoxime dehydratases
OxdAs are among the most recent proteins of this superfamily to receive structural and mechanistic attention.27,28 Like Clds and DyPs, they have a proximally histidine-ligated heme and a mostly-hydrophobic distal pocket. However, unlike Clds or DyPs, OxdAs are catalytically active in the ferrous form, and the substrate-bound ferric form of the protein is catalytically dead. OxdAs catalyze the conversion of an aliphatic aldoxime (R-CH=NOH) to the corresponding nitrile (R-C≡N) and H2O, and they are found only in select taxa. Among the Bacteria, they occur primarily in Actinobacteria and α, β, and γ Proteobacteria. Among the Fungi, they are found only in the Ascomycota and not in the filamentous Basidiomycota.
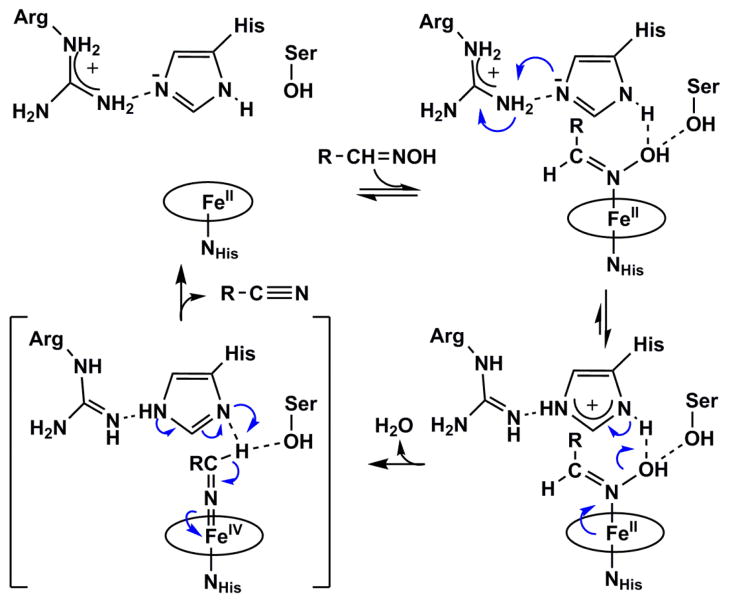
A structure-based mechanism (Figure 5C) was recently proposed (Scheme 1).28 The aldoxime (R-CH=NOH) substrate directly binds the heme iron through its nitrogen atom,27 where it forms hydrogen bonds to conserved serine and histidine residues through its –OH group. The histidine is adjacent to a conserved arginine which, like in the canonical peroxidases, is an arrangement that is proposed to stabilize the distal histidine in its neutral/deprotonated form. Heterolytic cleavage of the N-OH bond is facilitated by this histidine, which acts as an acid in the formation of H2O as a leaving group. The deprotonated/imidazolate ring then formed on the distal histidine is stabilized by partial or complete proton transfer from the nearby distal arginine. N-OH heterolysis is accompanied by the formal movement of two electrons from the ferrous heme and onto the coordinated nitrogen, forming an unusual FeIV=N=CH-R intermediate. This high-valent species is reminiscent of the FeIV=O (Compound II) intermediate formed in peroxidases and many heme-containing proteins. Proton donation from the intermediate to the distal histidine leads to the formation of the C≡N-R product and the regeneration of the ferrous heme.
Scheme 1. Structure-based mechanism for aldoxime dehydration catalyzed by OxdA.
Adapted from reference 27
While the distal serine, histidine, and arginine all have direct chemical roles in this proposed mechanism, the general hydrophobic nature of the pocket has been suggested to promote the evolution of H2O by entropic means. By the same token, the hydrogen bonding network anchoring the propionates and connecting the proximal and distal pockets have been proposed to modulate the electronic structure of the heme, allowing it to toggle between the Fe(II) and Fe(IV) states.79
Heme as a substrate and product
The CDE structural scaffold supports at least three types of heme-based catalysis. (See above.) Remarkably, it also uses heme derivatives as substrates and products in the HemQ and IsdG families:
HemQs
The HemQs are very close sequence relatives of the respiratory Clds – so much so that Pfam still groups the Clds and HemQs into a single family (Table 1), and network analysis resolves the two groups only at relatively high levels of stringency (Figure 7). These proteins are nonetheless clearly functionally distinct. This is made dramatically clear when comparing the reactivity of both proteins with ClO2−. Clds rapidly undergo thousands or tens of thousands of turnovers with this potent oxidant in order to generate O2.80 By contrast, the heme bound to the HemQ from Staphylococcus aureus is completely destroyed by exposure to just five equivalents of chlorite.81
The taxonomic distribution and genomic localizations of cld and hemQ genes reflect their distinct biological functions.3,82 Genes encoding respiratory Clds are found within perchlorate or chlorate reductase operons in Proteobacteria, where the cld gene can assume a number of orientations relative to the reductase.2 By contrast, genes encoding HemQ are found within heme biosynthesis operons in many gram-positive Actinobacteria,82 leading to the conclusion that the protein must be associated with heme metabolism. An association between the cld gene and the enzymes encoding the final two steps of heme biosynthesis was indeed demonstrated biochemically, leading to the renaming of the cld gene as hemQ.82 A ΔhemQ strain of Staphylococcus aureus was furthermore shown to be a heme auxotroph,81 in spite of having an apparently complete set of heme biosynthetic genes. What, then, could be the gene product’s biochemical function?
Using sophisticated bioinformatics methods and the breadth of currently available genome sequences, Dailey, Gerdes, et al. proposed a re-routing of several steps in heme biosynthesis in order to explain the function of HemQ.14 According to the well-known pathway from the textbooks, metal insertion into the porphyrin ring by the enzyme ferrochelatase is the final step in the biosynthesis of heme. While this pathway is still understood to be correct in many Bacteria and Eukaryotes, the Dailey group proposed that HemQ-catalyzes the double decarboxylation of iron-coproheme to yield heme as the final biosynthetic step in bacteria from the Firmicutes and Antinobacteria phyla.
A hemQ homolog lacking the signature distal arginine of Clds (Figures 5A and 5D) is also present in many facultatively or obligately aerobic Archaea. HemQ might provide an aerobic alternative to the AhbD enzyme in these species. AhbD also catalyzes the coproheme decarboxylation reaction in Archaea, but anaerobically via a radical S-adenosyl methionine dependent mechanism.83 Notably, the Cld/HemQ sequences coming from members of the Archaeal phylum Euryarchaeota, most of which are extreme halophiles and aerobes, comingle with the HemQ sequences from the phylum Firmicutes on sequence similarity networks, even at high stringency (Figure 7). Diverse members of the phylum Crenarchaeota as well as non-halophilic Euryarchaeota, by contrast, cluster together as a subgroup more closely related to the hemQs from Actinobacteria. Because representatives of both Bacterial phyla have been associated with heme metabolism, it seems likely that the proteins from both Archaeal phyla share this function as well. However, the occurrence of two major groups of HemQs suggests some degree of functional divergence between the two, which may relate to how the HemQs interact with other proteins in the heme biosynthetic pathway.
From a structural perspective, the selectivity of HemQs for the decarboxylation reaction, rather than O2 generation from chlorite, makes sense. A defining feature of Clds – the conserved distal arginine – is substituted by a charge-neutral glutamine, alanine, or serine in the HemQs.3 In the absence of a distal positive charge to stabilize an anionic leaving group, we expect the Cl-O bond of chlorite will not be disposed toward heterolytic cleavage,57 nor is there a means to recruit and stabilize anionic substrates.84 Instead, the neutral glutamine or serine could act as a hydrogen bond donor to stabilize an Fe(III)-OOH species, where it hypothetically would lack the electrostatic “pull” to promote heterolytic cleavage of the hydroperoxide O-O bond. The more neutral histidine ligand in the related Clds, relative to heme peroxidases, likewise argues against a substantial “push” toward bond heterolysis in trans. We therefore favor decarboxylation mechanisms that do not require the intermediacy of Compound I. These are currently under experimental investigation.
IsdGs
The final family to be described here is part of the larger ABM family of proteins, many of which have the unusual property of activating O2 in the absence of any metallo- or organic cofactor. 85–87 The substrates of many of these proteins themselves resemble (or in the case of IsdGs are) cofactors with well-described O2-activating capabilities. Enzymes like these blur the line between substrate and cofactor, as the substrate actively participates in its own demise. By the same token, Wilks et al. have aimed to distinguish enzymes whose primary biological function is to oxidatively degrade heme, and the wide array of heme-binding proteins which undergo some amount of unwanted heme degradation in the presence of their strongly oxidizing substrates/products (e.g., O2, O2−. H2O2, ClO−).88 The question being asked in either case is the same: when is heme a true substrate, and when is it a cofactor?
This question can be answered from both biological and chemical perspectives. From a biological standpoint, organisms which use heme as a nutritional iron source – for example, many pathogenic bacteria – require some means of removing iron from the tetrapyrrole. In that case, heme is a biological substrate of an enzymatic process. Until relatively recently, the only enzymes known to catalyze the removal of iron from heme were members of the heme oxygenase (HO) family (Pfam identifier PF01126).89 However, it became apparent in the early 2000s that many important pathogens that survive on heme did not possess an HO. This led to the discovery of the IsdG-family of proteins which, in Staphylococcus aureus, have been shown to act at the terminus of a cascade of proteins that strip heme from hemoglobin and shuttle it through the cell wall.90,91 Because they interface with a heme import system and are necessary for utilization of the iron from heme, these enzymes meet the same biological criteria as HOs for using heme as a substrate.
Chemically, IsdG-family proteins have been shown to degrade heme in the presence of O2, a reducing agent, and catalase.91 The inclusion of catalase to eliminate any H2O2 from the reaction is critical. Non-enzymatic destruction of heme has been shown to proceed with peroxide as the oxidant, while HO activates O2 at the ferrous heme.91 These observations suggested that IsdG is not merely a vessel but indeed an enzyme in which O2 is activated to degrade heme.
The organic end products of the IsdG reaction are distinct from both the non-enzymatic and the HO-mediated pathways for heme degradation.91–96 These observations, and the distinct sequence/structure of the IsdGs, indicate that heme degradation must also occur by a distinct mechanism. HOs catalyze 3 successive monooxygenase reactions. These reactions lead to heme hydroxylated at a meso-carbon, which is then converted to verdoheme and carbon monoxide (CO), and subsequently to the ring-opened product biliverdin (Scheme 2).97 O2 activation in the first and third reactions is understood to occur at the iron, and at the porphyrin ring in the second.98,99 In contrast, the IsdGs do not generate CO as a product but instead appear either to yield the more reduced C1 product, formaldehyde (CH2O) (Scheme 2, staphylobilins) or no C1 product at all (Scheme 2, mycobilins).95,96 This suggests that they also cannot generate verdoheme as a productive intermediate. Moreover, the end products of IsdG-family enzymes all incorporate three oxygen atoms instead of two (Scheme 2). The fact that IsdGs belong to a family known for cofactor-independent oxidation chemistry suggests that the enzyme itself could indeed facilitate O2 activation in a way that does not rely on iron.
Scheme 2. Reaction intermediates and products of heme oxygenase and IsdG-family enzymes.
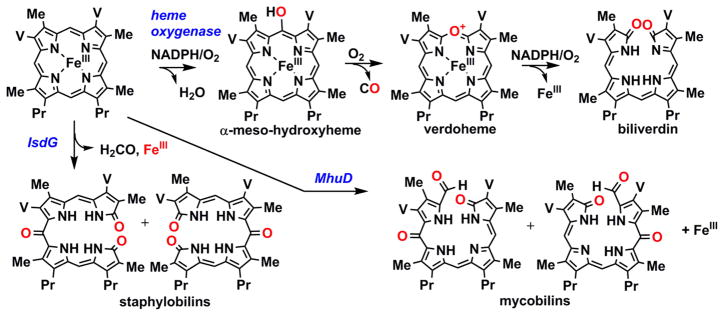
O2-derived oxygen atoms are shown in red. The heme side chains are abbreviated: Me = methyl; V = vinyl; Pr = propionate. Note that IsdG-family proteins yield products that are structural isomers, as shown. See references 73, 78, and 79.
Structurally, the IsdGs possess defining structural features that enable their heme-attacking chemistry. The most immediately noticeable of these is a heme that is strongly distorted from planarity, or ruffled. Ruffling is enforced by a tryptophan that is conserved family-wide. When it is mutated, the rate and extent of heme decomposition diminish sharply.100 Interestingly, the heme in OxdA, which is located in the same position in the pocket at IsdG’s but at a 180° rotation, is also somewhat ruffled (Figures 3F and 5E). Its structure shows a conserved tryptophan in the same position as IsdG’s (W172, Pseudomonas chlororaphis B23 OxdA numbering), though the residue’s role in that protein’s catalytic cycle has not been addressed. Like the other members of the superfamily, the IsdGs ligate the heme via a conserved histidine deriving from one of the alpha-helical strands of the pocket. The distal pocket is primarily hydrophobic, with a conserved asparagine lying close to the iron and serving an essential catalytic role. Notably, the well-described HOs have a network of water molecules in the distal pocket, which supply key hydrogen bonds and a conduit by which protons can move in and out of the active site.89 How IsdGs stabilize Fe/O2 intermediates without such a network, and how they convey molecules in/out of the pocket is not clear. Finally, many species possess not just one but two paralogous proteins from the IsdG family, differing from each other only subtly in sequence in structure. The major distinction between the two paralogs from S. aureus, which are called IsdG and IsdI, is found in a region called the “flexible loop.” This region is important for destabilizing the heme-free form of IsdG, but not IsdI. Hence, IsdI is understood to act as a workhorse enzyme, while IsdG has both sensor and enzymatic functions.101,102
Conclusions and perspectives
Discovering that one’s favorite molecule has a twin is not unlike the eerie experience of meeting a doppelgänger. We now see that the basic structural motif repeated many times over in Figure 2 reflects a common molecular heritage for the >20 families represented in Figure 1. From this common scaffold, at least a half dozen different heme-based chemistries have arisen. We have attempted here to identify important structural features essential for the functional diversity of this superfamily, whose story is just beginning to be told.
The CDE superfamily, is a diverse group of at least 20 protein families sharing a common α, β-barrel domain.
Of these, six different groups bind heme inside the barrel’s interior, using it alternately as a cofactor, substrate, or product.
At least two places in the pocket can be occupied by heme, in two distinct binding modes.
The active site residues distal to the heme in each of the six families are distinct.
At least one family, the HemQs, has an apparent structural means for heme entry and release.
Acknowledgments
Professor John Gerlt (University of Illinois Department of Biochemistry) and the Enzyme Function Initiative team (funded by National Institutes of Health grant U54GM093342-05) are gratefully acknowledged for providing training in the generation and manipulation of sequence similarity networks, for access to tools now publically available at the Enzyme Function Initiative website (enzymefunction.org), and for their generous assistance with the large sequence set required for generating Figure 1. We thank the National Institutes of Health for funding (R01GM090260) and Garrett Moraski for helpful conversations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maixner F, Wagner M, Luecker S, Pelletier E, Schmitz-Esser S, Hace K, Spieck E, Konrat R, Le Paslier D, Daims H. Environmental genomics reveals a functional chlorite dismutase in the nitrite-oxidizing bacterium ‘Candidatus Nitrospira defluvii’. Environmental microbiology. 2008;10:3043–3056. doi: 10.1111/j.1462-2920.2008.01646.x. [DOI] [PubMed] [Google Scholar]
- 2.Coates JD, Achenbach LA. Microbial perchlorate reduction: Rocket-fuelled metabolism. Nature Reviews Microbiology. 2004;2:569–580. doi: 10.1038/nrmicro926. [DOI] [PubMed] [Google Scholar]
- 3.Goblirsch B, Kurker RC, Streit BR, Wilmot CM, DuBois JL. Chlorite dismutases, DyPs, and EfeB: 3 microbial heme enzyme families comprise the CDE structural superfamily. Journal of Molecular Biology. 2011;408:379–398. doi: 10.1016/j.jmb.2011.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Geus DC, Thomassen EAJ, Hagedoorn PL, Pannu NS, van Duijn E, Abrahams JP. Crystal structure of chlorite dismutase, a detoxifying enzyme producing molecular oxygen. Journal of Molecular Biology. 2009;387:192–206. doi: 10.1016/j.jmb.2009.01.036. [DOI] [PubMed] [Google Scholar]
- 5.Goblirsch BR, Streit BR, DuBois JL, Wilmot CM. Structural features promoting dioxygen production by Dechloromonas aromatica chlorite dismutase. Journal of Biological Inorganic Chemistry. 2010;15:879–888. doi: 10.1007/s00775-010-0651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kostan J, Sjoeblom B, Maixner F, Mlynek G, Furtmueller PG, Obinger C, Wagner M, Daims H, Djinovic-Carugo K. Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”: Identification of a catalytically important amino acid residue. Journal of Structural Biology. 2010;172:331–342. doi: 10.1016/j.jsb.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Mlynek G, Sjoeblom B, Kostan J, Fuereder S, Maixner F, Gysel K, Furtmueller PG, Obinger C, Wagner M, Daims H, Djinovic-Carugo K. Unexpected diversity of chlorite dismutases: a catalytically efficient dimeric enzyme from Nitrobacter winogradsky. Journal of Bacteriology. 2011;193:2408–2417. doi: 10.1128/JB.01262-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugano Y, Muramatsu R, Ichiyanagi A, Sato T, Shoda M. DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family. Journal of Biological Chemistry. 2007;282:36652–36658. doi: 10.1074/jbc.M706996200. [DOI] [PubMed] [Google Scholar]
- 9.Cao J, Woodhall MR, Alvarez J, Cartron ML, Andrews SC. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E-coli O157: H7. Molecular Microbiology. 2007;65:857–875. doi: 10.1111/j.1365-2958.2007.05802.x. [DOI] [PubMed] [Google Scholar]
- 10.Letoffe S, Heuck G, Delepelaire P, Lange N, Wandersman C. Bacteria capture iron from heme by keeping tetrapyrrol skeleton intact. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11719–11724. doi: 10.1073/pnas.0903842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Du Q, Wang Z, Zhu D, Huang Y, Li N, Wei T, Xu S, Gu L. Crystal Structure and Biochemical Features of EfeB/YcdB from Escherichia coli O157 Asp(235) plays divergent roles in different enzyme-caltalyzed processes. Journal of Biological Chemistry. 2011;286:14922–14931. doi: 10.1074/jbc.M110.197780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miethke M, Monteferrante CG, Marahiel MA, van Dijl JM. The Bacillus subtilis EfeUOB transporter is essential for high-affinity acquisition of ferrous and ferric iron. Biochimica Et Biophysica Acta-Molecular Cell Research. 2013;1833:2267–2278. doi: 10.1016/j.bbamcr.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 13.Dailey HA, Septer AN, Daugherty L, Thames D, Gerdes S, Stabb EV, Dunn AK, Dailey TA, Phillips JD. The Escherichia coli protein YfeX functions as a porphyrinogen oxidase, not a heme dechelatase. Mbio. 2011;2:e00248–e00311. doi: 10.1128/mBio.00248-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dailey HA, Gerdes S, Dailey TA, Burch JS, Phillips J. Noncanonical coproporphyrin-dependent heme biosynthesis pathway that does not use protoporphyrin. Proc Natl Acad Sci. 2015;112:2210–2215. doi: 10.1073/pnas.1416285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farber G, Petsko G. The evolution of alpha-beta barrel enzymes. Trends in Biochemical Sciences. 1990;15(6):228–234. doi: 10.1016/0968-0004(90)90035-a. [DOI] [PubMed] [Google Scholar]
- 16.Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: Mechanistically diverse superfamilies and functionally distinct suprafamilies. Annual Review of Biochemistry. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 17.Chandonia JM, Brenner SE. The impact of structural genomics: Expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 18.Fox NK, Brenner SE, Chandonia JM. SCOPe: Structural classification of proteins-extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Research. 2014;42:D304–D309. doi: 10.1093/nar/gkt1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atkinson H, Morris J, Ferrin T, Babbitt P. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. Plos One. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyckoff EE, Mey AR, Leimbach A, Fisher CF, Payne SM. Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. Journal of Bacteriology. 2006;188:6515–6523. doi: 10.1128/JB.00626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zubieta C, Joseph R, Krishna SS, McMullan D, Kapoor M, Axelrod HL, Miller MD, Abdubek P, Acosta C, Astakhova T, Carlton D, Chiu HJ, Clayton T, Deller MC, Duan L, Elias Y, Elsliger MA, Feuerhelm J, Grzechnik SK, Hale J, Han GW, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kozbial P, Kumar A, Marciano D, Morse AT, Murphy KD, Nigoghossian E, Okach L, Oommachen S, Reyes R, Rife CL, Schimmel P, Trout CV, van den Bedem H, Weekes D, White A, Xu Q, Hodgson KO, Wooley J, Deacon AM, Godzik A, Lesley SA, Wilson IA. Identification and structural characterization of heme binding in a novel dye-decolorizing peroxidase, TyrA. Proteins-Structure Function and Bioinformatics. 2007;69:234–243. doi: 10.1002/prot.21673. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida T, Tsuge H, Hisabori T, Sugano Y. Crystal structures of dye-decolorizing peroxidase with ascorbic acid and 2,6-dimethoxyphenol. Febs Letters. 2012;586:4351–4356. doi: 10.1016/j.febslet.2012.10.049. [DOI] [PubMed] [Google Scholar]
- 24.Roberts JN, Singh R, Grigg JC, Murphy MEP, Bugg TDH, Eltis LD. Characterization of Dye-decolorizing peroxidases from Rhodococcus jostii RHA1. Biochemistry. 2011;50:5108–5119. doi: 10.1021/bi200427h. [DOI] [PubMed] [Google Scholar]
- 25.Singh R, Grigg JC, Armstrong Z, Murphy MEP, Eltis LD. Distal heme pocket residues of B-type Dye-decolorizing Peroxidase arginine but not aspartate is essential for peroxidase activity. Journal of Biological Chemistry. 2012;287:10623–10630. doi: 10.1074/jbc.M111.332171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strittmatter E, Liers C, Ullrich R, Wachter S, Hofrichter M, Plattner DA, Piontek K. First crystal structure of a fungal high-redox potential Dye-decolorizing Peroxidase substrate interaction sites and long range electron transfer. Journal of Biological Chemistry. 2013;288:4095–4102. doi: 10.1074/jbc.M112.400176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawai H, Sugimoto H, Kato Y, Asano Y, Shiro Y, Aono S. X-ray crystal structure of Michaelis complex of aldoxime dehydratase. Journal of Biological Chemistry. 2009;284:32089–32096. doi: 10.1074/jbc.M109.018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nomura J, Hashimoto H, Ohta T, Hashimoto Y, Wada K, Naruta Y, Oinuma K-i, Kobayashi M. Crystal structure of aldoxime dehydratase and its catalytic mechanism involved in carbon-nitrogen triple-bond synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2810–2815. doi: 10.1073/pnas.1200338110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zubieta C, Krishna SS, Kapoor M, Kozbial P, McMullan D, Axelrod HL, Miller MD, Abdubek P, Ambing E, Astakhova T, Carlton D, Chiu HJ, Clayton T, Deller MC, Duan L, Elsliger MA, Feuerhelm J, Grzechnik SK, Hale J, Hampton E, Han GW, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kumar A, Marciano D, Morse AT, Nigoghossian E, Okach L, Oommachen S, Reyes R, Rife CL, Schimmel P, van den Bedem H, Weekes D, White A, Xu Q, Hodgson KO, Wooley J, Deacon AM, Godzik A, Lesley SA, Wilson IA. Crystal structures of two novel dye-coloring peroxidases reveal a beta-barrel fold with a conserved heme-biding motif. Proteins. 2007;69:223–233. doi: 10.1002/prot.21550. [DOI] [PubMed] [Google Scholar]
- 30.Sugano Y, Yoshida T, Tsuge H. Dye-decolorizing peroxidase (DyP) complex with ascorbic acid. To be published. [Google Scholar]
- 31.Sugano Y, Yoshida T, Tsuge H. Dye-decolorizing peroxidase (DyP) complex with 2,6,-dimethoxyphenol. doi: 10.1016/j.febslet.2012.10.049. To be published. [DOI] [PubMed] [Google Scholar]
- 32.Bamford VA, Andrews SC, Watson KA. EfeB, the peroxidase component of the EfeUOB bacteria Fe(II) transport system also shows novel removal of iron from heme. To be published. [Google Scholar]
- 33.Lukk T, Hetta AMA, Jones A, Solbiati J, Majumdar S, Cronan JE, Gerlt JA, Nair SK. DyP-type peroxidases from Streptomyces and Thermobifida can modify organosolv lignin. To be published. [Google Scholar]
- 34.Tan K, Bigelow L, Bearden J, Joachimiak A. The crystal structure of a functionally unknown protein PEPE_1408 from Pediococcus pentosaceus ATCC 25745. To be published. [Google Scholar]
- 35.Chang C, Xu X, Savchnko A, Edwards A, Joachimiak A. Crystal structure of a putative chlorite dismutase TA0507. To be published. [Google Scholar]
- 36.Gilski M, Borek D, Chen Y, Collart F, Joachimiak A, Otwinowski Z. Crystal Structure of APC35880 protein of Bacillus Stearothermophilus. To be published. [Google Scholar]
- 37.Ebihara A, Okamoto A, Kousumi Y, Yamamoto H, Masui R, Ueyama N, Yokoyama S, Kuramitsu S. Structure-based functional identification of a novel heme-binding protein from Thermus thermophilus HB8. J Struct Funct Genomics. 2005;6:21–32. doi: 10.1007/s10969-005-1103-x. [DOI] [PubMed] [Google Scholar]
- 38.Hofbauer S, Gysel K, Bellei M, Hagmuller A, Schaffner I, Mlynek G, Kostan J, Pirker KF, Daims H, Furtmuller PG, Battistuzzi G, Dijnovic-Carugo K, Obinger C. Manipulatng conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity. Biochemistry. 2014;53:77–89. doi: 10.1021/bi401042z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofbauer S, Hagmuller A, Schaffner I, Mlynek G, Krutzler M, Stadlmayr G, Pirker KF, Obinger C, Daims H, Dijnovic-Carugo K, Furtmuller PG. Structure and heme-binding properties of HemQ (chlorite dismutase-like protein) from Listeria monocytogenes. Archives of Biochemistry and Biophysics. 2015 doi: 10.1016/j.abb.2015.01.010. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linde D, Pogni R, Cañellas M, Lucas F, Guallar V, Baratto MC, Sinicropi A, Sáez-Jiménez V, Coscolín C, Romero A, Medrano FJ, Ruiz-Dueñas FJ, Martínez AT. Catalytic surface radical in dye-decolorizing peroxidase: a computational, spectroscopic and site-directed mutagenesis study. Biochemical Journal. 2015;466:253–262. doi: 10.1042/BJ20141211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chim N, Iniguez A, Nguyen TQ, Goulding CW. Unusual diheme conformation of the heme-degrading protein from Mycobacterium tuberculosis. Journal of Molecular Biology. 2010;395:595–608. doi: 10.1016/j.jmb.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee WC, Reniere ML, Skaar EP, Murphy MEP. Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. Journal of Biological Chemistry. 2008;283:30957–30963. doi: 10.1074/jbc.M709486200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu R, Skaar EP, Zhang R, Joachimiak G, Gornicki P, Schneewind O, Joachimiak A. Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases. Molecular Microbiology. 2010;75:1529–1538. doi: 10.1074/jbc.M409526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graves AB, Morse RP, Chao A, Iniguez A, Goulding CW, Liptak MD. Crystallographic and spectroscopic insights into heme degradation by Mycobacterium tuberculosis MhuD. Inorganic Chemistry. 2014;53:5931–5940. doi: 10.1021/ic500033b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reniere ML, Ukpabi GN, Harry SR, Stec DF, Krull R, Wright DW, Bachmann BO, Murphy ME, Skaar EP. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Molecular Microbiology. 2010;75:1529–1538. doi: 10.1111/j.1365-2958.2010.07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katti S, Katz B, Wyckoff H. Crystal structure of muconolactone isomerase at 3.3A resolution. Journal of Molecular Biology. 1989;205:557–571. doi: 10.1016/0022-2836(89)90226-x. [DOI] [PubMed] [Google Scholar]
- 47.Ferraroni M, Kolomytseva M, Golovleva L, Scozzafava A. X-ray crystallographic and molecular docking studies on a unique chloromuconolactone dehalogenase from Rhodococcus opacus 1CP. Journal of Structural Biology. 2013;182:44–50. doi: 10.1016/j.jsb.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 48.Veith A, Urich T, Seyfarth K, Protze J, Frazao C, Kletzin A. Substrate pathways and mechanisms of inhibition in the sulfur oxygenase reductase of Acidianus ambivalens. Frontiers in Microbiology. 2011;2:37. doi: 10.3389/fmicb.2011.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marion GM, Catling DC, Zahnle KJ, Claire MW. Modeling aqueous perchlorate chemistries with applications to Mars. Icarus. 2010;207:675–685. [Google Scholar]
- 50.Rao B, Anderson TA, Redder A, Jackson WA. Perchlorate Formation by Ozone Oxidation of Aqueous Chlorine/Oxy-Chlorine Species: Role of ClxOy Radicals. Environmental Science and Technology. 2010;44:2961–2967. doi: 10.1021/es903065f. [DOI] [PubMed] [Google Scholar]
- 51.Coates JD, Michaelidou U, Bruce RA, O’Connor SM, Crespi JN, Achenbach LA. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Applied and Environmental Microbiology. 1999;65:5234–5241. doi: 10.1128/aem.65.12.5234-5241.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bender KS, Rice MR, Fugate WH, Coates JD, Achenbach LA. Metabolic primers for detection of (Per) chlorate-reducing bacteria in the environment and phylogenetic analysis of cld gene sequences. Applied and Environmental Microbiology. 2004;70:5651–5658. doi: 10.1128/AEM.70.9.5651-5658.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danielsson H, Stenklo TK, Karlsson J, Nilsson T. A gene cluster for chlorate metabolism in Ideonella dechloratans. Applied and Environmental Microbiology. 2003;69:5585–5592. doi: 10.1128/AEM.69.9.5585-5592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Streit BR, Blanc B, Lukat-Rodgers GS, Rodgers KR, DuBois JL. How active-site protonation state influences the reactivity and ligation of the heme in chlorite dismutase. Journal of the American Chemical Society. 2010;132:5711–5724. doi: 10.1021/ja9082182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teraoka J, Kitagawa T. Structural implication of the heme linked ionization of horseradish peroxidase probed by the Fe-histidine stretching Raman line. Journal of Biological Chemistry. 1981;256(8):3969–3977. [PubMed] [Google Scholar]
- 56.Blanc B, Rodgers KR, Lukat-Rodgers GS, DuBois JL. Understanding the roles of strictly conserved tryptophan residues in O-2 producing chlorite dismutases. Dalton Transactions. 2013;42:3156–3169. doi: 10.1039/c2dt32312e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayfield JA, Blanc B, Rodgers KR, Lukat-Rodgers GS, DuBois JL. Peroxidase-type reactions suggest a heterolytic/nucleophilic O-O joining mechanism in the heme-dependent chlorite dismutase. Biochemistry. 2013;52:6982–6994. doi: 10.1021/bi4005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.(a) Hofbauer S, Gysel K, Mlynek G, Kostan J, Hagmueller A, Daims H, Furtmueller PG, Djinovic-Carugo K, Obinger C. Impact of subunit and oligomeric structure on the thermal and conformational stability of chlorite dismutases. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2012;1824:1031–1038. doi: 10.1016/j.bbapap.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Clark IC, Melnyk RA, Engelbrektson A, Coates JD. Structure and evolution of chlorate reduction composite transposons. MBio. 2013;4:e00379–13. doi: 10.1128/mBio.00379-13. (b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stewart V. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiology Reviews. 1988;52:190–232. doi: 10.1128/mr.52.2.190-232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Celis AI, Geeraertz Z, Ngmenterebo D, Machovina MM, Kurker RC, Rajakumar K, Ivancich A, Rodgers KR, Lukat-Rodgers GS, DuBois JL. A dimeric chlorite dismutase exhibits O2-generating activity and acts as a chlorite antioxidant in Klebsiella pneumoniae MGH78578. Biochemistry. 2015;54:434–446. doi: 10.1021/bi501184c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rao B, Hatzinger P, Bohlke J, Sturchio N, Andraski B, Eckardt F, Jackson W. Natural chlorate in the environment: Application of a new IC-ESI/MS/MS method with a (ClO3-)-O-18 internal standard. Environmental Science & Technology. 2010;44:8429–8434. doi: 10.1021/es1024228. [DOI] [PubMed] [Google Scholar]
- 62.Sugano Y, Sasaki K, Shoda M. cDNA cloning and genetic analysis of a novel decolorizing enzyme, peroxidase gene dyp from Geotrichum candidum Dec1. Journal of Bioscience and Bioengineering. 1999;87:411–417. doi: 10.1016/s1389-1723(99)80087-5. [DOI] [PubMed] [Google Scholar]
- 63.Sugano Y, Nakano R, Sasaki K, Shoda M. Efficient heterologous expression in Aspergillus oryzae of a unique dye-decolorizing peroxidase, DyP, of Geotrichum candidum Dec1. Applied and Environmental Microbiology. 2000;66:1754–1758. doi: 10.1128/aem.66.4.1754-1758.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim SJ, Shoda M. Purification and characterization of a novel peroxidase from Geotrichum candidum Dec 1 involved in decolorization of dyes. Applied and Environmental Microbiology. 1999;65:1029–1035. doi: 10.1128/aem.65.3.1029-1035.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh R, Grigg JC, Qin W, Kadla JF, Murphy MEP, Eltis LD. Improved manganese-oxidizing activity of DypB, a peroxidase from a lignolytic bacterium. ACS Chemical Biology. 2013;8:700–706. doi: 10.1021/cb300608x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rahmanpour R, Bugg TD. Characterisation of Dyp-type peroxidases from Pseudomonas fluorescens Pf-5: Oxidation of Mn(II) and polymeric lignin by Dyp1B. Archives of Biochemistry and Biophysics. 2015 doi: 10.1016/j.abb.2014.12.022. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 67.Sugano Y, Matsushima Y, Shoda M. Complete decolorization of the anthraquinone dye Reactive blue 5 by the concerted action of two peroxidases from Thanatephorus cucumeris Dec 1. Applied Microbiology and Biotechnology. 2006;73:862–871. doi: 10.1007/s00253-006-0545-9. [DOI] [PubMed] [Google Scholar]
- 68.Ahmad M, Roberts JN, Hardiman EM, Singh R, Eltis LD, Bugg TDH. Identification of DypB from Rhodococcus jostii RHA1 as a Lignin Peroxidase. Biochemistry. 2011;50:5096–5107. doi: 10.1021/bi101892z. [DOI] [PubMed] [Google Scholar]
- 69.Ruiz-Duenas FJ, Lundell T, Floudas D, Nagy LG, Barrasa JM, Hibbett DS, Martinez AT. Lignin-degrading peroxidases in Polyporales: an evolutionary survey based on 10 sequenced genomes. Mycologia. 2013;105:1428–1444. doi: 10.3852/13-059. [DOI] [PubMed] [Google Scholar]
- 70.Rajasekaran MB, Nilapwar S, Andrews SC, Watson KA. EfeO-cupredoxins: major new members of the cupredoxin superfamily with roles in bacterial iron transport. Biometals. 2010;23:1–17. doi: 10.1007/s10534-009-9262-z. [DOI] [PubMed] [Google Scholar]
- 71.Sturm A, Schierhorn A, Lindenstrauss U, Lilie H, Brüser T. YcdB from Escherichia coli reveals a novel class of Tat-dependently translocated hemoproteins. Journal of Biological Chemistry. 2006;281:13972–13978. doi: 10.1074/jbc.M511891200. [DOI] [PubMed] [Google Scholar]
- 72.Turlin E, Debarbouille M, Augustyniak K, Gilles AM, Wandersman C. Staphylococcus aureus FepA and FepB proteins drive heme iron utilization in Escherichia coli. Plos One. 2013;8:e56529. doi: 10.1371/journal.pone.0056529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dunford HB. Heme Peroxidases. Wiley-VCH; New York: 1999. [Google Scholar]
- 74.Fürtmuller PG, Zederbauer M, Jantschko W, Helm J, Bogner M, Jakopitsch C, Obinger C. Active site structure and catalytic mechanisms of human peroxidases. Archives of Biochemistry and Biophysics. 2006;445:199–213. doi: 10.1016/j.abb.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 75.Poulos TL, Fenna RE. Peroxidases - Structure, function, and engineering. Metal Ions In Biological Systems. 1994;30:25–75. [Google Scholar]
- 76.Strittmatter E, Serrer K, Ullrich R, Hofrichter M, Piontek K, Schleicher E, Plattner DA. The toolbox of Auricularia auricula-judae dye-decolorizing peroxidase - Identification of three new potential substrate-interaction sites. Arch Biochem Biophys. 2014;S0003–9861:00432–00439. doi: 10.1016/j.abb.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 77.Erman JE, Vitello LB, Miller MA, Shaw A, Brown KA, Kraut J. Histidine-52 is a critical residue for rapid formation of cytochrome-C peroxidase compound-I. Biochemistry. 1993;32:9798–9806. doi: 10.1021/bi00088a035. [DOI] [PubMed] [Google Scholar]
- 78.Yoshida T, Tsuge H, Konno H, Hisabori T, Sugano Y. The catalytic mechanism of Dye-decolorizing Peroxidase DyP may require the swinging movement of an aspartic acid residue. FEBS J. 2011;278:2387–2394. doi: 10.1111/j.1742-4658.2011.08161.x. [DOI] [PubMed] [Google Scholar]
- 79.Pinakoulaki E, Koutsoupakis C, Sawai H, Pavlou A, Kato Y, Asano Y, Aono S. Aldoxime dehydratase: Probing the heme environment involved in the synthesis of the carbon-nitrogen triple bond. Journal of Physical Chemistry B. 2011;115:13012–13018. doi: 10.1021/jp205944e. [DOI] [PubMed] [Google Scholar]
- 80.Streit BR, DuBois JL. Chemical and steady-state kinetic analyses of a heterologously expressed heme dependent chlorite dismutase. Biochemistry. 2008;47:5271–5280. doi: 10.1021/bi800163x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mayfield JA, Hammer ND, Kurker RC, Chen TK, Ojha S, Skaar EP, DuBois JL. The chlorite dismutase (HemQ) from Staphylococcus aureus has a redox-sensitive heme and is associated with the small colony variant phenotype. The Journal of Biological Chemistry. 2013;288:23488–23504. doi: 10.1074/jbc.M112.442335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dailey TA, Boynton TO, Albetel AN, Gerdes S, Johnson MK, Dailey HA. Discovery and characterization of HemQ: An essential heme biosynthetic pathway component. Journal of Biological Chemistry. 2010;285:25978–25986. doi: 10.1074/jbc.M110.142604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.(a) Kuhner M, Haufschildt K, Neumann A, Storbeck S, Streif J, Layer G. The alternative route to heme in the methanogenic archaeaon Methanosarcina barkeri. Archaea. 2014:327637. doi: 10.1155/2014/327637. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bali S, Lawrence AD, Lobo SA, Saraiva LM, Golding BT, Palmer DJ, Howard MJ, Ferguson SJ, Warren MJ. Molecular hijacking of siroheme for the synthesis of heme and d(1) heme. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18260–18265. doi: 10.1073/pnas.1108228108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blanc B, Mayfield JA, McDonald CA, Lukat-Rodgers GS, Rodgers KR, DuBois JL. Understanding how the distal environment directs reactivity in chlorite dismutase: Spectroscopy and reactivity of Arg183 mutants. Biochemistry. 2012;51:1895–1910. doi: 10.1021/bi2017377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fetzner S, Steiner R. Cofactor-independent oxidases and oxygenases. Applied Microbiology and Biotechnology. 2010;86:791–804. doi: 10.1007/s00253-010-2455-0. [DOI] [PubMed] [Google Scholar]
- 86.Grocholski T, Koskiniemi H, Lindqvist Y, Mäntsälä P, Niemi J, Schneider G. Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism. Biochemistry. 2010;49:934–944. doi: 10.1021/bi901985b. [DOI] [PubMed] [Google Scholar]
- 87.Sciara G, Kendrew SG, Miele AE, Marsh NG, Federici L, Malatesta F, Schimperna G, Savino C, Vallone B. The structure of ActVA-Orf6: a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO Journal. 2003;22:205–215. doi: 10.1093/emboj/cdg031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wilks A, Heinzl G. Heme oxygenation and the widening paradigm of heme degradation. Archives of Biochemistry and Biophysics. 2014;544:87–95. doi: 10.1016/j.abb.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.De Montellano PRO, Wilks A. Heme oxygenase structure and mechanism. Advances in Inorganic Chemistry. 2001;5151:359–407. [Google Scholar]
- 90.Skaar EP, Gaspar AH, Schneewind O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. Journal of Biological Chemistry. 2004;279:436–443. doi: 10.1074/jbc.M307952200. [DOI] [PubMed] [Google Scholar]
- 91.Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annual Review of Microbiology. 2011;65:129–147. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sigman J, Wang X, Lu Y. Coupled oxidation of heme by myoglobin is mediated by exogenous peroxide. Journal of the American Chemical Society. 2001;123:6945–6946. doi: 10.1021/ja015776u. [DOI] [PubMed] [Google Scholar]
- 93.Ocarra P, Colleran E. Coupled oxidation of myoglobin with ascorbate as a model of haem breakdown in vivo. Biochemical Journal. 1969;115:13–14. doi: 10.1042/bj1150013pb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ocarra P, Colleran E. Methine-bridge specificity of coupled oxidation of myoglobin and haemoglobin with ascorbate. Biochemical Journal. 1970;119:42P–43P. doi: 10.1042/bj1190042p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Matsui T, Nambu S, Ono Y, Goulding CW, Tsurnoto K, Ikeda-Saito M. Heme degradation by Staphylococcus aureus IsdG and IsdI liberates formaldehyde rather than carbon monoxide. Biochemistry. 2013;52:3025–3027. doi: 10.1021/bi400382p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nambu S, Matsui T, Goulding CW, Takahashi S, Ikeda-Saito M. A New Way to Degrade Heme: the Mycobacterium tuberculosis enzyme MhuD catalyzes heme degradation without generating CO. Journal of Biological Chemistry. 2013;288:10101–10109. doi: 10.1074/jbc.M112.448399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ortiz de Montellano PR. Heme Oxygenase mechanism: Evidence for an electrophilic, ferric peroxidase species. Accounts of Chemical Research. 1998;31:543–549. [Google Scholar]
- 98.Matsui T, Iwasaki M, Sugiyama R, Unno M, Ikeda-Saito M. Dioxygen activation for the self-degradation of heme: reaction mechanism and regulation of heme oxygenase. Inorg Chem. 2010;49:3602–3609. doi: 10.1021/ic901869t. [DOI] [PubMed] [Google Scholar]
- 99.Liu Y, Moënne-Loccoz P, Loehr TM, Ortiz de Montellano PR. Heme oxygenase-1: intermediates in verdoheme formation and the requirement for reduction equivalents. J Biol Chem. 1997;272:6909–6917. doi: 10.1074/jbc.272.11.6909. [DOI] [PubMed] [Google Scholar]
- 100.Ukpabi G, Takayama S-iJ, Mauk AG, Murphy MEP. Inactivation of the Heme degrading enzyme IsdI by an active site substitution that diminishes heme ruffling. Journal of Biological Chemistry. 2012;287:34179–34188. doi: 10.1074/jbc.M112.393249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reniere ML, Skaar EP. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Molecular Microbiology. 2008;69:1304–1315. doi: 10.1111/j.1365-2958.2008.06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reniere ML, Haley KP, Skaar EP. The flexible loop of Staphylococcus aureus IsdG is required for its degradation in the absence of heme. Biochemistry. 2011;50:6730–6737. doi: 10.1021/bi200999q. [DOI] [PMC free article] [PubMed] [Google Scholar]