Abstract
To identify chromatin mechanisms of neuronal differentiation, we characterized chromatin accessibility and gene expression in cerebellar granule neurons (CGNs) of the developing mouse. We used DNase-seq to map accessibility of cis-regulatory elements and RNA-seq to profile transcript abundance across postnatal stages of neuronal differentiation in vivo and in culture. We observed thousands of chromatin accessibility changes as CGNs differentiated and verified by H3K27ac ChIP-seq, reporter gene assays, and CRISPR-mediated activation that many of these regions function as neuronal enhancers. Motif discovery within differentially accessible chromatin regions suggested a novel role for the Zic family of transcription factors in CGN maturation. We confirmed the association of Zic with these elements by ChIP-seq, and demonstrated by knockdown that Zic1/2 are required to coordinate mature neuronal gene expression patterns. Together these data reveal chromatin dynamics at thousands of gene regulatory elements that facilitate gene expression patterns necessary for neuronal differentiation and function.
Introduction
Specialized cell function requires precise control of gene expression patterns. Chromatin regulation plays a part in this process by establishing differential utilization of gene regulatory elements in cells of distinct fate lineages. Genome-wide studies indicate that cell-type specific differences in gene expression are highly correlated with differences in both accessibility and activation state of distal gene enhancers1,2. These data have led to the hypothesis that developmental regulation of chromatin at enhancer elements mediates the process of cellular differentiation3.
Neuronal differentiation is comprised of multiple steps, beginning with the commitment of neural stem cells to become specified neural progenitors, which then leave the cell cycle to become postmitotic neurons. Prenatal patterning of the brain is critically dependent on temporally and spatially-restricted expression of transcription factors that act at brain region-selective enhancer elements4,5. By contrast after birth sensory experience-driven synaptic activity plays an instructive role in initiating programs of gene expression that underlie neuronal maturation. This allows processes like synapse development and excitatory/inhibitory balance in neural circuits to be adapted to the environment6. However, whether chromatin-dependent regulation of enhancer function contributes to gene expression that mediates these later stages of neuronal maturation remains largely unknown.
To fill this gap in knowledge, we used the differentiation of cerebellar granule neurons (CGNs) in the postnatal mouse cerebellum to identify chromatin-based transcriptional mechanisms that drive the maturation of neuronal gene expression programs. CGNs, which comprise >99% of cerebellar neurons, are derived during early postnatal life from committed granule neuron precursors (GNPs) that proliferate in the outer portion of the external granular layer of the developing cerebellar cortex7. Upon exit from the cell cycle, GNPs differentiate into immature CGNs that migrate to the inner granular layer, where they form synaptic connections and then mature. These changes in CGN differentiation and function are accompanied by known changes in neuronal gene expression8. Importantly, primary GNPs isolated from the postnatal mouse brain recapitulate discrete and synchronized stages of CGN differentiation in culture, providing a means for experimental validation and genetic manipulation of gene regulatory mechanisms that mediate this process9,10.
DNase I hypersensitive (DHS) sites mark nucleosome-depleted regions that are universal hallmarks for gene regulatory elements, including promoters, enhancers, insulators, and most transcription factor binding sites11. We applied DNase-seq to globally map chromatin accessibility at three key stages in the development of mouse cerebellum. We report that widespread changes in chromatin accessibility occur as CGNs differentiate, marking dynamic enhancer elements that regulate the expression of genes necessary for proper neuronal function. Additionally, we used our identification of these regions to describe a novel role for the Zic transcription factors in coordinating gene expression programs required for neuronal maturation.
Results
Cerebellar development involves extensive chromatin remodeling
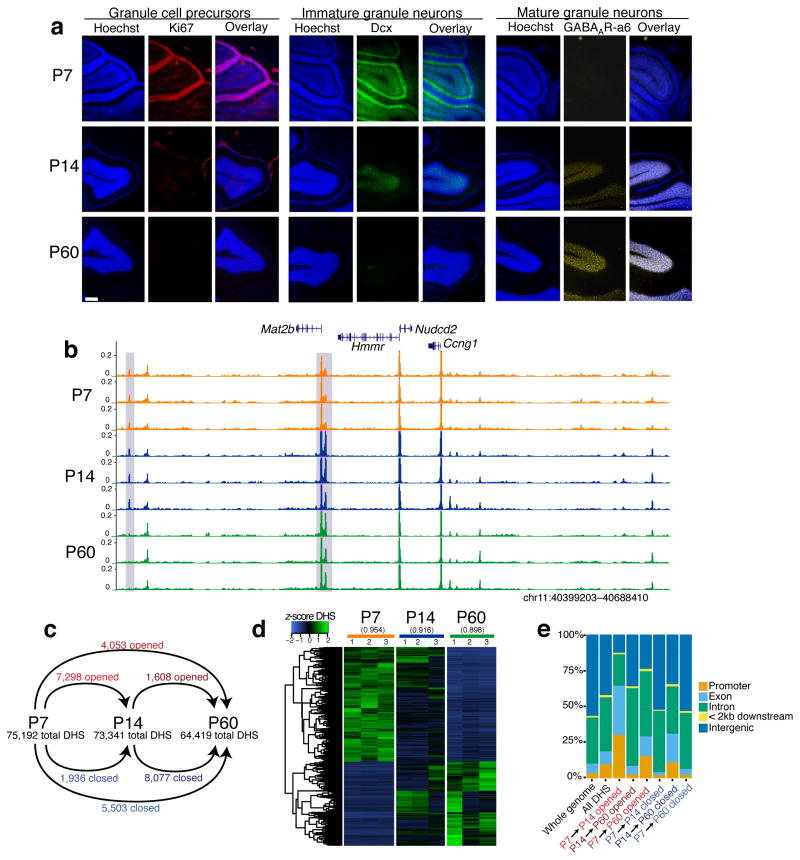
We used DNase-seq to globally map chromatin accessibility at three key stages in the development of the mouse cerebellum: 1) postnatal day 7 (P7), when the external granular layer (EGL) of the mouse cerebellar cortex has reached its maximal thickness12 due to the proliferation of granule neuron precursors (GNPs) (Fig. 1a); 2) P14, when newborn postmitotic CGNs begin to populate the internal granule layer (IGL)13; and 3) P60, when CGNs of the IGL express gene products that mediate mature synaptic functions8 (Fig. 1b, Supplementary Fig. 1a, Supplementary Table 1). We identified approximately 70,000 DHS sites at each of the three developmental stages, and found that these elements mapped to promoters, gene bodies, and intergenic regions (Supplementary Fig. 1b). The majority of DHS sites (~77%) are found outside of annotated gene promoter regions, highlighting the ability of DNase-seq to identify distal regulatory elements in the genome.
Figure 1. Postnatal cerebellar development involves extensive chromatin remodeling.
(a) Immunostaining of cerebellar cortex in sagittal section at postnatal day 7 (P7), 14 (P14), and 60 (P60). Ki67 marks active cell proliferation, doublecortin (Dcx) marks immature/migrating neurons, and GABAA receptor subunit α6 (GABAARα6) marks mature neurons. Nuclei stained with Hoechst (blue). White scale bar is 200 μm. This experiment was performed once. (b) Example region of mm9 genome showing reproducible DNase-seq chromatin accessibility profiles across biological triplicates of P7, P14, and P60 cerebella. Gray boxes denote DHS sites that change between developmental stages. Y-axes fixed across samples. (c) Number of total peak calls and significantly opened or closed DHS sites between P7, P14, and P60 cerebella pairwise comparisons (FDR < 0.05, n = 3 biological replicates of pooled cerebella). (d) Heatmap of DNase-seq accessibility across three timepoints for regions marked significant in DESeq analysis. Mean pairwise Pearson correlation between replicates listed above heatmap. (e) Distribution of gene features that opened, closed, and all identified DHS sites map to.
To determine how chromatin accessibility changes during cerebellar differentiation, we assessed quantitative differences in DHS signal intensity between the three stages. These analyses identified 24,886 total DHS sites that significantly “opened” or “closed” (gained or lost signal) during postnatal development (FDR < 0.05) (Fig. 1c,d; Supplementary Fig. 1c,d, Supplementary Table 2). Similar to all DHS sites, developmentally regulated DHS sites were found within both proximal gene promoters and at intronic, exonic, and intergenic regions (Fig. 1e). Interestingly, we find evidence of extensive chromatin remodeling between P14 and P60, even though these samples are primarily comprised of CGNs at two postmitotic stages of neuronal differentiation. These data indicate there is extensive chromatin remodeling over the course of neuronal differentiation in vivo both as proliferating neural progenitors leave the cell cycle and as postmitotic neurons mature.
Although an advantage of cerebellar cortex is that it is predominantly composed of granule neurons (~85%), this brain region also contains astrocytes (~15%) and other kinds of neurons such as Purkinje Cells (~0.2%)7 (Supplementary Fig. 2a,b). To determine the impact of these other cell types on our chromatin analyses, we first quantified aggregate DNase-seq signal at promoters of genes that are preferentially expressed in either CGNs, Purkinje neurons, or Bergmann glia of the adult mouse cerebellum14. DNase signal was greatest at promoters of genes preferentially expressed in adult CGNs, suggesting our DHS data represent chromatin state in this most abundant cell type (Supplementary Fig. 2c). Furthermore, when we considered the set of developmentally regulated DHS sites, these also had the strongest association with CGN enriched genes (Supplementary Fig. 2d). ChIP-seq datasets obtained from mouse cerebellum were similarly enriched at CGN gene promoters over other cell types (Supplementary Fig. 2e,f).
As an additional way to control for cellular heterogeneity, we purified GNPs from the P7 cerebellum and differentiated these cells to CGNs in culture. These cultures are highly enriched for CGNs and depleted ofglia and other kinds of neurons found in the cerebellar cortex in vivo15. Once plated, freshly dissociated GNPs rapidly exit the cell cycle16 and synchronously differentiate, displaying characteristic neuronal morphologies by 3 days in vitro (+3DIV) and forming synaptic connections by +7DIV17 (Supplementary Fig. 3a). We performed DNase-seq in triplicate on freshly isolated GNPs or GNPs cultured for 3 or 7 days, and we identified 28,119 differential DHS sites at FDR < 0.05 (Supplementary Fig. 3b–d, Supplementary Table 3). Similar to our findings from in vivo cerebellar samples, we found that opening and closing DHS sites in cultured CGNs are primarily non-promoter distal elements (Supplementary Fig. 3e). While the majority (10,115/11,657 = 86.8%) of changes in DHS site accessibility occurred as GNPs left the cell cycle to become newborn CGNs, we also observed 1,542 (13.2%) DHS site changes that occur between postmitotic +3 and +7DIV samples. These data again indicate that postmitotic neurons can undergo substantial chromatin accessibility changes as they establish synapses and mature.
Comparing developmental regulation of DHS sites between the in vivo and culture systems revealed most sites exhibit matched direction of change (Supplementary Fig. 4a), but the magnitude of accessibility changes was generally lower in the cultured neurons. However we also observed a cluster of DHS sites that closed gradually from P7 to P60 in vivo but became transiently more accessible at +3DIV before closing down at +7DIV in culture. Regulation of at least a subset of these sites accompanies transient increases in the expression of nearby genes. For example, Grin2b is a developmentally-regulated NMDA-type glutamate receptor subunit that is most highly expressed in the neonatal brain, where it plays a key role in the induction of long-term potentiation18. Two transiently opening DHS sites in an intron of the Grin2b gene parallel the transient increase in Grin2b mRNA expression at +3DIV in cultured CGNs (Supplementary Fig. 4b,c). These data suggest that in addition to reducing cellular heterogeneity, this culture system complements our in vivo differentiation analysis by improving temporal resolution of chromatin regulation during early steps in CGN differentiation.
Opening DHS sites mark mature CGN enhancers
To characterize the relationship between chromatin accessibility and regulation of gene expression in the developing cerebellum, we performed RNA-seq in triplicate from P7, P14, and P60 cerebellar cortex. Comparisons of gene expression levels between each stage confirmed robust transcriptional changes for 5,714 genes (FDR < 0.05) during postnatal cerebellar development (Supplementary Fig. 5a,b, Supplementary Table 4). By clustering patterns of differentially expressed genes, we confirmed a number of previously described granule neuron gene expression dynamics and recovered gene ontologies descriptive of the differentiation process19–22 (Supplementary Fig. 5c–g, Supplementary Table 5). As with our DNase-seq analysis we also performed RNA-seq on triplicate replicates of purified GNPs that were either freshly isolated or differentiated in culture for +3 or +7DIV. We found 1,972 differentially expressed genes in the cultures across this time course (Supplementary Fig. 6a), and principal components analysis of RNA-seq expression values placed +3 and +7DIV samples in between GNPs and P14 cerebellum along an axis consistent with developmental time (Supplementary Fig. 6b).
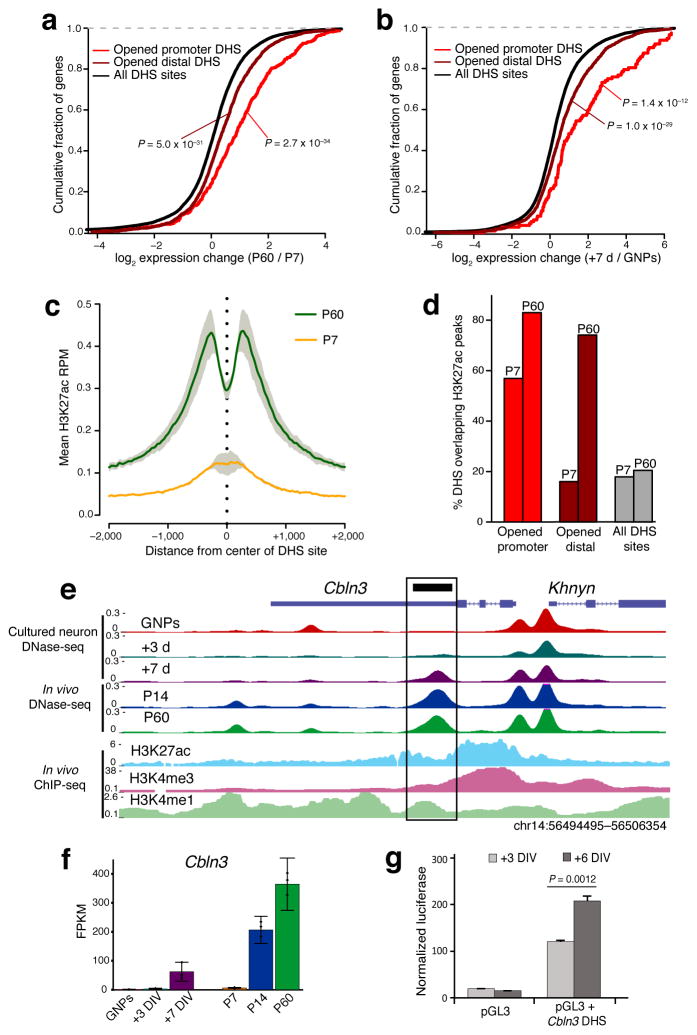
We postulated that the developmentally regulated chromatin accessibility changes we observed might mark a combination of promoter and enhancer elements that become activated at specific developmental stages to drive target gene expression. We first mapped each DHS site to its nearest gene and analyzed the relationship between that DHS site and the change in its associated gene expression value between P7 and P60. Whereas all DHS sites together exhibit a normal distribution of nearby gene expression changes centered on a fold-change of 1, both promoter and distal DHS sites that open up between P7 and P60 were significantly associated with genes that have higher expression levels at P60 (P= 2.7×10−34 and P = 5.0×10−31, respectively, Mann-Whitney test) (Fig. 2a). Similar to our in vivo data, we found a strong association between GNP to +7DIV opening DHS sites and increased nearby gene expression in cultured CGNs (Fig. 2b).
Figure 2. Opening DHS sites mark late-acting neuronal enhancers.
(a,b) Cumulative fraction of genes nearest to opened promoter-located, opened distal, and all identified DHS sites with given fold-change in RNA-seq expression from P7 to P60 cerebellum (a) or from GNPs to +7DIV (b). Rightward shift indicates genes increased in expression across developmental time. Significance assessed by Mann-Whitney U test. (n = 3 biological replicates of pooled cerebella). (c) Mean H3K27ac ChIP-seq signal (reads per million mapped) present at center of P7 to P60 opened DHS sites (4053 sites) in either P7 cerebellum (orange line) or P60 cerebellum (green line). Gray = s.e.m.. (n = 2 biological replicates of pooled cerebella). (d) Percent of opened promoter-located, opened distal, and all DHS sites overlapping H3K27ac peaks identified in P7 or P60 cerebellum. (e) UCSC browser image highlighting a DHS site found in the 3′UTR of Cbln3 (black box) that opens during development and overlaps H3K27ac and H3K4me1 enrichment in adult cerebellum. (f) RNA-seq expression of Cbln3 across developmental time in vivo and in cultured CGNs. DIV = Days In Vitro. Error bars = 95% CI. (g) Luciferase reporter assay for enhancer activity conferred by a DHS site in Cbln3 at +3DIV and +6DIV. P = 0.0012 by two-sided Student’s t-test (n = 3 transfections, t = 8.3). Error bars = s.e.m.
The functional identity of DHS sites (e.g. enhancers, promoters, and silencers) can be predicted by overlap with specific histone modifications23,24. With the knowledge that most DHS sites are located outside of proximal promoter regions, and that our opening sites correlated with increased gene expression, we asked if these sites colocalized with the enhancer mark H3K4me1 and the active enhancer/promoter modification H3K27ac. Comparing our developmentally-regulated DHS sites to available mouse ENCODE ChIP-seq data from P56 cerebellum25 showed an enriched overlap between our opened DHS sites and the H3K4me1 (~77%, P = 1.13×10−120, hypergeometric test) and H3K27ac (~50%, P = 8.82×10−88) histone marks (Supplementary Fig. 7a). The combination of these two histone marks was particularly enriched in non-promoter opening DHS sites (~45% overlap peaks of both marks), whereas promoter-located opening DHS sites were enriched for H3K27ac in combination with H3K4me3 (~40% overlap peaks of both marks). This indicates that DHS sites that open between P7 and P60 demarcate both active enhancers and promoters in adult cerebellum (Supplementary Fig. 7a). To determine whether the opening of DHS sites over developmental time was associated with changing of histone marks, we performed ChIP-seq for H3K27ac in P7 and P60 cerebellum in duplicate. We found a strong correlation (Pearson’s r = 0.538) between global H3K27ac and DHS signal changes in the developing cerebellum, indicating increased accessibility is associated with concomitant H3K27ac deposition (Supplementary Fig. 7b,c). Separating out H3K27ac peaks that map to proximal promoters further revealed that the global correlation is driven primarily by non-promoter sites (r = 0.278 promoters vs. r = 0.621 non-promoters) (Supplementary Fig 7d,e). Matching these observations, P7 to P60 opened DHS sites contained mean H3K27ac signal that increased dramatically over development (Fig. 2c). Furthermore, the set of opening DHS sites was markedly enriched for overlap with H3K27ac peaks compared to all DHS sites at P60 (Fig. 2d). Together these data suggest that increased chromatin accessibility is associated with the activation of enhancer elements that promote developmentally regulated increases in CGN gene expression.
To determine if the opening DHS sites indeed function as enhancers of mature CGN gene expression, we first used luciferase assays to test the function of a DHS site that becomes significantly more accessible both in vivo and in culture found in the 3′UTR of the cerebellin 3 precursor (Cbln3) gene, which encodes a secreted C1q-domain protein that modulates the formation, refinement, and maintenance of CGN to Purkinje cell synapses26,27 (Fig. 2e). This DHS site colocalizes with the enhancer histone modifications H3K27ac and H3K4me1 in adult cerebellum and correlates with increased Cbln3 expression over the course of CGN differentiation both in culture and in vivo (Fig. 2e,f). When cloned in front of a minimal promoter and transfected in CGNs, this element significantly increased luciferase expression (Fig. 2g). Furthermore, we found substantially more luciferase expression when we harvested neurons on +6DIV compared with +3DIV (P= 0.0012, Student’s t-test) (Fig. 2g), indicating that the activity of this enhancer increases with developmental maturation of the CGNs.
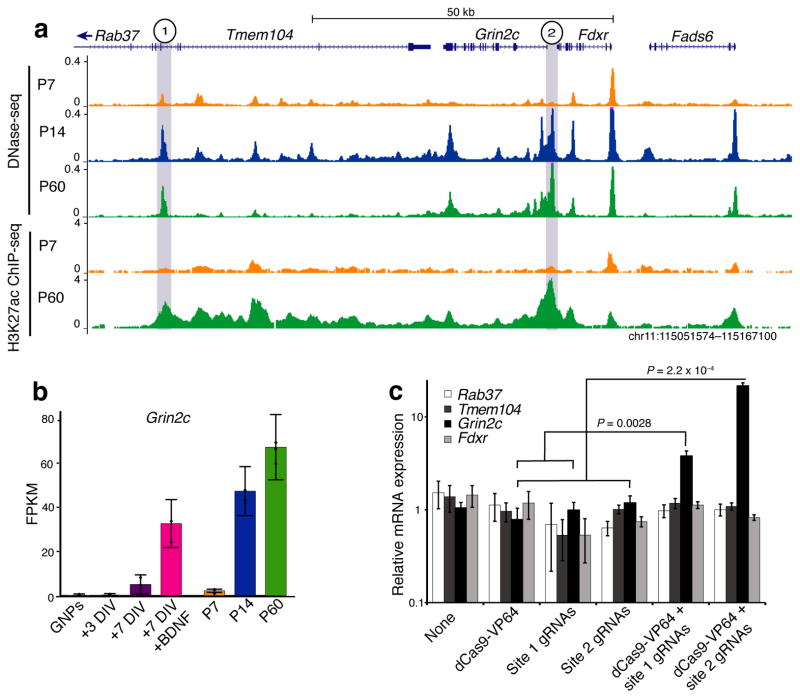
Next, to directly test the hypothesis that opening DHS sites represent enhancer elements in their native genomic context, we examined the locus surrounding the Grin2c gene on chromosome 11 in detail (Fig. 3a). Within this interval we identified two DHS sites that both open and gain H3K27ac signal across postnatal development – one just upstream of the Grin2c promoter and the second in an intron of the nearby Tmem104 gene. Of the four annotated genes in this region (Tmem104, Grin2c, Fdxr, Fads6), all are expressed in cerebellum, but only Grin2c expression is highly upregulated in differentiating CGNs, where it is a NMDA-type glutamate receptor subunit that mediates mature synaptic functions19 (Fig. 3b, Supplementary Table 6). Grin2c expression can also be robustly induced by culturing CGNs for seven days (+7DIV) in the presence of Brain-Derived Neurotrophic Factor (BDNF), offering an opportunity to experimentally test transcriptional mechanisms of Grin2c regulation28. Notably, BDNF-induced gene expression changes occur largely independent of changes in chromatin accessibility as we observed only 33 opened and 6 closed DHS sites in response to BDNF exposure (Supplementary Table 7).
Figure 3. CRISPR-VP64 based activation confirms enhancer activity of late-opening DHS sites nearby Grin2c.
(a) DNase-seq and H3K27ac ChIP-seq signal in vicinity of Grin2c with two DHS sites that open across development and increase in H3K27ac signal marked in gray. (b) RNA-seq expression of Grin2c across developmental time in vivo and in culture. BDNF = Brain-derived neurotrophic factor. Error bars = 95% CI. (c) Cultured CGNs were either left uninfected (none), or were infected with dCas9-VP64 activator, three gRNAs targeting site #1 from (a), three gRNAs targeting site #2, or a combination of dCas9-VP64 and one of the two sets of gRNAs on +1DIV, and harvested for qPCR on +7DIV. All data normalized to expression of Gapdh and scaled to average expression in control conditions (dCas9-VP64 or gRNAs only). P = 0.0028 for site 1 and P = 0.00022 for site 2 by two-sided Student’s t-test for dCas9-VP64 plus gRNAs vs. dCas9-VP64 or gRNAs alone (n = 5 for dCas9-VP64 with gRNAs, n = 8 for site 1 controls, n = 10 for site 2 controls; t = 5.5 for site 1, t = 12.3 for site 2). Error bars = s.e.m.
To determine whether the two identified DHS sites function as enhancers of Grin2c in their endogenous chromatin context, we used a CRISPR RNA-guided method to recruit a synthetic dCas9-linked transcriptional activator to each of the two putative enhancers flanking Grin2c in cultured CGNs29. Coinfection of dCas9-VP64 with gRNAs targeting either site was sufficient to induce a selective up-regulation of Grin2c expression compared with levels in control-infected neurons (Fig. 3c). By contrast, expression of three other nearby genes (Rab37, Tmem104, and Fdxr) remained unchanged. Together our CRISPR data demonstrate that two developmentally regulated DHS sites, including one located >50kb away from the Grin2c promoter, act as gene-specific enhancers, highlighting the power of our comparative DHS analysis to locate physiologically relevant enhancers of important neuronal genes.
Early-acting enhancers are deactivated prior to closing
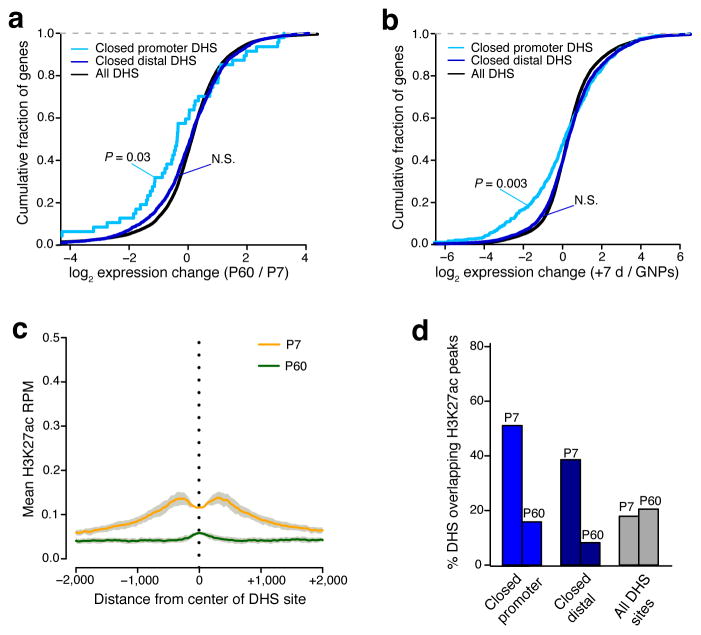
In contrast to the opening DHS sites, we observed only a modest association between DHS sites that close between P7 and P60 and the expression of their nearest gene (P = 0.03 promoter-located, N.S. for distal) or from GNPs to +7DIV in culture (P = 0.003 promoter-located, N.S. for distal) (Fig. 4a,b). As expected, closing DHS sites largely did not colocalize with the enhancer markers H3K4me1 and H3K27ac in P56 brain (Supplementary Fig. 7a). However, these regions did exhibit somewhat greater H3K27ac ChIP-seq signal in P7 cerebellum than at P60, with decreases in this active enhancer mark noted over the course of development at both promoter-located and distal DHS sites (Fig. 4c,d). These data suggest that at least a subset of the closing DHS sites are likely to be active enhancers in GNPs that are deactivated as CGNs differentiate. However given that we observed noticeably less H3K27ac enrichment at P7 for closing DHS sites compared with the enrichment of H3K27ac at P60 for opening DHS sites, we considered the possibility that the loss of chromatin accessibility may slowly follow the functional deactivation of enhancer elements.
Figure 4. A subset of closing DHS sites mark early postnatal enhancer elements.
(a,b) Cumulative fraction of genes nearest to closed promoter-located, closed distal, and all identified DHS sites with given fold-change in RNA-seq expression from P7 to P60 cerebellum (a) or from GNPs to +7DIV (b). Leftward shift indicates genes decreased in expression across developmental time. Significance assessed by Mann-Whitney U test. (c) Mean H3K27ac ChIP-seq signal (reads per million mapped) present at center of P7 to P60 closed DHS sites (5503 sites) in either P7 cerebellum (orange line) or P60 cerebellum (green line). Gray = s.e.m. (n = 2 biological replicates of pooled cerebella). (d) Percent of closed promoter-located, closed distal, and all DHS sites overlapping H3K27ac peaks identified in P7 or P60 cerebellum.
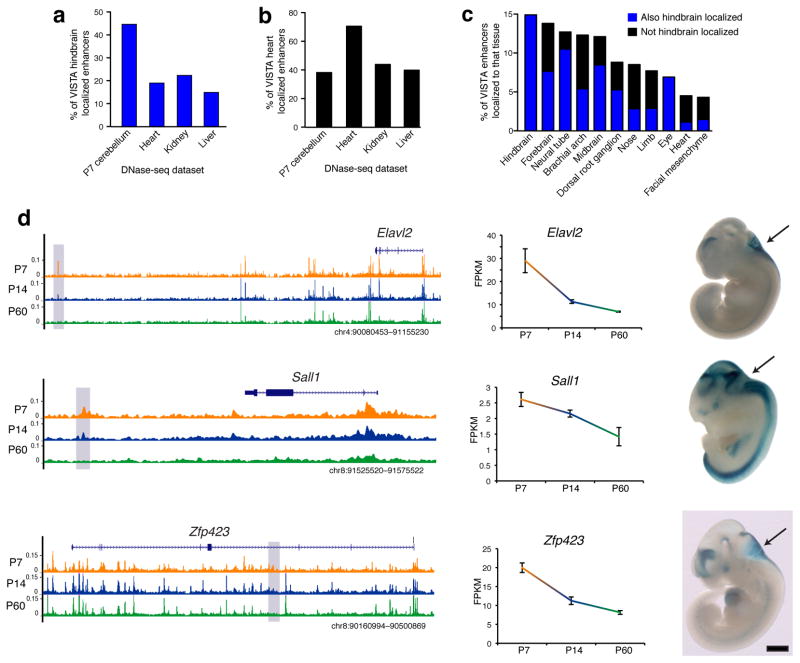
If this is the case, then the closing DHS sites may mark regions that function as enhancers in earlier stages in hindbrain development prior to the differentiation of GNPs. To test this hypothesis, we queried the VISTA Enhancer Browser, which contains enhancer elements that have been validated for their ability to drive tissue-restricted in vivo expression at embryonic day 11.5 (E11.5)30. At this age the proliferative neuroepithelial cells of the cerebellar primordium are found at the rostral end of the rhomencephalon surrounding the fourth ventricle7. We thus looked for overlap between our closing DHS sites and VISTA enhancers validated for their transcriptional activity in this region of the E11.5 embryo. We found that VISTA hindbrain-expressing enhancers overlapped more with P7 cerebellum DHS sites (108/242, 44.6%) compared to DHS sites from mouse heart (19%), kidney (22.3%), or liver (14.9%) (Fig. 5a). In contrast, VISTA heart enhancers overlapped more with mouse heart DHS sites compared to DHS sites from P7 cerebellum, kidney, or liver (Fig. 5b). Importantly, DHS sites that close in cerebellum between P7 and P60 displayed the highest overlap with enhancers that function in hindbrain (14.9%), forebrain (13.8%), and neural tube (12.7%) at E11.5, and a lower overlap with enhancers that function in non-neuronal tissues (Fig. 5c). This is in marked contrast with DHS sites that open between P7 and P60, which overlap only 1.7% of the E11.5 hindbrain enhancers (Supplementary Table 8).
Figure 5. Postnatal closing DHS sites are enriched for embryonic hindbrain enhancer activity.
Percent of hindbrain (a) or heart (b) localized VISTA Enhancer Browser staining (embryonic day 11.5) covered by all DHS sites identified in P7 cerebellum, heart, kidney, or liver mouse tissues (matching number of top peaks). (c) Percent of VISTA Enhancer Browser tissue-localized enhancers overlapped by DHS sites that close from P7 to P60. Tissues ordered by overlap rank. (d) Closed DHS sites located closest to the Elavl2, Sall1, and Zfp423 genes, all of which decrease in expression from P7 to P60 (center). Hindbrain expression is driven by each DHS site at E11.5, as seen with LacZ (blue) staining of embryos on right. Black scale bar = 1 mm. Images sourced from VISTA Enhancer Browser: hs643, hs152, and hs625.
Among the 36 DHS sites that both close during postnatal cerebellar development and drive hindbrain expression at E11.5, 13 (36%) were found nearest to a gene that displays significantly decreased expression from P7 to P60 (Fig. 5d, Supplementary Table 8). This list of associated genes included Elavl2 (HuB), which encodes an RNA-binding protein involved in neuronal development31, Sall1, which encodes a zinc-finger transcription factor, mutations in which are associated with developmental disorders and cognitive deficits in humans32, and Zfp423, which encodes a zinc-finger protein that is required for normal cerebellar development33. These data provide important experimental support for our hypothesis that many of the DHS sites that close during postnatal cerebellar development serve as enhancers of genes that function early in hindbrain development and further suggests that functional deactivation of these enhancers precedes DHS site closure.
Zics bind developmentally regulated DHS sites
Developmental regulation of chromatin architecture is thought to determine gene regulatory element accessibility to transcription factor binding1,34,35. We therefore sought to identify transcription factors whose regulatory functions may be dictated by chromatin accessibility changes by searching for enrichment of transcription factor motifs in DHS sites that opened or closed between P7 and P60 in vivo, as well as those that opened or closed over 7 days of GNP differentiation in culture.
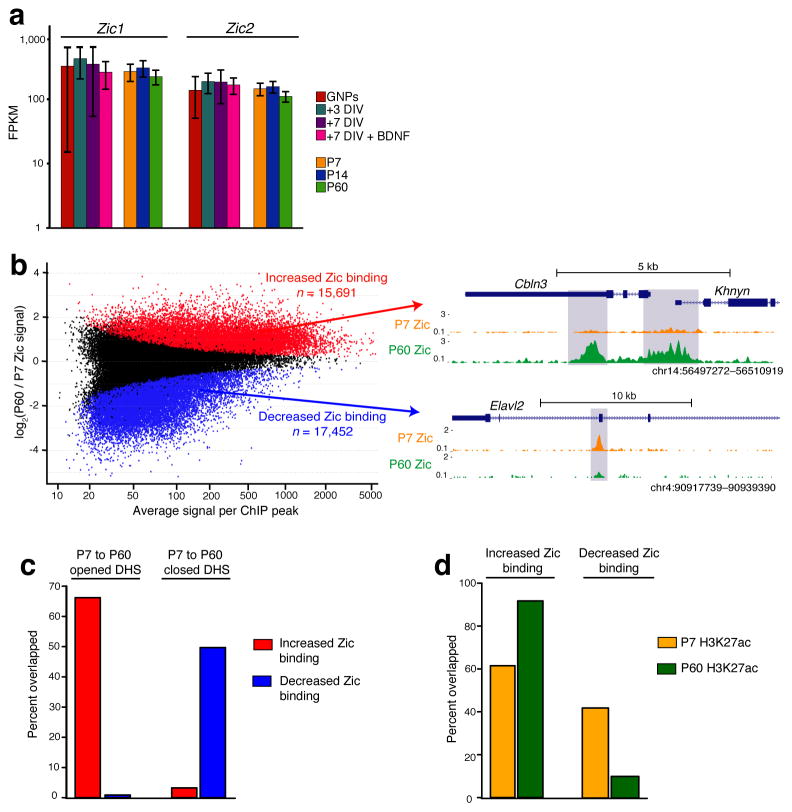
As expected, motifs for the MEF2 and NF1 families, which have been shown to play important roles in CGN differentiation9,36, were enriched in both opening and closing DHS sites (Supplementary Fig. 8). However we were surprised to find the zinc-finger in cerebellum (Zic) transcription factor family motifs enriched in the set of opening DHS sites. Mutations in the human ZIC genes have been associated with cerebellar development disorders, suggesting their importance in CGN differentiation, and mouse knockout studies have indicated that the Zics function in GNPs to prevent their premature exit from the cell cycle37,38. However the Zic transcription factors remain highly expressed in differentiated CGNs (Fig. 6a), raising the possibility that these factors also contribute to later stages in CGN maturation. Given our observation that the Zic motif was enriched in opening DHS sites, we hypothesized that the Zic transcription factors might change their gene targets over time by binding to developmentally regulated DHS sites.
Figure 6. Zic1/2 binding is highly dynamic across postnatal cerebellar development.
(a) Zic1 and Zic2 RNA-seq expression across in vivo and cultured CGN development (n = 3). (b) Zic binding changes between P7 and P60 cerebellum. Red points represent ChIP-seq peaks with significantly increased Zic binding from P7 to P60, blue points are peaks with decreased binding (FDR < 0.05, n = 2 biological replicates of pooled cerebella). UCSC genome browser images to the right show examples of increased and decreased Zic binding. Note the peak located in the 3′UTR of Cbln3 coincides with opened enhancer element tested in Fig. 2g. (c) Overlap between differential Zic binding sites and DHS sites that open or close from P7 to P60. (d) Overlap between differential Zic binding sites and H3K27ac ChIP-seq peaks identified in P7 or P60 cerebellum.
To determine if Zic transcription factors are differentially bound to DHS sites as CGNs differentiate, we performed ChIP-Seq from P7 or P60 cerebellum using an antibody specific for Zic1 and Zic239 (Supplementary Fig. 9a,b). As a control, no significant peaks were observed with IgG pulldown from P7 or P60 cerebellum, nor when we performed Zic ChIP-seq from P60 cortex, where Zic1/2 are not expressed (Supplementary Fig. 9c). Although 60% of ChIP-seq peaks overlapped between P7 and P60 (Supplementary Fig. 10a,b), we found over 15,000 peaks with significantly (FDR < 0.05) increased or decreased ChIP signal at P60 compared to P7 cerebellum (Fig. 6b, Supplementary Table 9). These data demonstrate that Zic transcription factors undergo dynamic changes in their DNA binding patterns in the developing cerebellum despite constant levels of expression.
Consistent with the enrichment of Zic binding motifs in opening DHS sites, we found that ~65% (2683/4053) of the opening DHS sites overlapped with Zic peaks that displayed stronger ChIP signal at P60. By contrast less than 1% (36/4053) of the opening DHS sites overlapped with Zic peaks with decreased ChIP signal across this time period (Fig. 6c). We also found nearly 50% (2735/5503) of closing DHS sites showed decreases in Zic binding, whereas only ~3% (179/5503) showed stronger Zic association over time. Increases and decreases in Zic binding over developmental time were correlated with H3K27ac signal, suggesting that Zic contributes to the enhancer activity of these elements (Fig. 6d). Taken together these data suggest that the chromatin-regulated binding of Zic transcription factors to enhancers may contribute to differential regulation of gene transcription during CGN differentiation.
Zics promote mature neuronal transcription
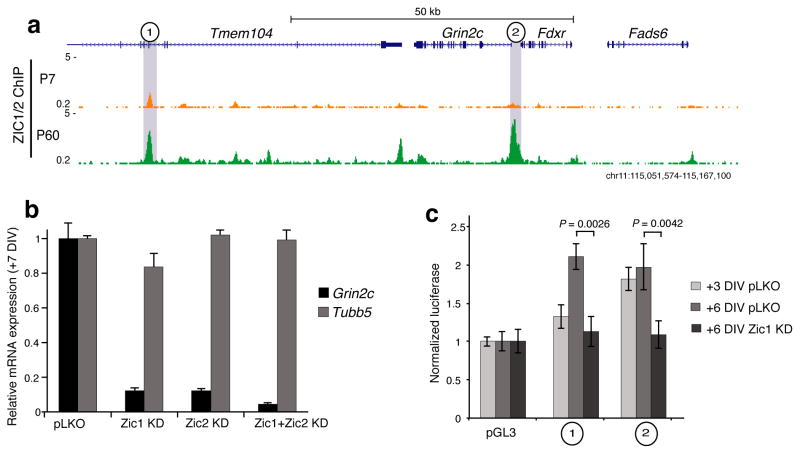
To test whether Zic is required for regulation of gene expression during CGN development, we returned to examining the ~120 kb region surrounding the Grin2c gene where we observed strong Zic ChIP-seq peaks in P60 cerebellum directly overlapping the two sites tested for enhancer function (Fig. 7a). To determine if Zic is required for the developmental up-regulation of Grin2c expression, we used lentiviral shRNAs to knockdown Zic1 and/or Zic2 expression in cultured CGNs (Supplementary Fig. 9a,b). Knockdown of either Zic1 or Zic2, or both together significantly reduced the expression of Grin2c compared with control-infected neurons (Fig. 7b). To test the role of Zic in the two DHS sites that we found to be Grin2c enhancers specifically, we cloned each element into a minimal promoter luciferase reporter plasmid, transfected them into cultured CGNs, and measured luciferase activity 3 or 6 days later. Both elements demonstrated enhancer activity at +6DIV (Fig. 7c). and knockdown of Zic1 significantly reduced the enhancer activity of both sites (P = 0.0026 and P = 0.0042, Student’s t-test) (Fig. 7c). Thus the developmentally regulated recruitment of Zic to these regions is functionally important for these elements to induce transcription.
Figure 7. Grin2c transcription is controlled by Zic1/2 stage-specific access to enhancer elements.
(a) Zic1/2 ChIP-seq signal in the vicinity of Grin2c. The two sites highlighted in gray increase accessibility, gain H3K27ac, and increase Zic1/2 affinity during development from P7 to P60 cerebellum. Also see Fig. 3a. (b) qRT-PCR Grin2c expression levels in culture at +7DIV following Zic1, Zic2, or combined Zic1 and Zic2 knockdown by shRNA, compared to empty vector (pLKO). Tubb5 used as control for total neuron number. Error bars = s.e.m. (n = 3 cultures). (c) Luciferase reporter assay for DHS sites highlighted in (a) in cultured neurons. Both sites are sufficient to confer enhancer activity that is reduced by Zic1 knockdown at +6DIV. Error bars = s.e.m. P = 0.0026 for site 1 and P = 0.0042 for site 2by two-sided Student’s t-test for Zic1 KD vs. pLKO control (n = 3 transfections, t = 6.7 and 5.8).
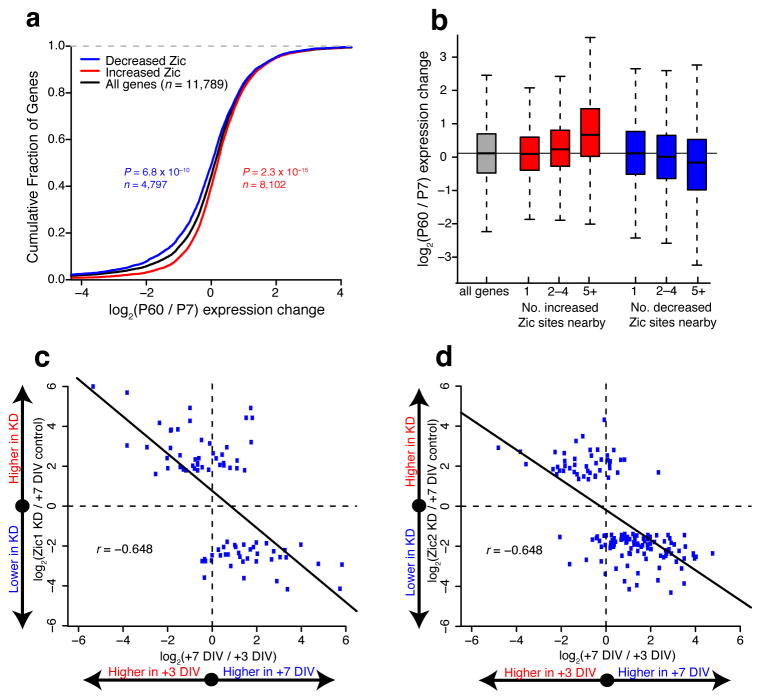
Finally, given the high degree of overlap between opening DHS sites and increased Zic1/2 binding as CGNs mature (Fig. 6c), we hypothesized that Zic might function to globally coordinate the maturation of gene expression programs in differentiating CGNs. Consistent with this hypothesis, we found a significant association between increased Zic binding and developmental up-regulation of nearby gene expression (Fig. 8a). The relationship between Zic binding and gene expression was strongest when multiple differential Zic binding sites mapped to a single gene (Fig. 8b). To determine the requirement for Zic in CGN differentiation, we used RNA-seq to analyze the effects of Zic1 or Zic2 knockdown on global gene expression in cultured CGNs. Knockdown of either Zic1 or Zic2 drove significant changes in the expression of 81 and 147 genes, respectively (FDR < 0.10) (Supplementary Table 10). To determine how this set of Zic-regulated genes was related to the gene expression changes that accompany CGN differentiation, we plotted the expression change of each Zic-regulated gene between +3DIV and +7DIV in control cultures versus the expression change for that same gene at +7DIV after knockdown of Zic1 (Fig. 8c) or Zic2 (Fig. 8d). These data reveal that genes that show reduced expression upon Zic1 or Zic2 knockdown tended to be up-regulated over the timecourse of CGN differentiation, whereas genes with higher expression following Zic1 or Zic2 knockdown tended to be down-regulated over differentiation. Thus the net effect of Zic1 or Zic2 knockdown is a shift in the gene expression program toward a less mature pattern. Overall these data indicate that Zic recruitment to regions of opened chromatin is essential for the maturation of gene expression programs in differentiating CGNs.
Figure 8. Zic1/2 promote the mature CGN transcriptional program.
(a) Relationship between Zic binding changes and nearby gene RNA-seq expression changes from P7 to P60. Significance of shifts assessed by two-sided Mann-Whitney U test. (b) Boxplots of gene expression change from P7 to P60 cerebellum binned by number of Zic binding sites associated with each gene. Having multiple Zic binding changes nearby more strongly associates with directional gene expression changes than single sites. (c,d) Relationship between cultured CGN development and Zic1 (c) or Zic2 (d) shRNA knockdown for genes marked significant in knockdown RNA-seq experiments (FDR < 0.10, n = 2 independent cultures). X-axis shows fold change in gene expression between +3DIV and +7DIV in cultured CGNs. Y-axis shows fold change in gene expression between control infected and Zic1 or Zic2 knockdown. Both knockdowns exhibit negative Pearson correlation coefficients (r).
Discussion
Mammalian genomes harbor over a million cis-regulatory elements marked by nucleosome-depleted DHS sites11. Previous studies comparing hundreds of diverse cell and tissue types demonstrate that the selective accessibility of these sites dictates cell type identity and function1,34,35. Developmentally regulated mammalian chromatin accessibility changes have previously been studied in the context of purified cell lineages like intestinal epithelium40 and CD4+ T cells41. Here we focused our analysis on a single neuronal cell lineage to map the in vivo chromatin accessibility changes that occur during development. Our data reveal that chromatin accessibility is highly dynamic across both early and postmitotic stages of neuronal maturation. We found accessibility to be strongly linked to H3K4me1 and H3K27ac deposition outside of promoters, indicating these chromatin accessibility changes identify regions that are enriched for poised and active enhancer elements. We demonstrated the value of identifying these dynamic DHS sites by predicting and validating a previously unknown role for the Zic transcription factors in neuronal maturation, thus enriching mechanistic understanding of brain development. The identification of postmitotic neuronal changes in enhancer accessibility and regulation is novel and raises the possibility that this form of chromatin plasticity could contribute to transcription-dependent forms of learning and memory in the adult brain.
We found that while DHS site opening frequently coincides with an increase both in H3K27ac signal and transcription of nearby genes, DHS site closing correlates weakly with decreasing gene expression and predominantly occurs at elements that already lack H3K27ac at P7. At least a subset of these closing elements function as enhancers in the cerebellar primordium during embryonic stages of brain development, suggesting that enhancer deactivation precedes the loss of accessibility. This evidence that an epigenetic signature is retained in the form of chromatin accessibility at some enhancers even though they are no longer active could permit the reactivation of gene expression programs otherwise thought to be cell-type or developmental stage-specific. Finally, we observed a significant difference in the magnitude to which DHS sites opened in CGNs that were differentiated in culture compared to in vivo. This suggests that full maturation of CGN chromatin accessibility requires cell non-autonomous factors not present outside the developing cerebellum. These external influences could include interactions with the Purkinje neurons, which are important for the proliferation and migration of CGNs, as well as patterned neural activity arising from sensory input to the intact brain, which plays a key role in refining cerebellar synapses42. Further elucidating the role of extrinsic factors in the regulation of chromatin accessibility is likely to uncover mechanisms of gene-environment interactions relevant to both normal and abnormal brain development.
Differential DHS analysis allowed us to predict and confirm that members of the Zic transcription factor family preferentially bind DHS sites that open during CGN differentiation. Mice lacking Zic1 alone or in combination with Zic2 have a small cerebellum, indicating a requirement for the Zics in the normal proliferation of cerebellar GNPs43. Heterozygous loss-of-function mutations in ZIC1 and ZIC4 have been identified in patients with Dandy-Walker malformation, a congenital malformation of the cerebellum37, further demonstrating fundamental roles for these transcription factors in brain development. However our evidence that Zics act in postmitotic CGNs to promote mature gene expression patterns was surprising in light of previous studies demonstrating a role for the Zics in preventing premature differentiation of dividing neuronal precursors43,44.
Zic family member expression remains constant throughout CGN development yet these factors have distinct genome binding profiles at P7 and P60. Our evidence that sites of regulated Zic binding overlap developmentally regulated DHS sites suggests that chromatin accessibility determines local Zic affinity. Developmental changes in chromatin accessibility are thought to be mediated by pioneer transcription factors that first recognizing their sequence-specific DNA binding sites and then recruit chromatin-remodeling complexes45. Our data show that many gene regulatory elements that recruit Zic binding between P7 andP60 already overlapped the active enhancer mark H3K27ac at P7, though they further increase in H3K27ac signal over development. These data raise the possibility that other developmentally-regulated factors bind these sites first, then recruit Zic. The switch in Zic binding sites may explain how Zics get repurposed for two very different functions - inhibition of premature differentiation in GNPs and promotion of maturation in post-mitotic CGNs - at distinct steps in the differentiation of the same cell lineage. Although the Zics are widely expressed in the developing brain, CGNs are one of the few kinds of neurons that maintain Zic expression in adulthood, suggesting other transcription factors may function at stage-specific enhancers to direct mature neuronal gene expression programs in other types of neurons.
Whereas previous studies have demonstrated that extracellular signal-regulated transcription factors can drive CGN maturation9,36,46,47, the contributions of chromatin accessibility dynamics to these processes have not been investigated. The idea that environmental stimuli can induce changes in chromatin accessibility has been called into question by studies that have shown that stimulus-dependent changes in gene transcription, for example those induced by glucocorticoid hormones48, or exposure of intestinal epithelia to microbiota49, can occur independent of changes in chromatin accessibility. However our evidence shows that DHS sites change substantially during the late stages of post-mitotic CGN maturation. Since these late stages of differentiation are primarily driven by the action of stimulus-regulated transcription factors9,36,46,47, this implies that at least a subset of environmental stimuli are capable of driving changes in chromatin accessibility. The identification of these environmental factors and how they interact with Zic and other transcription factors will be key to understanding neuronal development, plasticity, and disease.
Methods
Cerebellar immunostaining
C57BL/6Ncrl male or female mice (Charles River Labs) at P7, P14, or P60 were deeply anesthetized with isofluorane and their brains were dissected and flash frozen in OCT. Fresh frozen brains were cryostat sectioned in the sagittal plane at 18μm, then fixed on slides in 3% paraformaldehyde and rinsed with 1x phosphate-buffer saline (PBS). Slides were blocked in either 14% normal goat serum or 10% normal donkey serum (for primary antibodies raised in goat) and permeabilized in 0.3% Triton X-100 prior to antibody incubation. Hoechst dye was used to label nuclei for identification of anatomical landmarks. Images were captured on a Leica DMI4000 inverted fluorescence microscope at 10X or 20X magnification with a Leica DFC-365 camera using Metamorph 7.0 software (Molecular Devices). We used the following primary antibodies: mouse anti-Ki67 (1:50; BD Pharmingen 550609), goat anti-Dcx (1:100, Santa Cruz sc-8066), rabbit anti-GABAARα6 (1:1000, Millipore AB5610), rabbit anti-Zic1/2 (1:1000, Dr. Rosalind Segal, Harvard Medical School39), mouse anti-GFAP (1:100, Abcam cat. #ab10062), and rabbit anti-calbindin (1:1000, Swant cat. #CB38). Secondary antibodies were used at 1:500: donkey anti-goat Cy3 (Jackson Immunoresearch, 705-165-003), goat anti-rabbit Cy2 (115-225-146), goat anti-mouse Cy3 (115-165-146), and goat anti-mouse Cy5 (115-175-146). All animals were housed in groups of 4–5, given access to standard laboratory chow and water ad libitum and housed in a humidity and temperature-controlled room on a 14/10 h light/dark cycle. All experiments were conducted in accordance with an approved protocol from the Duke University Institutional Animal Care and Use Committee and guidelines from the National Institutes of Health for the Care and Use of Laboratory Animals.
Isolation of cerebellar nuclei
For in vivo samples, C57BL/6Ncrl male and female mice at P7, P14, or P60 were deeply anesthetized with isofluorane and decapitated for brain harvesting. The cerebellar cortex was dissected, flash frozen, and stored at −80°C. For CGN cultures (described below), neurons were scraped into PBS and harvested by centrifugation at 1000 × g for 5 min. Nuclei were extracted essentially as previously described50. Briefly, each cerebellum was dounced in 5mL 2M sucrose, 1mM MgCl2 and poured through a 100μM filter. Filtered tissue was ultracentrifuged in 15 mL of 2M sucrose, 1mM MgCl2 at 65,000 × g for 80 min. Supernatant was removed and the tight nuclear pellet was resuspended by pipetting in 1 mL buffer RSB from the DNase-seq protocol51. Nuclei were quantified by hemocytometer, yielding approximately 5–10 million per adult cerebellum (~30 mg tissue).
DNase-seq library generation
DNase-seq libraries were constructed from nuclear preparations as previously described51. Briefly, 15–30 million nuclei were digested with a range of recombinant DNase I enzyme (Roche) concentrations (between 1.2 and 12U) for 15 minutes at 37°C in 120uL 1X DNase buffer. Digestions were checked by pulse field gel electrophoresis and material was pooled from 3 different DNase concentrations (extent of digestion matched between samples) in equimolar amounts following blunt-ending reactions. Following ligation to adapters, MmeI digestion, streptavidin bead-based enrichment, and 14 cycles of PCR amplification, each library was sequenced for either 36 cycles on an Illumina GAIIx machine, or 50 cycles on a Hi-Seq 2000 platform to an average depth of 115 million aligned reads per sample. Nuclei from cerebellae of 4–6 mice or ~20 million cultured CGNs were pooled for each biological replicate, and three independent biological replicates were analyzed for each timepoint.
RNA isolation and sequencing
RNA was isolated from P7, P14, or P60 C57BL/6 mouse cerebellar cortex or cultured mouse CGNs using the Absolutely RNA kit (Agilent). For in vivo samples, RNA from at least two cerebellae was pooled for each biological replicate, and in culture a single litter constituted a biological replicate of cultured CGNs. Three independent biological replicates were analyzed for each timepoint. 2μg of total RNA was used for standard TruSeq library preparation with polyA selection (performed by the Duke Sequencing and Analysis Core Resource). Libraries were subjected to 50 bp paired-end Illumina Hi-Seq 2000 sequencing, with the exception of Zic knockdown experiments, in which biological duplicates were sequenced as single-end 50 bp.
Cerebellar granule neuron culture
Granule neuron precursors were isolated and cultured as previously described52. Briefly, cerebellae from a litter of P7 mice were chopped into small pieces then digested at 37°C for 30 min in 10 U/ml papain (Worthington), 200g/ml L-cysteine (Sigma), and 250 U/ml DNase (Sigma) in PBS. The tissue was mechanically triturated in PBS with trypsin inhibitor and 250 U/ml DNase then centrifuged and resuspended in PBS/BSA. The cell suspension was passed through a cell strainer (Becton Dickinson), layered on a step gradient of 35% and 65% Percoll (Sigma), and centrifuged at 2500 rpm for 12 min at room temperature. Granule Neuron Precursors (GNPs) were harvested from the 35/65% interface, washed in PBS/BSA, resuspended in Neurobasal medium with B27 supplements and 2% fetal bovine serum and plated on poly-D-lysine coated dishes. In some cases as noted in the text, Brain-Derived Neurotrophic Factor (Peprotech) was added at 50 ng/mL on +4DIV.
CGN Immunostaining
For immunostaining, freshly isolated GNPs were plated on glass coverslips coated with poly-D-lysine and laminin in Neurobasal media with B27 supplements and 2% serum as above. Cells were then either fixed in 4% paraformaldehyde immediately (+0DIV; GNPs) or allowed to differentiate for 3 or 7 DIV. Cells were blocked in 10% goat serum and permeabilized with 0.3% tritonX-100 prior to primary antibody application overnight. We used the following primary antibodies: mouse anti-Ki67 (1:50, BD Pharmingen 550609), chicken anti-MAP2 (1:200, Millipore AB5543), and rabbit anti-synaptophysin (1:500, Millipore MAB5258). Secondary antibodies labeled with Cy2, Cy3, or Cy5 were used at 1:500 (see Cerebellar Immunostaining). Hoechst dye was used to label nuclei for cellular identification. Images were captured on a Leica DMI4000 inverted fluorescence microscope at 20X or 40X magnification with a Leica DFC-365 camera using Metamorph 7.0 software.
Luciferase reporter assays
Putative enhancers were cloned in the vector pGL3 (Promega). For Cbln3, we cloned mm9 region chr14:56500910–56501851, at the BglII site upstream of luciferase. For the two regions in the Grin2c interval on Chr. 11, we cloned the following mm9 regions at the BamHI or SalI sites downstream of the luc+ coding sequence: Site #1: chr11:115064061–115064510; Site #2: chr11:115128431–115128780. Luciferase reporters were transfected into cultured CGNs on +1DIV by calcium phosphate precipitation as described53. Cotransfection of TK-renilla luciferase (Promega, Madison, WI) was used to control for transfection efficiency and sample handling. As described in the text, in some cases plasmids encoding shRNAs targeting Zic1 in the vector pLKO.1 or empty pLKO.1 were cotransfected with the luciferase plasmids. Data presented are the average of at least three measurements from each of at least two independent experiments.
Chromatin Immunoprecipitation
For ChIP we pooled cerebellum or cortex from three P7 mice or two P60 mice for each biological replicate. Brain samples were dounced in 1% formaldehyde PBS buffer and kept at room temperature for 15 minutes, washed twice with cold PBS, then lysed in 600μl lysis buffer (1% SDS, 10mM EDTA, and 50mM Tris pH 8.1). The crosslinked material was sonicated with a Bioruptor (Diagenode) with 30 seconds on/off cycles to an average size range of 150–350bp as visualized by agarose gel electrophoresis. Sonicated supernatants were diluted 10 fold in dilution buffer (0.01%SDS, 1.1% Triton X-100, 1.2mM EDTA, 16.7mM Tris-HCL, pH 8.1, 167mM NaCl) before immunoprecipitation. 6μL of antibody (anti-Zic 1/2 C-terminus, courtesy of Dr. Rosalind Segal, Harvard Medical School39, total rabbit IgG, Millipore 12-370, or anti histone H3K27Ac, Abcam Ab472) was first incubated with 100 μL of Dynabeads Protein G (Invitrogen 10004D) beads for 4 hours at 4°C, then the antibody conjugate was added to 6ml of cell lysis for overnight IP. Standard TruSeq adapters were ligated for library preparation, which for Zic used the MicroPlex Library Preparation Kit (Diagenode), and for H3K27ac used the NEB Library Preparation Kit (NEB 6240S and NEB E7335s). 50bp single-end sequencing was performed at the Duke Sequencing and Analysis Core Resource on a Hi-Seq 2000 machine. Two independent biological replicates were performed for each antibody, developmental timepoint, and brain region.
RNA interference
For the knockdown of Zic1 and Zic2 transcription factors, we purchased shRNAs targeting mouse Zic1 and Zic2 that were cloned in the lentiviral vector pLKO.1 (Thermo Scientific). Zic1 shRNA (TRCN0000096685), Zic2 shRNA (TRCN0000095251). Empty pLKO.1 was used as the control. Viral shRNAs were packaged as lentivirus in HEK293T cells (ATCC) following standard procedures. Neurons were infected with lentivirus at a multiplicity of infection of 1 on +1DIV, and samples were harvested for RNA or protein analysis on +7DIV.
Quantitative RT-PCR
800ng of total RNA was used for reverse transcription with oligo dT primers and Superscript II (Invitrogen). Quantitative SYBR green PCR was performed on an ABI 7300 real-time PCR machine (Applied Biosystems) using the following intron-spanning primers (IDT): Grin2b, F: GAGCATAATCACCCGCATCT, R: AAGGCACCGTGTCCGTATCC; Zic1, F: CAGCCCGCGATCCGAGCACTATG, R: GCAGCCCTCGAACTCGCACTTGAA; Zic2, F: GGCGGCGCAGCTCCACAACCAGTA, R: TTGCCACAGCCCGGGAAAGGACAG; Grin2c, F: TGTGGGCCTTCTTCGCTGTCATCT, R: TGCCATTAGGTACCGTGCCAAAAC; Tubb5, F: GGGAGGTGATAAGCGATGAA, R: CCCAGGTTCTAGATCCACCA; Gapdh, F: CATGGCCTTCCGTGTTCCT, R: TGATGTCATCATACTTGGCAGGTT. Each sample was measured in triplicate, and all data were normalized to the expression of the housekeeping gene Gapdh.
Western blotting
1 million CGNs or 10mg cerebellar or non-cerebellar brain tissue was lysed in RIPA buffer to a final concentration of 10mg/mL, then disrupted by sonication. 100μg total lysate was run for SDS-PAGE and transferred to nitrocellulose for western blotting. Actin was used as a loading control. Primary antibodies were mouse anti-Zic1/2 C-terminus39, mouse anti-Actin (1:10,000; Millipore MAB1501). Secondary antibodies were goat anti-rabbit 770 (Biotium 20078) and goat anti-mouse 680 (Biotium 20065). Bands were visualized with fluorescent secondary antibodies using the Odyssey imaging system (LI-COR Bioscience).
Cas9-based RNA-guided gene activation
The CRISPR web interface from the Zhang lab at MIT (http://crispr.mit.edu) was used to design CRISPR guide RNA (gRNA) followed by a PAM site (NGG) that have minimal off-target matches. We selected three gRNA sequences for each element tested (site #1: chr11:115064061–115064510; site #2: chr11:115128431–115128780). Oligonucleotides containing target sequences were obtained from IDTand cloned at the BsmBI site in a FUGW-based U6 chimeric gRNA expression vector co-expressing GFP. Site 1-1: GCTCCACACCTGCGGAGGTA, Site 1-2: GCACGCAAACCTCTACTGGGC, Site 1–3: GTTAGAAAGAAGGGCGTACGG, Site 2-1: GCACAGCCCCGCCGCGGGGCG, Site 2-2: GAGGGGCCAGCTAGGAGCAC, Site 2–3: GAGATGGAGCTCGCCACCGC. We used an enzymatically dead Cas9-VP64 fusion protein cloned in a lentiviral backbone with T2A expression of GFP (pLV hUbC-dCas9VP64-T2A-GFP; Addgene plasmid #53192). Constructs were packaged as lentivirus in HEK293T cells and CGNs were infected on +1DIV with either dCas9-VP64 alone, a pooled set of the three gRNA viruses targeting one of the elements, or dCas9-VP64 together with one of the gRNA pools. RNA was harvested on +7DIV for cDNA synthesis and quantitative PCR. For each treatment we analyzed at least three samples from at least two independent experiments. In addition to Grin2c and Gapdh primers above, we used the following primers: Rab37, F: ACTGGCATGAACGTGGAGTT, R: TAGGGGGTCACACAAAGGAG; Tmem104, F: AAGTAACCTCTTGGGCCAGC, R: CACCAGCCCGACATAAGGAG; Fdxr, F: CATTTACACAGACAGCCCGC, R: CTGCTCCATAACTCAGCACCA.
Bioinformatic analysis
DNase-seq alignments, peak calling, and differential signal tests
DNase-seq reads were trimmed to the first 20 bp (fixed insert length generated by MmeI digest following first linker ligation in library preparation) and aligned to the NCBI37/mm9 reference genome using BWA54, allowing a single mismatch and mapping up to 4 locations. Alignments were filtered for PCR artifacts and normalized, smoothened coverage tracks and discrete peak calls were generated by F-seq55. DHS sites were annotated to genic features using the mm9 UCSC Genes knownGene table. The R package DESeq v1.8.3 was used for signal normalization and calling differential DHS sites (exact test on negative binomial distributions) between conditions to take into account signal-to-noise ratios and variance between replicates across sequencing depth56. Raw read counts (defined by the first 5-prime base of each read) summed in the 250 bp windowed union of top 100,000 DHS peaks identified in the samples under consideration served as input to DESeq analysis. 250 bp windows with 50bp overlaps were chosen for increased resolution within peak calls.
RNA-seq alignments and differential expression tests
Following quality score-based trimming and adapter filtering, reads were aligned to the UCSC Genes mm9 reference transcriptome and then NCBI37/mm9 reference genome using Tophat with options –x 4 and –n 2 57. All transcriptome assemblies produced by the alignments were combined with Cuffmerge prior to differential testing. Cuffdiff58 was used for pairwise tests of differential expression between normalized gene read counts of Fragments Per Kilobase of exon per Million fragments mapped (FPKM) with the default significance threshold of FDR < 0.05 used for all analyses, except for comparisons with Zic knockdown experiments, which used FDR < 0.10. Normalized UCSC browser coverage was generated by conversion of bam format alignments to bp resolution bigWig files and scaling by total number of mapped reads.
RNA-seq expression pattern clustering
RNA-seq data was analyzed in part with the cummeRbund R package (http://compbio.mit.edu/cummeRbund/) operating on Cuffdiff ouputs. For cluster profiles, all significantly differential genes from the in vivo time course were clustered by JS distance to a priori defined expression vectors (for example, [0, 1, 0] for elevated expression in P14 relative to P7 and P60). The 100 best match genes from each vector were plotted and subjected to the Database for Annotation, Visualization and Integrated Discovery (DAVID) to find enriched Cellular Component and Biological Process gene ontologies59. Principal components analyses (PCA) of gene expression were performed in cummeRbund.
Relationship between differential DHS and gene expression changes
To link differential DHS sites with gene expression changes, each DHS site was associated with its nearest UCSC mm9 reference gene. The fold change in each gene’s FPKM values from P7 to P60 for in vivo, or GNPs to +7DIV for cultured neurons were calculated. The distribution of fold changes was plotted as a cumulative function and two-sided Mann-Whitney tests between distributions assessed significance. DHS sites mapping within 2kb of an annotated TSS were considered promoter-located and all other DHS sites were considered distal.
ENCODE histone mark ChIP-seq datasets
H3K27ac, H3K4me1, and H3K4me3 ChIP-seq data from P56 C57BL/6 cerebellum were retrieved from the Mouse ENCODE project August 2012 release from Ludwig Institute for Cancer Research (Bing Ren lab). Available peak call files were used for determining overlap fractions with DHS sites in this study.
Cell-type enriched gene expression comparisons
Purkinje cell, Bergmann glia, and granule neuron (CGN) enriched gene lists were obtained from14. Sequenced RNA abundances captured with translating polysomes (TRAP-seq) in each cell type were compared with the DESeq package. This included 922 Purkinje cell, 2084 Bergmann glia, and 986 CGN enriched genes. For beanplots of DNase-seq, H3K27ac, and H3K4me3 signal, total read counts were extracted for each gene within 2kb of the TSSand normalized by total aligned reads in that sample. Replicates were summed. Genes with FPKM between 0 and 1 were considered “low” expression, and FPKM > 1 considered “high”. Significance assessed with two-sided Mann-Whitney test. To associate cell type enriched genes with differential DHS sites, differential DHS sites between P7 and P60 cerebellum were each assigned a single nearest gene and the percent overlap of this list with cell type enriched genes that also displayed differential expression between P7 and P60 was computed. These percentages were compared to the percent overlap obtained from random sampling from all expressed genes (10,000 iterations).
Motif-finding in DHS sites
Enriched motifs in differential DHS sites were identified using MEME-ChIP60 with a 3rd order Markov model background of all DHS sites. The top 10 Discriminative DNA Motif Discovery (DREME) hits by e-value were selected and aligned to known motifs contained in the JASPAR vertebrates and UniPROBE mouse databases.
Vista Enhancer Browser Analyses
The VISTA Enhancer Browser was accessed October 10, 2013 while containing 1940 total tested putative enhancer elements30. The top 75,192 DHS sites from P7 cerebellum, adult mouse heart, liver, and kidney DNase-seq were overlapped with all VISTA tested elements that showed reproducible staining in hindbrain or heart. All VISTA tested elements overlapping P7 DHS sites are provided in Supplementary Table 8. When associating VISTA elements with genes, we used the nearest reference gene and ignored any gene annotation that showed zero expression by RNA-seq across time points.
H3K27ac and Zic ChIP-seq analysis
50 bp SR ChIP-seq reads were aligned to the NCBI37/mm9 reference genome with Bowtie, allowing up to 2 mismatches and requiring unique alignments. bigWig coverage tracks were normalized by total number of mapped reads. Captured fragment length shifts were estimated for each library by cross-strand correlation maxima61,62 and used for peak calling with MACS v1.4.263 with matched input controls and default P < 0.00001 threshold for each replicate. Cross-strand correlation maxima were also used for quality control to assess signal to noise ratio of each ChIP-seq experiment and samples displaying low scores were omitted. For differential Zic binding detection, raw read counts mapped to the union of Zic ChIP-seq peak calls for all P7 and P60 replicates were used as input to DESeq56. Differential binding events between P7 and P60 were determined by DESeq negative binomial test at a FDR < 0.05 threshold. To correlate with expression changes observed, Zic binding sites were linked to the nearest expressed gene.
Statistical analysis
Individual statistical tests employed are noted in figure legends. Two-sided unpaired t-tests were used for comparisons of normal distributions and Mann-Whitney or exact tests were used for non-normally distributed sequencing data. No statistical methods were used to predetermine sample sizes but our sample sizes are similar to those reported in previous studies14,49,64. Data collection and analysis were not performed blind to the conditions of the experiments. Order of sequencing sample collection was random but no other randomization of data was performed.
A Supplementary Methods Checklist is available.
Supplementary Material
Acknowledgments
We thank the Duke Genome Sequencing & Analysis Core for sequencing the RNA-seq, ChIP-seq, and DNase-seq libraries. This work was supported by the Duke Institute for Brain Sciences, NIH grant 1R21NS084336 (A.E.W. and G.E.C.), and NIH grant R01DA036865 (C.A.G. and G.E.C).
Footnotes
Author Contributions: A.E.W., G.E.C., C.L.F., R.W. and F.L. designed the study. F.L., C.L.F., R.W., L.S., and A.S. performed DNase-seq and ChIP-seq experiments. M.T.B., M.G.Y., C.M.V., and C.A.G. designed and performed targeted enhancer function assays. F.L. performed Zic knockdown experiments. C.L.F. performed all bioinformatic analyses. C.L.F., A.E.W., and G.E.C. wrote the manuscript with input from all authors.
Accession codes:
Sequencing data have been deposited at Gene Expression Omnibus (GEO) under accession number GSE60731.
References
- 1.Song L, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine M, Cattoglio C, Tjian R. Looping back to leap forward: transcription enters a new era. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 5.Pattabiraman K, et al. Transcriptional regulation of enhancers active in protodomains of the developing cerebral cortex. Neuron. 2014;82:989–1003. doi: 10.1016/j.neuron.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altman J, Bayer SA. Development of the cerebellar system: in relation to its evolution, structure, and functions. CRC Press; 1997. [Google Scholar]
- 8.Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- 9.Ding B, et al. Temporal regulation of nuclear factor one occupancy by calcineurin/NFAT governs a voltage-sensitive developmental switch in late maturing neurons. J Neurosci Off J Soc Neurosci. 2013;33:2860–2872. doi: 10.1523/JNEUROSCI.3533-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhar SG, et al. Changing patterns of gene expression define four stages of cerebellar granule neuron differentiation. Dev Camb Engl. 1993;117:97–104. doi: 10.1242/dev.117.1.97. [DOI] [PubMed] [Google Scholar]
- 11.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altman J. Autoradiographic and histological studies of postnatal neurogenesis. 3. Dating the time of production and onset of differentiation of cerebellar microneurons in rats. J Comp Neurol. 1969;136:269–293. doi: 10.1002/cne.901360303. [DOI] [PubMed] [Google Scholar]
- 13.Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 14.Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatten ME. Neuronal regulation of astroglial morphology and proliferation in vitro. J Cell Biol. 1985;100:384–396. doi: 10.1083/jcb.100.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogarty MP, Emmenegger BA, Grasfeder LL, Oliver TG, Wechsler-Reya RJ. Fibroblast growth factor blocks Sonic hedgehog signaling in neuronal precursors and tumor cells. Proc Natl Acad Sci U S A. 2007;104:2973–2978. doi: 10.1073/pnas.0605770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fletcher TL, De Camilli P, Banker G. Synaptogenesis in hippocampal cultures: evidence indicating that axons and dendrites become competent to form synapses at different stages of neuronal development. J Neurosci Off J Soc Neurosci. 1994;14:6695–6706. doi: 10.1523/JNEUROSCI.14-11-06695.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yashiro K, Philpot BD. Regulation of NMDA receptor subunit expression and its implications for LTD, LTP, and metaplasticity. Neuropharmacology. 2008;55:1081–1094. doi: 10.1016/j.neuropharm.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farrant M, Feldmeyer D, Takahashi T, Cull-Candy SG. NMDA-receptor channel diversity in the developing cerebellum. Nature. 1994;368:335–339. doi: 10.1038/368335a0. [DOI] [PubMed] [Google Scholar]
- 20.Ruppert C, Goldowitz D, Wille W. Proto-oncogene c-myc is expressed in cerebellar neurons at different developmental stages. EMBO J. 1986;5:1897–1901. doi: 10.1002/j.1460-2075.1986.tb04442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eroglu C, et al. Gabapentin receptor alpha2delta-1 is a neuronal thrombospondin receptor responsible for excitatory CNS synaptogenesis. Cell. 2009;139:380–392. doi: 10.1016/j.cell.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez C, Tatard VM, Bertrand N, Dahmane N. Differential modulation of Sonic-hedgehog-induced cerebellar granule cell precursor proliferation by the IGF signaling network. Dev Neurosci. 2010;32:59–70. doi: 10.1159/000274458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mouse ENCODE Consortium et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bao D, et al. Cbln1 is essential for interaction-dependent secretion of Cbln3. Mol Cell Biol. 2006;26:9327–9337. doi: 10.1128/MCB.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirai H, et al. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat Neurosci. 2005;8:1534–1541. doi: 10.1038/nn1576. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki K, Sato M, Morishima Y, Nakanishi S. Neuronal depolarization controls brain-derived neurotrophic factor-induced upregulation of NR2C NMDA receptor via calcineurin signaling. J Neurosci Off J Soc Neurosci. 2005;25:9535–9543. doi: 10.1523/JNEUROSCI.2191-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hambardzumyan D, et al. AUF1 and Hu proteins in the developing rat brain: implication in the proliferation and differentiation of neural progenitors. J Neurosci Res. 2009;87:1296–1309. doi: 10.1002/jnr.21957. [DOI] [PubMed] [Google Scholar]
- 32.Vodopiutz J, et al. Homozygous SALL1 mutation causes a novel multiple congenital anomaly-mental retardation syndrome. J Pediatr. 2013;162:612–617. doi: 10.1016/j.jpeds.2012.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warming S, Rachel RA, Jenkins NA, Copeland NG. Zfp423 is required for normal cerebellar development. Mol Cell Biol. 2006;26:6913–6922. doi: 10.1128/MCB.02255-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stergachis AB, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheffield NC, et al. Patterns of regulatory activity across diverse human cell types predict tissue identity, transcription factor binding, and long-range interactions. Genome Res. 2013;23:777–788. doi: 10.1101/gr.152140.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shalizi A, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 37.Grinberg I, et al. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat Genet. 2004;36:1053–1055. doi: 10.1038/ng1420. [DOI] [PubMed] [Google Scholar]
- 38.Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci. 2004;26:205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Borghesani PR, et al. BDNF stimulates migration of cerebellar granule cells. Dev Camb Engl. 2002;129:1435–1442. doi: 10.1242/dev.129.6.1435. [DOI] [PubMed] [Google Scholar]
- 40.Kim TH, et al. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506:511–515. doi: 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Samstein RM, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sotelo C. Cellular and genetic regulation of the development of the cerebellar system. Prog Neurobiol. 2004;72:295–339. doi: 10.1016/j.pneurobio.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 43.Aruga J, Inoue T, Hoshino J, Mikoshiba K. Zic2 controls cerebellar development in cooperation with Zic1. J Neurosci Off J Soc Neurosci. 2002;22:218–225. doi: 10.1523/JNEUROSCI.22-01-00218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aruga J, et al. Mouse Zic1 is involved in cerebellar development. J Neurosci Off J Soc Neurosci. 1998;18:284–293. doi: 10.1523/JNEUROSCI.18-01-00284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ben-Arie N, et al. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 47.Flora A, Klisch TJ, Schuster G, Zoghbi HY. Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science. 2009;326:1424–1427. doi: 10.1126/science.1181453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Camp JG, et al. Microbiota modulate transcription in the intestinal epithelium without remodeling the accessible chromatin landscape. Genome Res. 2014 doi: 10.1101/gr.165845.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas JO, Thompson RJ. Variation in chromatin structure in two cell types from the same tissue: a short DNA repeat length in cerebral cortex neurons. Cell. 1977;10:633–640. doi: 10.1016/0092-8674(77)90096-4. [DOI] [PubMed] [Google Scholar]
- 51.Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5384. pdb.prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kokubo M, et al. BDNF-mediated cerebellar granule cell development is impaired in mice null for CaMKK2 or CaMKIV. J Neurosci Off J Soc Neurosci. 2009;29:8901–8913. doi: 10.1523/JNEUROSCI.0040-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tao X, West AE, Chen WG, Corfas G, Greenberg ME. A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron. 2002;33:383–395. doi: 10.1016/s0896-6273(01)00561-x. [DOI] [PubMed] [Google Scholar]
- 54.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinforma Oxf Engl. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinforma Oxf Engl. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 60.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinforma Oxf Engl. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paige SL, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.