Abstract
Myosins and tropomyosins represent two cytoskeletal proteins that often work together with actin filaments in contractile and motile cellular processes. While the specialized role of tropomyosin in striated muscle myosin-II regulation is well characterized, its role in non-muscle myosin regulation is poorly understood. We previously showed that fission yeast tropomyosin (Cdc8p) positively regulates myosin-II (Myo2p) and myosin-V (Myo52p) motors. To understand the broader implications of this regulation we examined the role of two mammalian tropomyosins (Tpm3.1cy/Tm5NM1 and Tpm4.2cy/Tm4) recently implicated in cancer cell proliferation and metastasis. Like Cdc8p, the Tpm3.1cy and Tpm4.2cy isoforms significantly enhance Myo2p and Myo52p motor activity, converting non-processive Myo52p molecules into processive motors that can walk along actin tracks as single molecules. In contrast to the positive regulation of Myo2p and Myo52p, Cdc8p and the mammalian tropomyosins potently inhibited skeletal muscle myosin-II, while having negligible effects on the highly processive mammalian myosin-Va. In support of a conserved role for certain tropomyosins in regulating non-muscle actomyosin structures, Tpm3.1cy supported normal contractile ring function in fission yeast. Our work reveals that actomyosin regulation by tropomyosin is dependent on the myosin isoform, highlighting a general role for specific isoforms of tropomyosin in sorting myosin motor outputs.
Keywords: actin, tropomyosin isoforms, myosin isoforms, motor sorting, regulation
INTRODUCTION
Tropomyosins form strand-like molecules made up of parallel dimeric coiled-coils (Hitchcock-DeGregori 2008). The association of tropomyosin with actin promotes filament stability and is known to regulate associations with actin-binding proteins such as myosin motors (Barua et al. 2014; Barua et al. 2012; Bryce et al. 2003; Clayton et al. 2010; Coulton et al. 2010; Fanning et al. 1994; Lehrer 1994; Stark et al. 2010; Tang and Ostap 2001), filament cross-linkers (Clayton et al. 2010; Creed et al. 2008; Skau and Kovar 2010), filament severing factors (Bernstein and Bamburg 1992; Fan et al. 2008; Fattoum et al. 1983; Ishikawa et al. 1989a; Ishikawa et al. 1989b; Nakano and Mabuchi 2006; Ono and Ono 2002), filament cappers (Fowler et al. 1993; Weber et al. 1994), and actin nucleation factors (Blanchoin et al. 2001; Skau et al. 2009; Wawro et al. 2007). The mechanism by which tropomyosin regulates myosin has been well-studied in muscle, yet we are only now beginning to learn how tropomyosin regulates myosin function in non-muscle cells. There is still much to be learned in this area given the multiple classes of myosins and >40 tropomyosin isoforms expressed in mammalian cells (Gunning et al. 2005).
Fission yeast has recently proved to be a useful model with which to study tropomyosin function owing to its tractable genetics, well-defined actin cytoskeleton, and established actin biochemistry (Kovar et al. 2011). Fission yeast tropomyosin (Cdc8p) is restricted to the unbranched formin-mediated actin filaments that make up the contractile rings used in cytokinesis and the actin cables used in intracellular transport and maintenance of cell shape (Balasubramanian et al. 1992; Johnson et al. 2014; Kovar et al. 2011; Lo Presti et al. 2012). By contrast, Cdc8p is absent from the Arp2/3-mediated branched actin utilized in endocytic patches (Balasubramanian et al. 1992; Kovar et al. 2011). Each of the three fission yeast actin structures associate with different myosins (East and Mulvihill 2011; Kovar et al. 2011), and previous work with Cdc8p highlighted a role for tropomyosin in the regulation of these actin-based motors. As opposed to limiting access to the actin track as in muscle (Lehrer 1994), decoration of actin with Cdc8p positively regulates myosin-II (Myo2p) activity and function at contractile rings (Stark et al. 2010). In addition, myosin-V (Myo52p) activity and transport along cables is favored by Cdc8p (Clayton et al. 2014; Clayton et al. 2010). In contrast to Myo2p and Myo52p, decoration of actin with Cdc8p inhibits myosin-I (Myo1p) in vitro (Clayton et al. 2010). This negative regulation presumably helps to restrict Myo1p motor activity to the branched actin networks (that lack Cdc8p) at endocytic patches.
Tropomyosins form filaments that associate weakly with actin, forming a co-polymer in which tropomyosin wraps longitudinally along both sides of the actin filament (Gunning et al. 2005). In such copolymers each tropomyosin covers 4–7 actin monomers depending on the tropomyosin isoform in play (Gunning et al. 2005). Neighboring tropomyosins associate with one another via end-to-end interactions between their N- and C-termini (Hitchcock-DeGregori 2008). These interactions account for cooperative tropomyosin-actin associations and facilitate tropomyosin self-assembly at low ionic strength (Tobacman 2008). Mutational analysis of the Cdc8p N-terminus revealed a role for this region in promoting actin association and Cdc8p polymer formation (East et al. 2011).
Interestingly, Cdc8p can be acetylated on its N-terminal methionine, a modification that alters actin-tropomyosisn structure, enhances actin associations, and contributes to Cdc8p function in the cell (Coulton et al. 2010; Skoumpla et al. 2007). Acetylated Cdc8p predominates at the contractile ring and is important for myosin-II localization and ring function, whereas unacetylated Cdc8p predominates at actin cables and faithfully supports Myo52p transport in the cell (Coulton et al. 2010). Thus, two specific forms of tropomyosin are generated from one isoform in fission yeast. The choice of fission yeast actinnucleation factor (i.e. the formin isoform) distinguishes which of the two forms of Cdc8p are recruited to the two different unbranched actin filaments structures in the cell (Johnson et al. 2014).
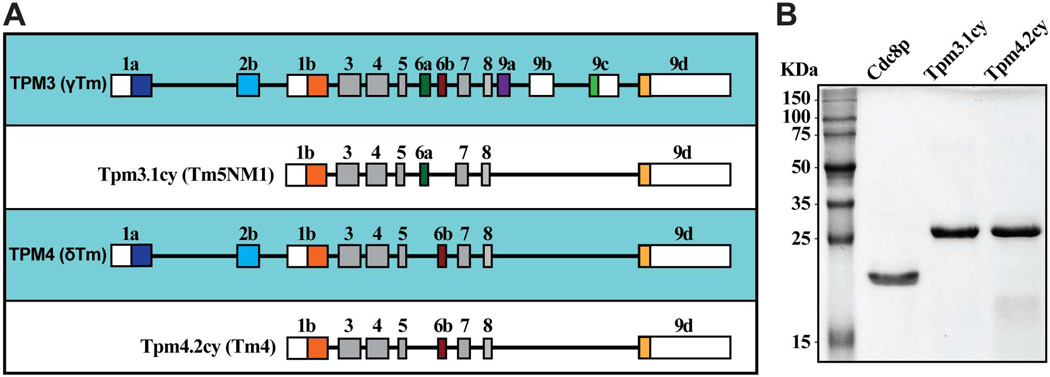
In contrast to fission yeast, a large variety of tropomyosins are expressed in mammalian cells, primarily generated through alternative splicing of a uniform set of exons from four different genes (TPM1-TPM4) (Geeves et al. 2014; Gunning et al. 2005). Some isoforms are expressed in a tissue-specific manner and are classified as striated muscle-specific (from skeletal and cardiac muscle tissues), smooth muscle-specific, or brain-specific. The remaining isoforms are referred to as ‘cytoplasmic’, being distributed amongst all other cell types. The different tropomyosins can also be broken down into two general classes based on sequence length. Low molecular weight (LMW) tropomyosins are ~248 amino acids in length and incorporate exon 1b of the TPM gene (Figure 1A); whereas high molecular weight (HMW) tropomyosins are ~284 amino acids long and utilize exons 1a and 2a (or 2b) (Figure 1A).
Figure 1. Tropomyosins under study.
A) Schematic illustration summarizing exon usage by mammalian tropomyosin TPM3 and TPM4 gene products Tpm3.1cy and Tpm4.2cy. While the two proteins are the products of two different genes, their exon usage makes these two LMW tropomyosins very similar. The only obvious difference being that Tpm3.1cy employs exon 6a, while Tpm4.2cy employs exon 6b. B) Purified protein samples following SDS-PAGE and gel staining with Coomassie Blue. Molecular weights are included on the left. Cdc8p and the mammalian tropomyosins were purified from bacteria (acetylation-mimicking forms including the Ala-Ser dipeptide are shown).
In this study we compared the influence of Cdc8p and two LMW cytoplasmic tropomyosins implicated in cancer cell growth and metastasis on four different myosins (Myo2p, Myo52p, skeletal muscle myosin-II, and chick brain myosin-Va). A combination of in vitro and in vivo approaches suggest that the ability of tropomyosin to positively regulate non-muscle myosins Myo2p and Myo52p is not restricted to Cdc8p. Moreover, the effects of tropomyosin on myosin activity differ depending on the myosin isoform involved underscoring a general concept in tropomyosin-mediated myosin regulation. Such regulation has the potential to provide tight specification of both non-muscle and muscle actomyosin function in complex cellular environments. This work has broad implications with regard to sorting of actomyosin function in cancer cells and developing muscle tissue.
RESULTS
Selecting representative mammalian tropomyosin isoforms
We wished to compare the role of fission yeast Cdc8p with non-muscle mammalian tropomyosins previously implicated in myosin function in cancer cells. We chose to focus on two LMW tropomyosin isoforms (Tpm3.1cy/Tm5NM1 and Tpm4.2cy/Tm4) originating from the TPM3 and TPM4 genes respectively (Figure 1A). We are employing the latest nomenclature to describe mammalian tropomyosin isoforms (Geeves et al. 2014). As such Tpm3.1cy/Tm5NM1 and Tpm4.2cy/Tm4 will be referred to as Tpm3.1cy and Tpm4.2cy respectively from here on in.
Our choice of tropomyosins reflects information gleaned from previous work. Tpm3.1cy and Tpm4.2cy were shown to regulate stress fiber formation and myosin-II recruitment to fibers in B35 neuroblastoma and U2OS osteosarcoma cells respectively (Bryce et al. 2003; Tojkander et al. 2011). Furthermore, Tpm3.1cy, its closely related isoform Tpm3.2cy, and Tpm4.2cy represent the predominant LMW isoforms found in primary tumors and tumor cell lines (Stehn et al. 2006). Interestingly, Tpm3.1cy and Tpm3.2cy were recently shown to be essential for neuroblastoma cell proliferation, and represent the only two tropomyosin isoforms up-regulated upon transformation of both primary immortalized human BJ fibroblasts and human melanoma cells (Stehn et al. 2013). Previous studies revealed that overexpression of Tpm3.1cy generates actin stress fibers exhibiting reduced susceptibility to traditional actin-targeting drugs, suggesting that tropomyosin composition can influence drug action (Creed et al. 2008). Importantly, recently developed anti-cancer compounds targeting Tpm3.1cy and Tpm3.2cy are showing promise based on their ability to inhibit tumor cell motility and growth (Stehn et al. 2013).
N-terminal acetylation of tropomyosin has been shown to be important for both Cdc8p (Coulton et al. 2010; Skoumpla et al. 2007) and striated muscle α-tropomyosin/Tpm1.1st (Heald and Hitchcock-DeGregori 1988; Hitchcock-DeGregori and Heald 1987; Urbancikova and Hitchcock-DeGregori 1994) function. While post-translational modification offers another potential interesting avenue of regulation, we largely focused on the function of acetylated-mimicking forms of Cdc8p, Tpm3.1cy, and Tpm4.2cy purified following over-expression in E. coli (Figure 1B). Chicken skeletal muscle actin represents the standard actin isoform employed in all our in vitro studies.
Differential regulation of Myo2p and skeletal muscle myosin-II by mammalian tropomyosins
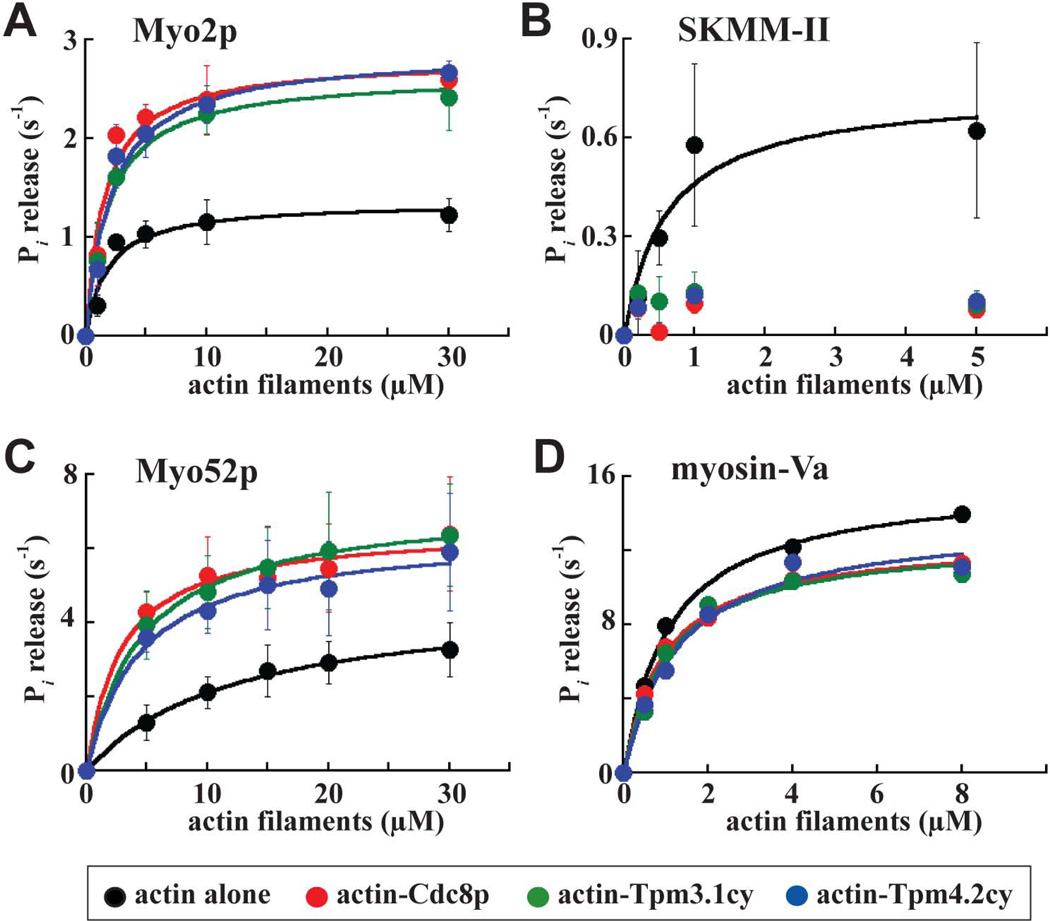
We compared the ability of Cdc8p, Tpm3.1cy, and Tpm4.2cy to regulate full-length fission yeast myosin-II (Myo2p) and full-length skeletal muscle myosin-II (SKMM-II) using bulk actin-activated ATPase and ensemble actin filament gliding assays. The activities of Myo2p and SKMM-II were compared in an identical fashion in both assays. The concentration of myosin used in the ATPase assays was generally much lower (0.03–0.09 µM) than the various concentrations of actin/actin-tropomyosin filaments present (0.1–30 µM). In contrast, the concentration of myosin (5 nM) employed in the filament gliding assays was closer to the actin/actin-tropomyosin filament concentration used (25 nM).
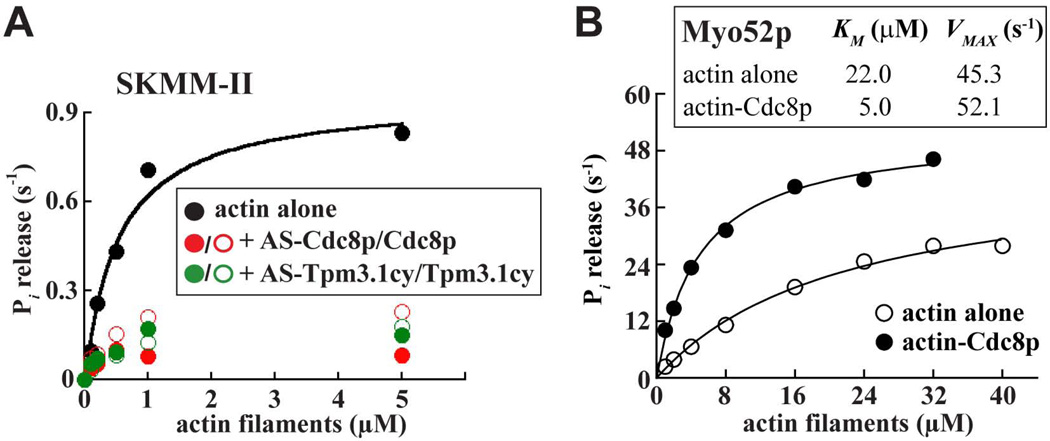
Michaelis-Menten analysis was employed to compare the ATPase activity of Myo2p and SKMM-II with actin or actin-tropomyosin filaments. Cdc8p increased the maximal ATPase rate (VMAX) and the overall catalytic efficiency of Myo2p approximately 2-fold (Figure 2A; Table I). Tpm3.1cy and Tpm4.2cy also enhanced Myo2p ATPase activity with a similar potency (Figure 2A; Table I). In stark contrast to the positive effect of tropomyosins on Myo2p, Cdc8p and both cytoplasmic tropomyosins inhibited the ATPase activity of SKMM-II (Figure 2B; Table I). This inhibition does not appear to depend on the acetylation status of the tropomyosins since absence of the acetylation-mimicking Ala-Ser dipeptide (on the N-termini of Cdc8p or Tpm3.1cy) did not significantly influence the ability of the tropomyosins to inhibit SKMM-II activity (Figure 3A).
Figure 2. Regulation of Myo2p, SKMM-II, Myo52p, and myosin-Va motor activity by tropomyosin.
The actin-activated Mg2+-ATPase activity of Myo2p, SKMM-II, Myo52p, and myosin-Va was measured as a function of actin or actin-tropomyosin concentration. Basal myosin ATPase activities and background Pi from actin or actin-tropomyosin were subtracted from all measurements. Each average curve fit presented was generated from average values obtained from 4 (Myo2p and Myo52p), 3 (SKMM-II), and 1 (myosin-Va) dataset(s). A) Myo2p, B) SKMM-II, C) Myo52p, and D) myosin-Va ATPase activities compared with and without Cdc8p, Tpm3.1cy, and Tpm4.2cy. All the values are summarized in Table I. The myosin concentrations in these assays were: 30 nM (Myo2p), 90 nM (SKMM-II), 10 nM (Myo52p), and 14 nM (myosin-Va). Actin filaments were pre-incubated with tropomyosin at a molar ratio of 2 actin:1 tropomyosin. The Myo2p, SKMM-II, and Myo52p curves were generated from data measured using a malachite green-based Pi detection assay. The myosin-Va curves were generated from data measured using an NADH-linked-/ATP-regenerating-based ATPase assay. Myo52p curves generated using the ATP-regenerating-based assay are provided in Figure 3B.
Table I. Effect of Cdc8p and the mammalian tropomyosins on the motor activity and motility of Myo2p, SKMM-II, Myo52p, and myosin-Va.
Table summarizes how tropomyosins Cdc8p, Tpm3.1cy, and Tpm4.2cy influence myosin motor performance.
| Myosin and actin track |
aActin-activated ATPase | Actin filament gliding | |||
|---|---|---|---|---|---|
|
VMAX (s−1) |
KM (µm) |
bcatalytic efficiency |
cspeed (µm·s−1) |
dmotility efficiency |
|
| Myo2p | |||||
| actin alone | 1.4 ± 0.1 | 1.8 ± 0.6 | 0.77 | 0.48 ± 0.06 | 1.00 |
| actin-Cdc8p | 2.8 ± 0.2 | 1.5 ± 0.5 | 1.87 | 0.36 ± 0.03 | 1.64 |
| actin-Tpm3.1cy | 2.7 ± 0.2 | 1.8 ± 0.3 | 1.50 | 0.38 ± 0.04 | 1.80 |
| actin-Tpm4.2cy | 2.9 ± 0.2 | 2.1 ± 0.5 | 1.38 | 0.41 ± 0.05 | 1.62 |
| SKMM-II | |||||
| actin alone | 0.74 ± 0.12 | 0.6 ± 0.3 | 1.23 | 2.85 ± 0.84 | 1.00 |
| actin-Cdc8p | ND | ND | ND | 0.61 ± 0.23 | 0.16 |
| actin-Tpm3.1cy | ND | ND | ND | 1.59 ± 0.62 | 0.20 |
| actin-Tpm4.2cy | ND | ND | ND | 0.97 ± 0.44 | 0.07 |
| Myo52p | |||||
| actin alone | 4.7 ± 0.2 | 9.3 ± 0.9 | 0.48 | 0.84 ± 0.19 | 1.00 |
| actin-Cdc8p | 6.6 ± 0.4 | 2.8 ± 0.8 | 2.33 | 0.47 ± 0.11 | 1.19 |
| actin-Tpm3.1cy | 7.2 ± 0.2 | 4.5 ± 0.4 | 1.60 | 0.56 ± 0.10 | 1.37 |
| actin-Tpm4.2cy | 6.4 ± 0.4 | 4.4 ± 1.0 | 1.46 | 0.54 ± 0.15 | 1.18 |
| Myosin-Va | |||||
| actin alone | 16.3 | 1.2 | 13.6 | 0.23 ± 0.05 | 1.00 |
| actin-Cdc8p | 12.7 | 1.0 | 12.7 | 0.23 ± 0.04 | 1.16 |
| actin-Tpm3.1cy | 12.7 | 1.0 | 12.7 | 0.26 ± 0.08 | 1.06 |
| actin-Tpm4.2cy | 13.7 | 1.3 | 10.5 | 0.26 ± 0.05 | 1.22 |
Myo2p, SKMM-II, Myo52p, and myosin-Va actin-activated ATPase values were generated from the average curves presented in Figure 2A–D. Assays utilized a malachite green-based Pi detection system, except for myosin-Va where an ATP-regenerating-based ATPase assay was employed (see Supplemental Procedures for further details). Datasets: n=4 (Myo2p and Myo52p), n=3 (SKMM-II), and n=1 (myosin-Va).
Catalytic efficiency = VMAX/KM.
n=50–150. Paired Student’s t-tests were used to test the significance of the tropomyosin-mediated changes in actin filament gliding rate versus actin alone (p <0.05 indicates a significant difference between datasets). All tropomyosins significantly reduced the gliding rates of Myo2p (Cdc8p, <0.0001; Tpm3.1cy, <0.0001; Tpm4.2cy, <0.0001), SKMM-II (Cdc8p, <0.0001; and Tpm3.1cy, <0.0001; Tpm4.2cy, <0.0001), and Myo52p (Cdc8p, <0.0001; and Tpm3.1cy, <0.0001; Tpm4.2cy, <0.0001). However, in the case of myosin-Va, two out of the three tropomyosins tested (Tpm3.1cy and Tpm4.2cy) yielded a significant increase in gliding rate, while Cdc8p had no significant effect (Cdc8p, 0.756; Tpm3.1cy, 0.036; Tpm4.2cy, 0.0006). We performed additional statistical analyses to gauge any significant differences between the effects of the three different tropomyosins on motility. For Myo2p, there was significant difference for Cdc8p vs. Tpm3.1cy (0.02) and Cdc8p vs. Tpm4.2cy (0.0015), but no significant difference for Tpm3.1cy vs. Tpm4.2cy (0.15). For SKMM-II, there were significant differences between all of the tropomyosins (<0.0001 for all combinations). For Myo52p, there was significant difference for Cdc8p vs. Tpm3.1cy (0.0002) and Cdc8p vs. Tpm4.2cy (0.0139), but no significant difference for Tpm3.1cy vs. Tpm4.2cy (0.0786). For myosin-Va, there was significant difference for Cdc8p vs. Tpm3.1cy (<0.0001) and Tpm3.1cy vs. Tpm4.2cy (0.0004), but no significant difference for Cdc8p vs. Tpm4.2cy (0.183).
Relative number of motile (myosin-driven) actin filaments/total actin filaments. A total of five different movie fields were scored for each sample at a set concentration of myosin. The relative number of motile filaments scored for actin alone was set to 1.0.
Figure 3. Assessment of tropomyosin acetylation on SKMM-II inhibition and Cdc8p–mediated activation of Myo52p in actin-activated ATPase assays.
A) The influence of acetylated-mimicking versus unacetylated forms of both Cdc8p and Tpm3.1cy on the actin-activated ATPase activity of SKMM-II were compared in parallel (in the same fashion as the SKMM-II experiments described in Figure 2). The fitted values for each condition are the averages from two independent experiments. B) The Myo52p VMAX values obtained from ATPase assays employing malachite green-based Pi detection (Figure 2C; Table I) were lower than what we would predict from the speeds at which single Myo52p molecules move on Cdc8p-decorated actin tracks (Clayton et al. 2014). Assuming Myo52p has a 36 nm step size similar to other myosin-Vs (Trybus 2008), a single Myo52p molecule must take ~35 steps per second to achieve the average speed (1.25 µm/s) of Myo52p on Cdc8p-actin tracks (Clayton et al. 2014). This rate of 35 s−1 is ~5-fold higher than what we obtained (~7 s−1) from our ATPase data (Table I). In light of these observations we employed an alternative assay that utilizes an ATP-regenerating system to avoid any inhibitory build-up of ADP. This assay revealed a higher VMAX of 52 s−1 for Myo52p, closer to the 35 s−1 predicted from single molecule studies. Nevertheless, the general trend of tropomyosin-mediated activation of Myo52p remained the same: the presence of Cdc8p on actin increased the VMAX from 45 to 52 s−1 (compared to 4.7 to 6.6 s−1 in our original assays; Table I) and significantly reduced the KM from 22 to 5 µM (compared to 9.3 to 2.8 s−1 in our original assays; Table I). The steady-state actin-activated ATPase activity of Myo52p was measured as a function of actin concentration (actin alone: open circles; actin plus Cdc8p: filled circles). Myo52p was included in the assays at a concentration of 10 nM. Lines are fits to Michaelis-Menten kinetics. The measurements and fitted values (shown inset) are the averages from two independent experiments.
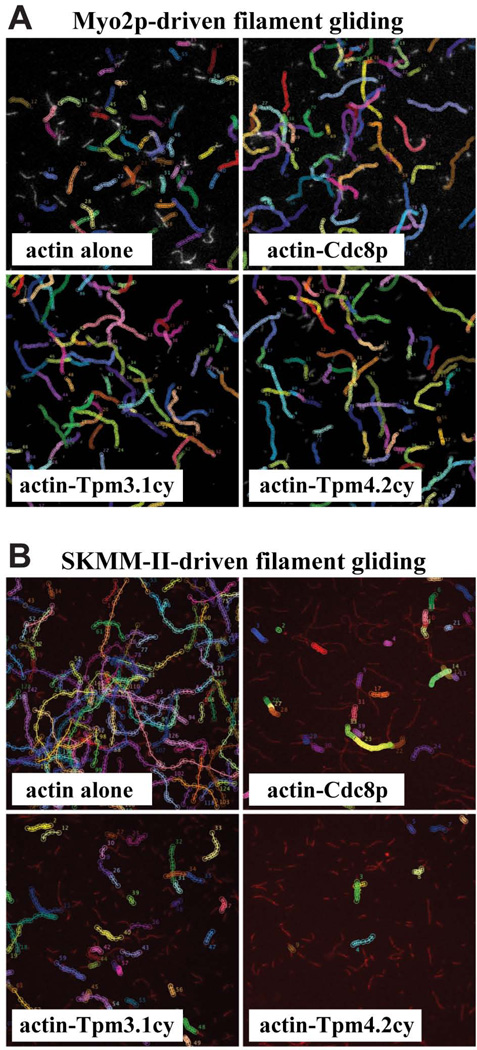
Decoration of actin filaments with Cdc8p led to a significant drop in the rate of Myo2p–driven actin filament gliding, an effect that was also observed with Tpm3.1cy and Tpm4.2cy (Table I). This drop in rate was statistically more penetrant for actin-Cdc8p cf. the mammalian tropomyosins (Table I). Based on past studies with Myo2p and Cdc8p (Stark et al. 2010), this tropomyosin-mediated reduction in speed does not reflect inhibition per se, more so it reflects a Cdc8p-mediated bias for the strong actin-bound ADP or apo states of Myo2p (Stark et al. 2010), states which are rate-limiting for myosin motility (Siemankowski et al. 1985). Consistent with such a positive effect, the presence of Cdc8p, Tpm3.1cy, and Tpm4.2cy increased the persistence and efficiency of Myo2p–driven filament gliding (Figure 4A; Table I).
Figure 4. Regulation of Myo2p- and SKMM-II-driven actin filament gliding by tropomyosin.
The efficiency of myosin-driven actin filament gliding is portrayed using representative fields of (rhodamine-phalloidin labeled) filaments generated from time-lapse epi-fluorescence microscopy experiments. Images shown are maximum projections of 60 frames captured every second for both Myo2p (A) and SKMM-II (B). Myo2p and SKMM-II were employed at a working concentration of 5 nM (the same concentration used to generate the filament gliding data presented in Table I). The trajectory of filament movement over time is tracked and summarized by marking (with colored circles for each frame) the leading ends of any filament exhibiting motility during the movies. Trajectories of filament motility were compared for bare actin and the indicated tropomyosin-decorated filaments. A) The motility events for Myo2p with bare actin filaments are a little faster (circles slightly further apart) yet shorter in duration (shorter trajectory length) compared with the events tracked with actin-tropomyosin filaments. This reflects the slightly slower yet more continuous and efficient filament motility observed for Myo2p with tropomyosin-actin filaments. (Table I). B) The motility events for SKMM-II with bare actin filaments are much faster (circles an obvious distance apart), more numerous, and more longer lived compared with the events tracked with actin-tropomyosin filaments. This reflects the much slower and much less efficient filament motility observed with tropomyosin-actin filaments (where many more filaments associated with the coverslip surface are non-motile or fail to sustain persistent motility) (Table I). The speeds and efficiencies of SKMM-II-driven actin filament gliding were also compared with and without tropomyosins at higher SKMM-II concentrations (Figure 5A–B).
In contrast to the results obtained with Myo2p, decoration of actin with Cdc8p or the mammalian tropomyosins led to a much bigger reduction in SKMM-II-driven actin filament gliding speed (Table I). While these reduced speeds varied statistically (between the different tropomyosins tested), they all reflected tropomyosin-mediated inhibition as the number of motile filaments and motility efficiency of SKMM-II was greatly diminished in the presence of each tropomyosin (Figure 4B; Table I).
Recent studies have indicated that the use of saturating amounts of SKMM-II avoids tropomyosin-mediated inhibition of filament gliding (Barua et al. 2014). Nevertheless, we found that the inhibitory effects of the tropomyosins on motility speed and efficiency were still maintained when saturating concentrations of SKMM-II were employed (Figure 5A–B). These contrasting results may reflect differences in the way SKMM-II preparations were attached to the slides in motility chambers. Barua et al. employed high salt to facilitate attachment of individual SKMM-II molecules, whereas we employed lower concentrations of salt to pre-assemble SKMM-II filament ensembles prior to attachment.
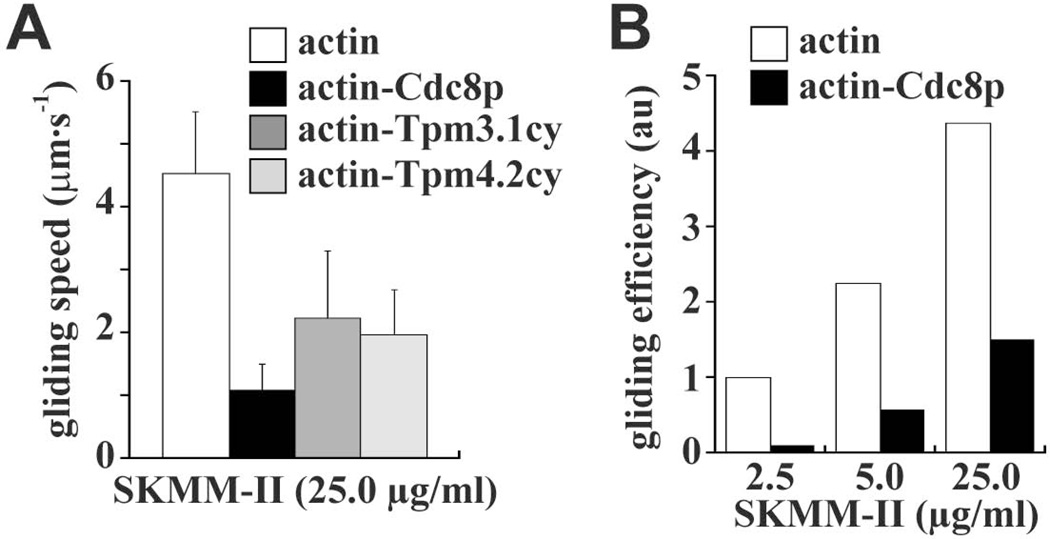
Figure 5. Effect of tropomyosin on SKMM-II-driven actin filament gliding in relation to myosin concentration.
The working concentration of SKMM-II employed in our actin filament gliding assays (Figure 4B; Table I) was 5 nM (2.5 µg/ml). We also compared the motility activity of SKMM-II at higher myosin concentrations. A) The effect of saturating SKMM-II (50 nM/25.0 µg/ml) was tested with bare actin filaments versus Cdc8p-, Tpm3.1cy-, or Tpm4.2cy-decorated filaments. The histogram summarizes the SKMM-II filament gliding rates (n=30–70 filaments per experiment). B) Histogram summarizing SKMM-II gliding efficiency versus myosin concentration with bare actin filaments versus Cdc8p-decorated filaments. Gliding efficiency reflects the number of motile (myosin-driven) actin filaments/total actin filaments. A total of 2–5 different movie fields were scored. The relative number of motile filaments scored for actin alone at 2.5 µg/ml was set to 1.0.
In summary, while Cdc8p and the mammalian tropomyosins had an obvious positive effect on non-muscle Myo2p motor function, these same tropomyosins potently inhibited SKMM-II motor function in the very same assays.
Differential regulation of Myo52p and myosin-Va by mammalian tropomyosins
Myosin-Vs are transporters that typically operate as processive motors capable of taking multiple steps along actin filaments without falling off. Consequently, most myosin-V isoforms studied to date are high duty ratio motors, meaning that they spend a large proportion of their ATPase cycle in the strong actin-bound state. The high duty ratio and dimeric nature of these molecules facilitates processive, hand-over-hand walking along actin (De La Cruz et al. 1999; Mehta et al. 1999). Mammalian myosin-Va represents a well-studied and highly processive form of myosin-V (De La Cruz et al. 1999; Mehta et al. 1999). In contrast to myosin-Va, fission yeast Myo52p has a relatively low duty ratio for a myosin-V and is non-processive (Clayton et al. 2014; Clayton et al. 2010). Myo52p processivity and recruitment to actin cables in the cell relies on Cdc8p (Clayton et al. 2014). We wished to compare the ability of Cdc8p and the mammalian tropomyosins to regulate Myo52p and myosin-Va actin-activated ATPase activity and motility. Truncated heavy meromyosin (HMM) forms of Myo52p and myosin-Va were used that lack the C-terminal cargo-binding globular tail domain. These HMM forms represent dimeric myosin-V molecules possessing the motor domain, the light chain-binding region, and the coiled-coil region.
Like Cdc8p, Tpm3.1cy, and Tpm4.2cy enhanced the activity of Myo52p motors in ATPase assays, as evidenced by a lower KM, higher VMAX, and greater overall catalytic efficiency compared to bare actin filaments (Figure 2C; Table I). Similar changes in Myo52p KM and VMAX values were also obtained in the presence of Cdc8p when using an alternative ATPase assay that utilizes an ATP-regenerating system (Figure 3B). This system avoids inhibitory build-up of ADP, which is common with high duty ratio motors like myosin-V. This assay revealed higher VMAX values for Myo52p, values closer to those one would predict from the Myo52p single molecule motility speeds previously reported from our laboratory (Clayton et al. 2014) (Figure 3B).
We compared the effect of Cdc8p and the mammalian tropomyosins on myosin-Va actin-activated ATPase activity utilizing the ATP-regenerating system (Figure 2D). In contrast to the obvious effects seen with Myo52p (where the tropomyosins increased VMAX, reduced KM, and increased catalytic efficiency), the tropomyosins had little effect on myosin-Va activity. The presence of the three different tropomyosins (if anything) only slightly reduced the VMAX of myosin-Va, while having little effect on KM and catalytic efficiency values (Figure 2D; Table I).
Similar to the effects of Cdc8p on Myo2p motility (Table I), decoration of actin with Cdc8p significantly reduced the speed of Myo52p motility in filament gliding assays (Table I). Consistent with the positive effect of the mammalian tropomyosins on Myo52p ATPase activity (Figure 2C; Table I), Tpm3.1cy, and Tpm4.2cy had the same effect as Cdc8p and significantly reduced Myo52p actin gliding rate (Table I). As seen with Myo2p, this drop in Myo52p motility rate was statistically more penetrant for actin-Cdc8p cf. the mammalian tropomyosins (Table I). As with Myo2p, we predict that the reduced rates of gliding reflect the ability of tropomyosin to favor movement into the strong actin-bound state. In terms of motility efficiency, all three tropomyosins increased the frequency of Myo52p–driven filament gliding events (Table I), similar to the effect of the tropomyosins on Myo2p motility (Table I).
In contrast to Myo52p, decoration of actin with Cdc8p or the mammalian tropomyosins had minor effects on myosin-Va-driven filament gliding (Table I), consistent with the negligible effects of these tropomyosins on myosin-Va ATPase activity (Figure 2D). While decoration of actin with Cdc8p did not alter myosin-Va gliding speed, decoration with Tpm3.1cy or Tpm4.2cy yielded minor increases in gliding speed (Table I). The motility efficiency of myosin-Va was slightly improved in the presence of both Cdc8p and Tpm4.2cy, a feature which may reflect the wider distribution of motility rates observed with actin-Tpm3.1cy (Table I).
In summary, Cdc8p and the mammalian tropomyosins had an obvious positive effect on the non-processive Myo52p motor, whereas analysis of the same tropomyosins in the same assays revealed only minimal effects on the highly processive myosin-Va motor. Our previous work (Clayton et al. 2014) and the new findings with the mammalian tropomyosins implies that Cdc8p, Tpm3.1cy, and Tpm4.2cy favor the strong actin-bound state of myosin, which in turn favors an increase in the duty ratio of non-processive myosin-Vs.
Activation of fission yeast myosin-V processivity by mammalian tropomyosins
We previously used total internal reflection fluorescence (TIRF) microscopy to show that Cdc8p is required for single Myo52p motors to move processively along actin filaments (Clayton et al. 2014). We repeated these experiments to test whether the mammalian tropomyosins could also regulate Myo52p processivity. The Myo52p–HMM employed had a C-terminal biotin tag for attachment to streptavidin-coated fluorescent quantum dots (Qdots) (Clayton et al. 2014). Fluorescently-labeled actin filaments were attached to a glass coverslip, and a solution containing Myo52p-labeled with Qdots was applied. We assume our labeling approach achieves single molecule resolution based on optimization in previous studies (Clayton et al. 2014; Hodges et al. 2009; Hodges et al. 2012).
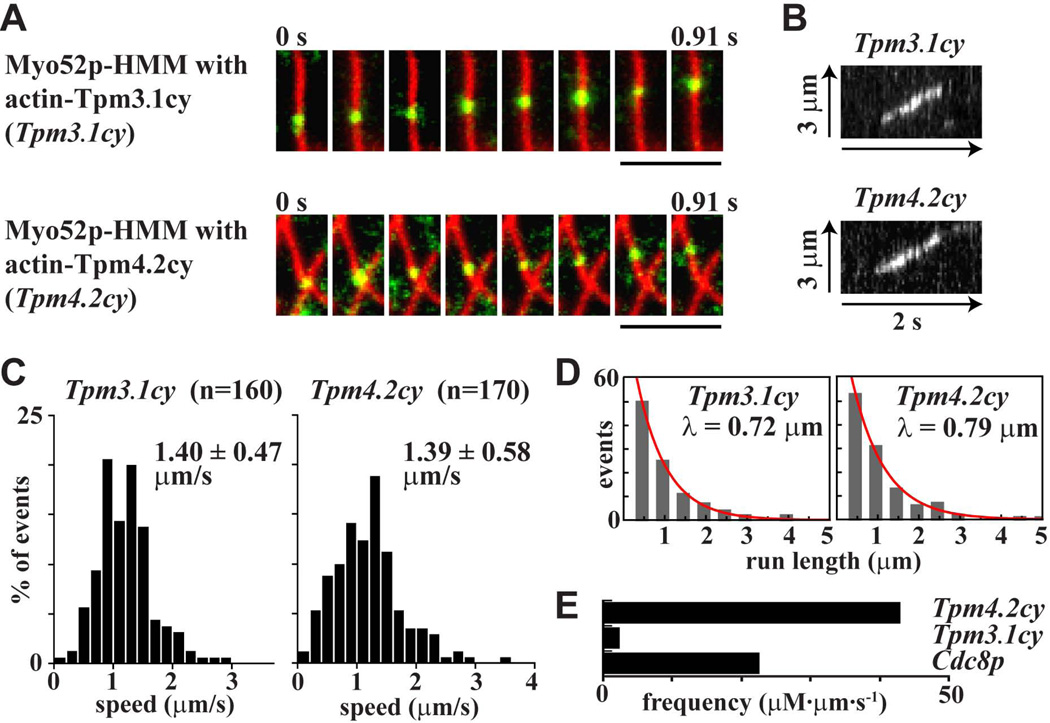
Consistent with our ATPase and filament gliding results, Tpm3.1cy and Tpm4.2cy behaved like Cdc8p and activated efficient processive movements of Myo52p along actin filaments (Figure 6A–B; see Movies S1 and S2 in supplementary material). The speeds and run lengths of these processive movements along Tpm3.1cy- and Tpm4.2cy-decorated actin tracks were in fact a little higher (Figure 6C–D) than those previously reported for Cdc8p–decorated tracks (speed: 1.25 ± 0.43 µm·s−1; run length: 0.64 µm) (Clayton et al. 2014). The extent to which the different tropomyosin isoforms supported processivity varied with respect to run frequency. The run frequency with Tpm4.2cy was higher than with Cdc8p, while the frequency with Tpm3.1cy was lower than Cdc8p (Figure 6E). In summary, the ability of Cdc8p to convert Myo52p to a processive motor is not restricted to yeast tropomyosin. This processivity can be accounted for by a tropomyosin-mediated increase in the myosin-V duty ratio.
Figure 6. Regulation of Myo52p processivity by tropomyosin.
Processive movement of Myo52p-HMM molecules coupled to Qdots along tropomyosin-decorated rhodamine phalloidin-labeled actin filaments using TIRF microscopy. A) Movement of a Myo52p-Qdot (green) along Tpm3.1cy- and Tpm4.2cy-decorated actin filaments (red). Bars: 4 µm. Representative events are presented in Movies S1 and S2. B) Kymographs of processive runs of Myo52p-HMM along actin filaments decorated with Tpm3.1cy and Tpm4.2cy. C) Histograms of the speed distributions for processive Myo52p runs along actin filaments decorated with Tpm3.1cy and Tpm4.2cy. D) Run length histograms for Myo52p movements along actin filaments decorated with Tpm3.1cy and Tpm4.2cy. The red curves show exponential fits (y = Ae-x/λ) to determine the run length λ. E) Histogram comparing the frequencies of processive Myo52p runs on actin filaments decorated with Cdc8p, Tpm3.1cy or Tpm4.2cy. Frequency values reflect the number of Myo52p events (runs) observed per µm of actin-tropomyosin filament track available over time. Final values incorporate the myosin concentration to yield frequency as µM Myo52p/µm actin/second.
Mammalian tropomyosin Tpm3.1cy can function in place of fission yeast Cdc8p in vivo
Fission yeast actomyosin contractile ring assembly, cytokinesis, and cell growth relies on both Myo2p motor activity and Cdc8p (Balasubramanian et al. 1992; Coffman et al. 2009; Kitayama et al. 1997; Lord et al. 2005; May et al. 1997; Stark et al. 2010). Likewise, recent studies highlighted an essential role for Tpm3.1cy in cancer cell proliferation (Stehn et al. 2013). Given that Tpm3.1cy and Tpm4.2cy can function like Cdc8p in their ability to positively regulate Myo2p activity in vitro (Table I), we wished to test whether either of these tropomyosins could function in place of Cdc8p in vivo. The ability of mammalian tropomyosin to support Cdc8p function is not unprecedented given that rat HMW tropomyosin Tm2 (Tpm1.6cy) can functionally substitute for Cdc8p in fission yeast (Balasubramanian et al. 1992).
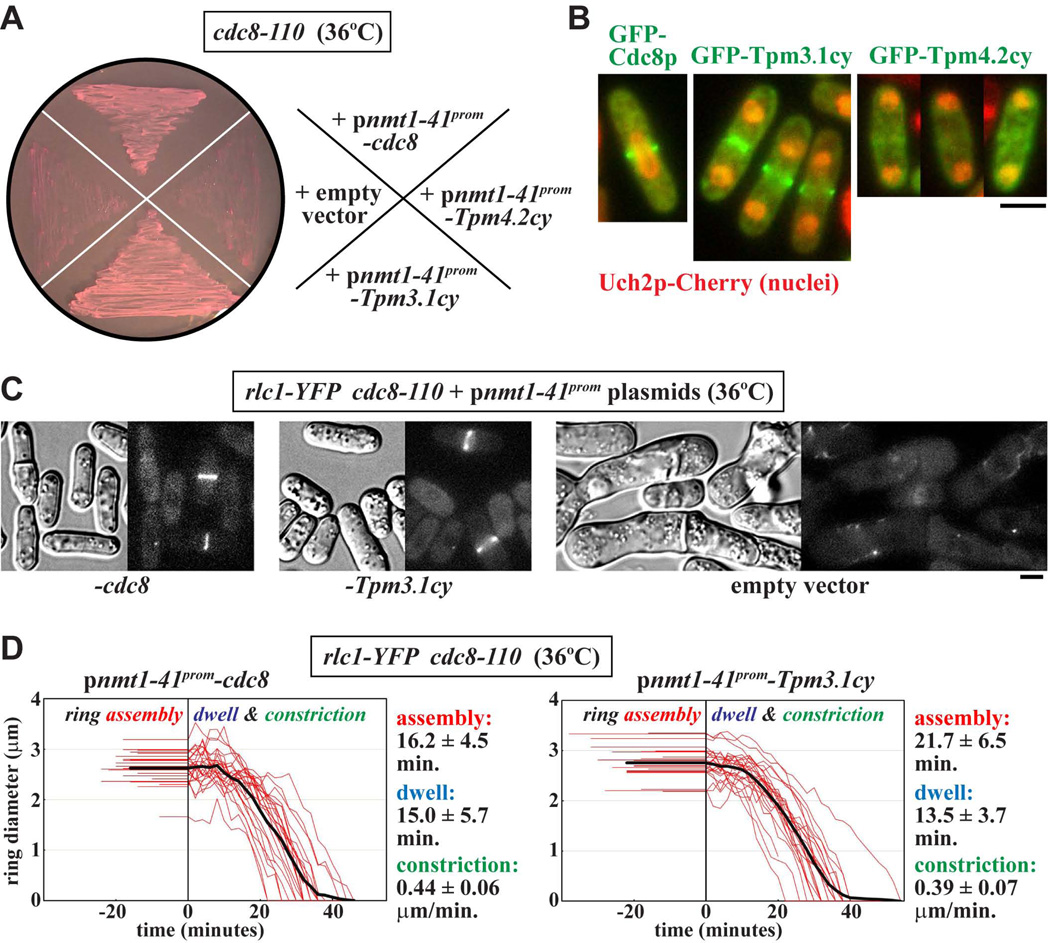
We tested whether Tpm3.1cy or Tpm4.2cy could rescue the lethal cytokinesis defects associated with loss of Cdc8p function in the temperature-sensitive cdc8–110 mutant. While cdc8–110 cells carrying plasmids expressing (untagged) Cdc8p, Tpm3.1cy, or Tpm4.2cy all grew at the permissive growth temperature (25°C), only mutant cells expressing Cdc8p or Tpm3.1cy could sustain growth at the restrictive temperature (36°C) (Figure 7A). Although GFP-Cdc8p and GFP-Tpm3.1cy were routinely found to concentrate at rings during cytokinesis, screening of many cells failed to detect any fluorescently-labeled rings upon GFP-Tpm4.2cy expression. Thus, Tpm4.2cy may simply fail to rescue cytokinesis in the cdc8–110 background owing to its failure to effectively target actin in the contractile ring (Figure 7B).
Figure 7. Mammalian tropomyosin Tpm3.1cy rescues Cdc8p function in vivo.
The ability of Tpm3.1cy or Tpm4.2cy to substitute for Cdc8p during cytokinesis was tested in fission yeast. Tropomyosin (either untagged or GFP-tagged) expression was driven from the inducible nmt41x promoter in fission yeast plasmids (see Experimental Procedures for details). A) Temperature-sensitive cdc8-110 cells were transformed with various ura4+ plasmids and isolated under permissive growth conditions (25°C) on EMM-Ura minimal media plates. The plate shown indicates the ability/inability of the different plasmids to rescue Cdc8p function. Here transformants were grown on EMM-Ura plates at the restrictive temperature (36°C) to attenuate Cdc8-110p function and select for rescue. The plates contained 5 µg/ml phloxin B, a pink dye that accumulates inside dead cells (note the darker pink color of the cell streaks that failed to grow). Plasmids harbored untagged cdc8 (positive control), cDNAs for Tpm3.1cy or Tpm4.2cy, or lacked an insert altogether (empty vector, negative control). B) Images of wild-type cells expressing GFP-tagged versions of Cdc8p, Tpm3.1cy, or Tpm4.2cy were captured during cytokinesis. The strain background includes a uch2-mCherry fusion integrated into the chromosome to mark dividing/divided nuclei. Cells were grown in EMM-Ura minimal media at 30°C. Scale bar: 4 µm. C) DIC and fluorescence images of cdc8-110 cells expressing untagged Cdc8p (left) or Tpm3.1cy (center). Control cells carrying an empty vector are also shown (right). Cells were grown at 36°C in EMM-Ura minimal media, and Rlc1p-YFP contractile ring structures visualized via an integrated rlc1-mYFP fusion. Scale bar: 4 µm. D) Plots charting the assembly, dwell, and constriction phases of individual ring diameter traces (red, n=25) from cdc8-110 cells expressing untagged Cdc8p (left) or Tpm3.1cy (right). The thick black lines reflect average fits of all the individual ring traces. Ring dynamics were recorded using time-lapse epi-fluorescence microscopy of Rlc1p-YFP. For each plot the length of flat line traces shown at negative time represent the assembly phase, while the line traces shown at positive time correspond to ring diameter during the dwell (pre-constriction) and constriction phases. Cells were grown as described in C. Average assembly times, dwell times, and (circumferential) constriction rates are included to the right of each plot.
While contractile rings failed to assemble at 36°C in cdc8–110 cells harboring an empty vector control or Tpm4.2cy, cells expressing Cdc8p or Tpm3.1cy supported ring assembly and cytokinesis (Figure 7C). Cdc8p and Tpm3.1cy restored normal actomyosin ring dynamics in cdc8–110 cells grown at the restrictive temperature (Figure 7D). The ring dynamics observed for cells expressing Cdc8p or Tpm3.1cy were similar to those of wild-type cells we have previously examined under these higher temperature conditions (Pollard et al. 2012). Thus, at least in the case of Tpm3.1cy, mammalian tropomyosin function is suitably conserved to sustain actomyosin ring function in fission yeast cells lacking their functional endogenous tropomyosin.
DISCUSSION
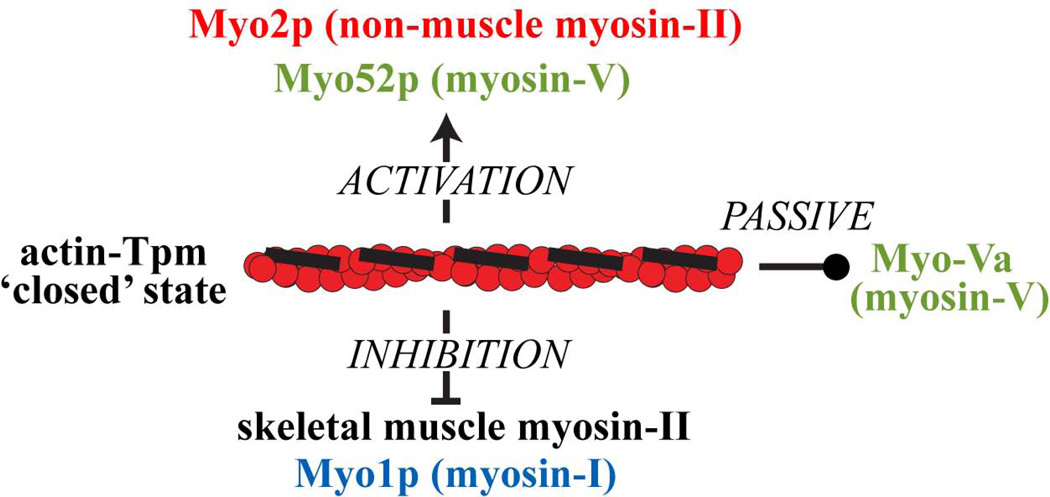
Fission yeast tropomyosin Cdc8p localizes to contractile rings and actin cables where it positively regulates myosin-II and myosin-V respectively (Clayton et al. 2014; Clayton et al. 2010; Coulton et al. 2010; Stark et al. 2010). Such regulation has the potential to contribute to the spatial control of actomyosin function in more complex cellular environments where multiple isoforms of tropomyosin and myosin abound. In this study we demonstrate that Cdc8p and two mammalian non-muscle LMW tropomyosins (Tpm3.1cy and Tpm4.2cy) differentially regulate myosin-IIs and myosin-Vs in a myosin isoform-specific manner (Figure 8). We propose that myosin isoform-specific regulation reflects a general feature of certain non-muscle tropomyosins.
Figure 8. Model depicting differential regulation of myosins by the ‘closed’ actin-tropomyosin state.
The ‘closed’ state reflects the position of Cdc8p and other tropomyosins bound along actin filaments in the absence of troponin. While referred to as ‘closed’, this state can act positively, negatively, or passively on myosins depending on the myosin isoform. Such features offer the potential for tight spatial regulation of myosin motor activity in complex cellular environments.
Interestingly, Tpm3.1cy successfully substituted for Cdc8p in vivo, supporting cytokinesis and cell growth. This finding suggests that Tpm3.1cy also shares functional similarity with the HMW isoform Tm2 (Tpm1.6cy) - the mammalian tropomyosin shown to substitute for Cdc8p in the original study on fission yeast tropomyosin (Balasubramanian et al. 1992). Tpm3.1cy is essential for cancer cell proliferation and motility, and is beginning to show promise as a target for anti-cancer compounds (Stehn et al. 2013). The ability of Tpm3.1cy to positively regulate non-muscle myosin function may contribute to its role in cancer cell viability. In contrast to Tpm3.1cy, Tpm4.2cy could not function in place of Cdc8p in vivo. Given that Tpm3.1cy and Tpm4.2cy exhibit similar behaviors to Cdc8p in their abilities to regulate myosins, the short-coming of Tpm4.2cy in vivo may simply reflect differences relating to other key actin-binding proteins besides myosin. For example, unlike Cdc8p, Tpm4.2cy may not correctly protect actin filaments from severing by fission yeast ADF/cofilin Adf1p (Skau and Kovar 2010), and/or fail to recognize and associate with formin- (Cdc12p-) nucleated actin filaments (Johnson et al. 2014).
Positive regulation of non-muscle myosin motors by tropomyosins
We previously demonstrated that Cdc8p promotes fission yeast Myo2p and Myo52p function by favoring movement into the strong actin-bound ADP/apo states (Clayton et al. 2010; Stark et al. 2010). This regulation increases the duty ratio of both myosins (Clayton et al., 2010; Stark et al., 2010), converting the non-processive Myo52p into a processive motor that can walk along the actin track as a single molecule (Clayton et al. 2014). Both of the mammalian tropomyosins tested were also capable of positively regulating Myo2p and Myo52p motor function. Both of these cytoplasmic tropomyosins converted Myo52p into a processive motor underscoring a conserved mechanism of regulation centered on positive modulation of the motor duty ratio.
Collectively, our findings suggest that positive non-muscle actomyosin regulation represents a conserved function of specific isoforms of tropomyosin. This is surprising given the large evolutionary distance between yeast and mammals reflected by the limited homology between tropomyosins from these two distinct systems. This regulation most likely reflects some conserved features of the tropomyosin sequence in general, which appear to extend from human to yeast tropomyosins (Cranz-Mileva et al. 2013). Striated muscle α-tropomyosin/Tpm1.1st possesses a conserved periodic series of acidic and basic residues that establish association with actin (Barua et al. 2013; Hitchcock-DeGregori et al. 2002; Singh and Hitchcock-DeGregori 2007), while other conserved acidic residues on the tropomyosin surface influence SKMM-II regulation (Barua et al. 2012; Oguchi et al. 2011). These conserved acidic and basic residues are often found at the b, c, and f positions of the heptad repeats in the tropomyosin coiled-coil. These three positions are not involved with the folding and stability of the coiled-coil and are consequently available for association with other proteins. Similar conserved acidic and basic residues are also found at b, c, and f positions in Cdc8p and other fungal tropomyosins (Cranz-Mileva et al. 2013). Interestingly, amino acid substitutions at some of these sites in Cdc8p were recently found to compromise actin association and in vivo function (Cranz-Mileva et al. 2013).
We speculate that the mammalian cytoplasmic tropomyosins we have studied can work like Cdc8p and facilitate positive regulation of myosins in mammals and other higher eukaryotes. Targets of such regulation likely include non-muscle myosin-IIs, myosin-Vs, and potentially other classes of myosins that have yet to be tested. It will be interesting to further test the generality of this idea in the future. The influence here of other mammalian tropomyosins, such as other non-muscle LMW isoforms, non-muscle HMW isoforms, or striated muscle Tm1.1st may differ from Tpm3.1cy and Tpm4.2cy.
Importance of the myosin isoform in tropomyosin-mediated regulation
The ability of Cdc8p and mammalian tropomyosins to promote non-muscle myosin function breaks with the established role of tropomyosin in gating muscle myosin-II activity (Lehrer 1994). In striated muscle, the troponin-tropomyosin complex initially assumes the ‘blocked’ orientation on the actin filament which prevents interactions between myosin heads and actin. Ca2+-binding by troponin shifts tropomyosin into the ‘closed’ position which allows limited binding between myosin heads and actin. However, once a myosin head binds actin it promotes further displacement of the tropomyosin molecule creating the ‘open’ state facilitating cooperative binding of myosin heads nearby. Nevertheless, tropomyosins were obviously present in other cells and tissues long before muscle evolved. Thus, the role of tropomyosin in muscle is highly specialized and represents the exception rather than the rule.
In the absence of troponin and Ca2+, tropomyosin-actin assumes the ‘closed’ conformation (Lehman et al. 1994; Lehman et al. 2000). Electron microscopy of actin filaments decorated with Cdc8p revealed that fission yeast tropomyosin also assumes the ‘closed’ conformation (Skoumpla et al. 2007). We found that the presence of Cdc8p or mammalian tropomyosins inhibited SKMM-II ATPase and actin filament gliding activities, despite the ability of these same tropomyosins to activate Myo2p and Myo52p under the same conditions. SKMM-II did show some residual activity in the presence of the tropomyosins in filament gliding assays, where the higher SKMM-II to actin ratios employed in such assays presumably facilitated some cooperative binding in this assay. Nevertheless, significant tropomyosin-mediated inhibition of SKMM-II was always observed even when these saturating concentrations of this myosin were employed in motility assays (Figure 5A–B).
In some ways our results are consistent with previous studies assessing the influence of tropomyosins on SKMM-II activity (Fanning et al. 1994). As we found with Tpm3.1cy and Tpm4.2cy, Fanning et al. saw that the presence of striated muscle Tm1.1st and non-muscle HMW isoform Tpm4.1cy inhibited SKMM-II activity. In contrast to our findings, Fanning et al. observed increased SKMM-II activity in the presence of Tpm4.2cy. While the root of this discrepancy is not yet clear, it should be noted that our current study employed full-length SKMM-II that was included in experiments under low, physiological salt concentrations - conditions that yield SKMM-II filament formation. On the other hand, Fanning et al. employed the truncated S1 (sub-fragment 1) form of SKMM-II, which is unable to self-assemble at any salt concentration (as it only includes the motor domain and light chain-binding region of the myosin-II molecule). Nevertheless, consistent with the conflicting results of Fanning et al., Barua et al. (2014) observed increased rates of SKMM-II-driven actin filament gliding in the presence of both Tpm3.1cy and Tpm4.2cy (as pointed out earlier in our Results). However, unlike in our experiments, Barua et al. applied SKMM-II to motility chambers at a high salt concentration to facilitate antibody-based attachment of individual myosin molecules. We propose that the tropomyosins under study favor inhibition of SKMM-II ensembles, an effect that appears to be reversed when SKMM-II is prepared in its soluble, monomeric form.
While Cdc8p, Tpm3.1cy, and Tpm4.2cy promote Myo52p function, none of the tropomyosins tested had a significant impact on the highly processive myosin-Va. Given that most class-V myosins studied to date are intrinsically processive, the ability of tropomyosins to positively regulate myosin-V may be restricted to non-processive motors such as Myo52p (Clayton et al. 2014), budding yeast myosin-V (Hodges et al. 2012), and other non-processive myosin-Vs found in higher eukaryotes (Takagi et al. 2008; Toth et al. 2005; Watanabe et al. 2008).
In summary, tropomyosin-mediated regulation of SKMM-II and the non-muscle myosins underscores how actin-tropomyosin can specify different motor outputs (Figure 8). The ‘closed’ actin-tropomyosin conformation appears to be capable of myosin activation (Myo2p and Myo52p) or inhibition (SKMM-II and type-I myosins), while at the same time acting passively on other myosins (myosin-Va). This differential regulation has broad implications for understanding how myosin activity can be sorted at specific tropomyosin-actin structures in time and space in complex systems, such as developing muscle tissue or metastatic cancer cells.
MATERIALS AND METHODS
Bacterial plasmids
Tropomyosin constructs were used to over-express acetylated-mimicking forms of the protein in E. coli. The cdc8 gene was PCR-amplified from S. pombe cDNA using the following primers: 5’ NdeI-cdc8 CAATCATATGGCTAGCATGGATA-AGCTTAGAGAG (the NdeI site is italicized, and the start codon of the cdc8 ORF is underlined) and 3’ BamHI-cdc8 CAATGGATCCCTACAAGTCC-TCAAGAGCTT (the BamHI site is italicized, and the cdc8 stop codon is underlined). The 5’ primer includes encodes an Ala-Ser (acetylation-mimicking) dipeptide immediately upstream of the cdc8 start codon. The start codon for the recombinant protein actually lies in the second half of the NdeI site allowing for incorporation of the Ala-Ser codons. Mammalian tropomyosin constructs were made in an identical fashion using human Tpm3.1cy and Tpm4.2cy cDNAs as templates. Cdc8p and Tpm3.1cy plasmids lacking the Ala-Ser dipeptide were also constructed. Each oligonucleotide was individually cloned into the pET3 vector and fidelities confirmed by DNA sequencing.
Fission yeast plasmids and strains
Untagged and N-terminally GFP-tagged versions of Cdc8p and the mammalian tropomyosins were expressed in fission yeast strains using the pREP-41x and pDS573a-41x vectors respectively. These ura4+ plasmids utilize the inducible nmt1–41x (medium strength) promoter which drives expression in the absence of thiamine. The cdc8 gene was PCR-amplified from S. pombe cDNA using the following primers: 5’ NotI-cdc8 GCGG-CCGCATGGATAAGCTTAGAGAGGTATGAG (the NotI site is italicized, and the start codon of the cdc8 ORF is underlined) and 3’ SalI-cdc8 GT-CGACCTACAAATCCTCAAGAGCTTGGTGA-AC (the SalI site is italicized, and the cdc8 stop codon is underlined). The cdc8 gene was inserted into NotI/SalI linearized pREP-41x and pDS573a-41x. Mammalian tropomyosin constructs were made in an identical fashion using human Tpm3.1cy and Tpm4.2cy cDNAs as templates. The fidelities of inserts was confirmed by DNA sequencing.
Untagged tropomyosins were expressed in a temperature-sensitive cdc8–110 mutant (h− leu1–32 ura4-D18 his3-D1 ade6-M216 cdc8–110 rlc1-mYFP:kanR) mutant, while GFP-tagged tropomyosins were expressed in wild-type (h− leu1–32 ura4-D18 his3-D1 ade6-M216 uch2-mCherry:natR) cells. The rlc1-mYFP and uch2-mCherry genomic integrations were constructed by standard techniques using the appropriate kanamycin-resistant kanR and nourthreothricin-resistant natR casettes (Bahler et al. 1998; Snaith et al. 2010).
Protein purification
Full-length fission yeast Myo2p was over-expressed and purified as previously described (Lord and Pollard 2004). Chicken skeletal muscle myosin-II was purified using standard techniques (Kielley and Harrington 1960). Recombinant HMM forms of fission yeast Myo52p and mammalian (chick brain) myosin-Va were over-expressed in the Baculovirus/Sf9 insect cell system and purified as previously described (Clayton et al. 2014; Krementsov et al. 2004). Biotin was included in the cell culture media to ensure full biotinylation of biotin-binding tags engineered at the C-terminus of the HMMs. Additional myosin-V light chains were purified following over-expression in bacteria (see below) (Clayton et al. 2014; Krementsov et al. 2004). Chicken skeletal muscle actin was employed throughout and purified from acetone powder as previously described (Spudich and Watt 1971).
Tropomyosin and myosin-V light chain (Cam1p, Cdc4p, and CaMΔall) proteins were produced in E. coli BL21 DE3 competent cells. Cultures were grown in LB media at 37°C and protein over-expression induced at an OD600 of 0.5 by addition of 0.4 mM IPTG. Cells were harvested following overnight induction at 25°C. Proteins were purified as described previously (Krementsov et al. 2004; Pruyne et al. 1998). Cell pellets were resuspended in lysis buffer (100 mM NaCl, 10 mM imidazole, pH 7.2, 2 mM EDTA, and 1 mM DTT) and sonicated. Lysates were boiled for 10 minutes while stirring, then pelleted to remove denatured protein and other debri. Soluble tropomyosins (or light chains) were precipitated from the supernatant by lowering the pH to 0.2 units below the iso-electric point of the relevant protein. The protein precipitate was resuspended in 50 mM NaCl, 1 mM DTT, 10 mM imidazole, pH 7.4 and dialyzed for three rounds (3 × 1 L) against the same buffer, followed by addition of 5 µg/ml leupeptin.
Actin-activated ATPase assays
Standard actin-activated ATPase assays were carried out at room temperature using the Malachite green assay to quantitate Pi release (Henkel et al. 1988). This approach was employed to assay Myo2p, SKMM-II, and Myo52p-HMM. Reactions were carried out in 2 mM Tris-HCl, 10 mM imidazole, pH 7.2, 60 mM KCl, 0.1 mM CaCl2, 3 mM MgCl2, 2 mM ATP, and 1 mM DTT. Colorimetric measurements were taken at 595 nm in a plate reader. In addition, actin-activated ATPase assays were carried out at room temperature using an ATP-regenerating system to assay Myo52p-HMM and myosin-Va-HMM. Reactions in this NADH-linked assay were carried out in 10 mM imidazole, pH 7.4, 50 mM KCl, 4 mM MgCl2, 1 mM EGTA, and 1 mM DTT. Reactions were initiated by the addition of 2 mM Mg2+-ATP. The buffers also contained an ATP regenerating system (0.5 mM phosphoenolpyruvate, 100 units/ml pyruvate kinase), 0.2 mM NADH, and 20 units/ml lactate dehydrogenase. The rate of the reaction was measured from the decrease in absorbance at 340 nm.
The assays included 30 nM Myo2p, 90 nM SKMM-II, 10 nM Myo52p–HMM (with excess 350 nM Cam1p and Cdc4p light chains), or 14 nM myosin-Va-HMM (with excess 350 nM calmodulin), plus 0–30 µM actin/actin-tropomyosin filaments. Actin was pre-incubated with the different tropomyosins at a 2:1 molar ratio 60 minutes prior to dilution. Curves were fit to Michaelis-Menten kinetics using Kaleidagraph software.
In vitro actin filament gliding assays
We used motility assays based on an established protocol (Kron and Spudich 1986). Myosins under study were delivered into motility chambers at the following concentrations: Myo2p, 5 nM; SKMM-II, 5 nM; Myo52p-HMM, 27 nM; and myosin-Va, 21 nM. Higher concentrations of SKMM-II were also tested (10 and 50 nM). 5 µM stocks of fluorescent actin were polymerized by addition of 50 mM KCl and 1 mM MgCl2 incubated for 30 minutes, and then labeled with 5 µM of rhodamine phalloidin for 30 minutes. After myosins were adhered to the surface of a nitrocellulose-coated cover-slip for 10 min, the chamber was washed: a) 3x with Motility Buffer (25 mM imidazole, pH 7.4, 50 mM KCl, 1mM EGTA, 4 mM MgCl2, 2 mM DTT) plus 0.5 mg/ml BSA. Excess light chains were also included in this buffer for experiments with Myo52p-HMM (14 µM of Cam1p and Cdc4p) and myosin-Va-HMM (14 µM of calmodulin), b) 3x with Motility Buffer alone, c) 2x with Motility Buffer containing 1 µM of vortexed (30 s) unlabeled actin filaments, d) 3x with Motility Buffer plus 1 mM ATP, e) 2x with Motility Buffer plus 25 nM rhodamine phalloidin-labeled actin filaments and oxygen scavengers (50 µg/ml catalase, 130 µg/ml glucose oxidase, and 3 mg/ml glucose), f) twice with Motility Buffer plus 20 mM DTT, 0.5 % methyl-cellulose, and scavengers, and g) twice with Motility Buffer plus 20 mM DTT, 0.5 % methyl-cellulose, 1.5 mM ATP and scavengers. To limit actin-tropomyosin disscoaition, washes (e-g) included 10 µM tropomyosin for assays with decorated filaments. Filaments were observed at room temperature by epi-fluorescence microscopy and recorded at intervals of 1 s for 1 minute. Image J software was used to calculate filament velocities from time-lapse series (n=35–150 filaments).
Tracking Myo52p-Qdot motility by TIRF microscopy
Myo52p-HMM (0.2 µM) was mixed with a 2-fold molar excess of actin and 2 mM Mg2+-ATP, and centrifuged for 20 minutes at 400,000 g to remove any myosin that was unable to dissociate from actin in the presence of ATP. To investigate processivity of individual myosin V motors, Myo52p was then mixed with a 10-fold molar excess of 655 nm streptavidin-coated Qdots (Invitrogen). In principle, a small fraction of Qdots will have two or more motors bound, but previous controls demonstrated that Qdots are driven primarily by a single motor at this mixing ratio (Hodges et al. 2012). Flow cells made from glass coverslips were prepared by introducing the following solutions into the flow cell: 0.1 mg/ml N-ethylmaleimide-modified skeletal muscle myosin (5 min incubation), 5x rinse of 1 mg/ml BSA (2 min), 2 µM rhodamine-phalloidin-labeled chicken skeletal muscle actin filaments (plus or minus 1 µM tropomyosin) (2–5 min), 5x rinse with motility buffer, and, finally 0.2 nM Myo52p in motility buffer with 2 mM MgATP. Motility buffer consists of 50 mM KCl, 25 mM imidazole, pH 7.4, 4 mM MgCl2, 1 mM EGTA, 50 mM DTT, 1 mg/ml BSA, 3.5 µM Cam1p, 3.5 µM Cdc4p, and an oxygen-scavenging system (3 mg/ml glucose, 0.1 mg/ml glucose oxidase, and 0.18 mg/ml catalase). For experiments with tropomyosin, 1 µM tropomyosin was included in the motility buffer to prevent dissociation from actin.
Through-the-objective TIRF microscopy was performed at room temperature using the Nikon Eclipse Ti-U microscope equipped with a 100x Plan Apo objective lens (1.49 NA) and auxiliary 1.5x magnification. Fluorophores were excited with a 473-nm (Qdots) laser line and images were obtained using the XR/Turbo-Z camera (Stanford Photonics) running Piper Control software (v2.3.39). The pixel resolution was 95.0 nm, and data were collected at 5–30 frames per second. Qdot movement along rhodamine-labeled actin filaments was tracked by hand using ImageJ. For each event, we required Qdot-labeled Myo52p to move continuously for at least 3 frames to qualify as a run. Runs that artificially terminated by running off the end of an actin filament were not included in the run length analysis.
Live cell imaging
DIC and epi-fluorescence cell images were captured using a Nikon TE2000-E2 inverted microscope with motorized fluorescence filter turret and a Plan Apo 60x (1.45 NA) objective. Fluorescence utilized an EXFO X-CITE 120 illuminator. NIS Elements software was used to control the microscope, two Uniblitz shutters, a Photometrics CoolSNAP HQ2 14-bit camera, and auto-focusing. Still images (GFP-tropomyosins, Rlc1-YFP) or time-lapse images (Rlc1p–YFP) of contractile rings were generated using the appropriate filters. For ring movies, images were captured every 2 minutes and auto-focusing was performed on the DIC channel before each capture. Cell suspensions (3 µl) were mounted on flat 30 µl EMM media pads (solidified by 1% agarose) prepared on the slide surface. VALAP (1:1:1 vasoline, lanolin, and paraffin) was used to seal slides and cover-slips. Images were captured using Nikon ND software and analysis of ring dynamics was performed using Image J and Microsoft Excel software. Ring dynamics were quantified by assessing individual phases: assembly, time taken for Rlc1p-YFP to compact into a mature ring following its appearance as a broad band of nodes; dwell, time from completion of ring compaction until initiation of constriction; constriction, change in ring circumference over time. Dwell times and constriction initiation were discerned by plotting ring diameter over time for each ring, and circumferential ring constriction rates were derived from the slopes of these plots.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Justine Stehn and Peter Gunning (University of New South Wales, Sydney, Australia) for providing the mammalian tropomyosin cDNAs. We thank Elena Krementsova and Kathy Trybus (University of Vermont, Burlington, VT, USA) for providing myosin-Va. We are grateful to Kelly Begin (University of Vermont, Burlington, VT, USA) for providing chicken skeletal muscle myosin-II. We thank Dan McCollum (University of Massachusetts Medical School, Worcester, MA) for the cdc8-110 strain. We are also very grateful to Guy Kennedy and David Warshaw (University of Vermont, Burlington, VT, USA) for providing access to TIRF microscopy facilities. This work was supported by a National Institutes of Health grant (GM097193) to ML.
REFERENCES
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14(10):943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Helfman DM, Hemmingsen SM. A new tropomyosin essential for cytokinesis in the fission yeast S. pombe. Nature. 1992;360(6399):84–87. doi: 10.1038/360084a0. [DOI] [PubMed] [Google Scholar]
- Barua B, Fagnant PM, Winkelmann DA, Trybus KM, Hitchcock-DeGregori SE. A periodic pattern of evolutionarily conserved basic and acidic residues constitutes the binding interface of actin-tropomyosin. J Biol Chem. 2013;288(14):9602–9609. doi: 10.1074/jbc.M113.451161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua B, Nagy A, Sellers JR, Hitchcock-DeGregori SE. Regulation of nonmuscle myosin II by tropomyosin. Biochemistry. 2014;53(24):4015–4024. doi: 10.1021/bi500162z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua B, Winkelmann DA, White HD, Hitchcock-DeGregori SE. Regulation of actin-myosin interaction by conserved periodic sites of tropomyosin. Proc Natl Acad Sci U S A. 2012;109(45):18425–18430. doi: 10.1073/pnas.1212754109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. Actin in emerging neurites is recruited from a monomer pool. Mol Neurobiol. 1992;6(2–3):95–106. doi: 10.1007/BF02780546. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11(16):1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- Bryce NS, Schevzov G, Ferguson V, Percival JM, Lin JJ, Matsumura F, Bamburg JR, Jeffrey PL, Hardeman EC, Gunning P, et al. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol Biol Cell. 2003;14(3):1002–1016. doi: 10.1091/mbc.E02-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton JE, Pollard LW, Sckolnick M, Bookwalter CS, Hodges AR, Trybus KM, Lord M. Fission yeast tropomyosin specifies directed transport of myosin-V along actin cables. Mol Biol Cell. 2014;25(1):66–75. doi: 10.1091/mbc.E13-04-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton JE, Sammons MR, Stark BC, Hodges AR, Lord M. Differential regulation of unconventional fission yeast myosins via the actin track. Curr Biol. 2010;20(16):1423–1431. doi: 10.1016/j.cub.2010.07.026. [DOI] [PubMed] [Google Scholar]
- Coffman VC, Nile AH, Lee IJ, Liu H, Wu JQ. Roles of formin nodes and myosin motor activity in Mid1p–dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20(24):5195–5210. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulton AT, East DA, Galinska-Rakoczy A, Lehman W, Mulvihill DP. The recruitment of acetylated and unacetylated tropomyosin to distinct actin polymers permits the discrete regulation of specific myosins in fission yeast. J Cell Sci. 2010;123(Pt 19):3235–3243. doi: 10.1242/jcs.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranz-Mileva S, Pamula MC, Barua B, Desai B, Hong YH, Russell J, Trent R, Wang J, Walworth NC, Hitchcock-DeGregori SE. A molecular evolution approach to study the roles of tropomyosin in fission yeast. PLoS One. 2013;8(10):e76726. doi: 10.1371/journal.pone.0076726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creed SJ, Bryce N, Naumanen P, Weinberger R, Lappalainen P, Stehn J, Gunning P. Tropomyosin isoforms define distinct microfilament populations with different drug susceptibility. Eur J Cell Biol. 2008;87(8–9):709–720. doi: 10.1016/j.ejcb.2008.03.004. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc Natl Acad Sci U S A. 1999;96(24):13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East DA, Mulvihill DP. Regulation and function of the fission yeast myosins. J Cell Sci. 2011;124(Pt 9):1383–1390. doi: 10.1242/jcs.078527. [DOI] [PubMed] [Google Scholar]
- East DA, Sousa D, Martin SR, Edwards TA, Lehman W, Mulvihill DP. Altering the stability of the Cdc8 overlap region modulates the ability of this tropomyosin to bind co-operatively to actin and regulate myosin. Biochem J. 2011;438(2):265–273. doi: 10.1042/BJ20101316. [DOI] [PubMed] [Google Scholar]
- Fan X, Martin-Brown S, Florens L, Li R. Intrinsic capability of budding yeast cofilin to promote turnover of tropomyosin-bound actin filaments. PLoS One. 2008;3(11):e3641. doi: 10.1371/journal.pone.0003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning AS, Wolenski JS, Mooseker MS, Izant JG. Differential regulation of skeletal muscle myosin-II and brush border myosin-I enzymology and mechanochemistry by bacterially produced tropomyosin isoforms. Cell Motil Cytoskeleton. 1994;29(1):29–45. doi: 10.1002/cm.970290104. [DOI] [PubMed] [Google Scholar]
- Fattoum A, Hartwig JH, Stossel TP. Isolation and some structural and functional properties of macrophage tropomyosin. Biochemistry. 1983;22(5):1187–1193. doi: 10.1021/bi00274a031. [DOI] [PubMed] [Google Scholar]
- Fowler VM, Sussmann MA, Miller PG, Flucher BE, Daniels MP. Tropomodulin is associated with the free (pointed) ends of the thin filaments in rat skeletal muscle. J Cell Biol. 1993;120(2):411–420. doi: 10.1083/jcb.120.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves MA, Hitchcock-DeGregori SE, Gunning PW. A systematic nomenclature for mammalian tropomyosin isoforms. J Muscle Res Cell Motil. 2014 doi: 10.1007/s10974-014-9389-6. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15(6):333–341. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Heald RW, Hitchcock-DeGregori SE. The structure of the amino terminus of tropomyosin is critical for binding to actin in the absence and presence of troponin. J Biol Chem. 1988;263(11):5254–5259. [PubMed] [Google Scholar]
- Henkel RD, VandeBerg JL, Walsh RA. A microassay for ATPase. Anal Biochem. 1988;169(2):312–318. doi: 10.1016/0003-2697(88)90290-4. [DOI] [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE. Tropomyosin: function follows structure. Adv Exp Med Biol. 2008;644:60–72. doi: 10.1007/978-0-387-85766-4_5. [DOI] [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE, Heald RW. Altered actin and troponin binding of amino-terminal variants of chicken striated muscle alpha-tropomyosin expressed in Escherichia coli. J Biol Chem. 1987;262(20):9730–9735. [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE, Song Y, Greenfield NJ. Functions of tropomyosin's periodic repeats. Biochemistry. 2002;41(50):15036–15044. doi: 10.1021/bi026519k. [DOI] [PubMed] [Google Scholar]
- Hodges AR, Bookwalter CS, Krementsova EB, Trybus KM. A nonprocessive class V myosin drives cargo processively when a kinesin- related protein is a passenger. Curr Biol. 2009;19(24):2121–2125. doi: 10.1016/j.cub.2009.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges AR, Krementsova EB, Bookwalter CS, Fagnant PM, Sladewski TE, Trybus KM. Tropomyosin is essential for processive movement of a class V Myosin from budding yeast. Curr Biol. 2012;22(15):1410–1416. doi: 10.1016/j.cub.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Yamashiro S, Matsumura F. Annealing of gelsolin-severed actin fragments by tropomyosin in the presence of Ca2+. Potentiation of the annealing process by caldesmon. J Biol Chem. 1989a;264(28):16764–16770. [PubMed] [Google Scholar]
- Ishikawa R, Yamashiro S, Matsumura F. Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin. Potentiation of protective ability of tropomyosins by 83-kDa nonmuscle caldesmon. J Biol Chem. 1989b;264(13):7490–7497. [PubMed] [Google Scholar]
- Johnson M, East DA, Mulvihill DP. Formins determine the functional properties of actin filaments in yeast. Curr Biol. 2014;24(13):1525–1530. doi: 10.1016/j.cub.2014.05.034. [DOI] [PubMed] [Google Scholar]
- Kielley WW, Harrington WF. A model for the myosin molecule. Biochim Biophys Acta. 1960;41:401–421. doi: 10.1016/0006-3002(60)90037-8. [DOI] [PubMed] [Google Scholar]
- Kitayama C, Sugimoto A, Yamamoto M. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J Cell Biol. 1997;137(6):1309–1319. doi: 10.1083/jcb.137.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Sirotkin V, Lord M. Three's company: the fission yeast actin cytoskeleton. Trends Cell Biol. 2011;21(3):177–187. doi: 10.1016/j.tcb.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krementsov DN, Krementsova EB, Trybus KM. Myosin V: regulation by calcium, calmodulin, and the tail domain. J Cell Biol. 2004;164(6):877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc Natl Acad Sci U S A. 1986;83(17):6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman W, Craig R, Vibert P. Ca(2+)-induced tropomyosin movement in Limulus thin filaments revealed by three-dimensional reconstruction. Nature. 1994;368(6466):65–67. doi: 10.1038/368065a0. [DOI] [PubMed] [Google Scholar]
- Lehman W, Hatch V, Korman V, Rosol M, Thomas L, Maytum R, Geeves MA, Van Eyk JE, Tobacman LS, Craig R. Tropomyosin and actin isoforms modulate the localization of tropomyosin strands on actin filaments. J Mol Biol. 2000;302(3):593–606. doi: 10.1006/jmbi.2000.4080. [DOI] [PubMed] [Google Scholar]
- Lehrer SS. The regulatory switch of the muscle thin filament: Ca2+ or myosin heads? J Muscle Res Cell Motil. 1994;15(3):232–236. doi: 10.1007/BF00123476. [DOI] [PubMed] [Google Scholar]
- Lo Presti L, Chang F, Martin SG. Myosin Vs organize actin cables in fission yeast. Mol Biol Cell. 2012;23(23):4579–4591. doi: 10.1091/mbc.E12-07-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M, Laves E, Pollard TD. Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast. Mol Biol Cell. 2005;16(11):5346–5355. doi: 10.1091/mbc.E05-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M, Pollard TD. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J Cell Biol. 2004;167(2):315–325. doi: 10.1083/jcb.200404045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May KM, Watts FZ, Jones N, Hyams JS. Type II myosin involved in cytokinesis in the fission yeast, Schizosaccharomyces pombe. Cell Motil Cytoskeleton. 1997;38(4):385–396. doi: 10.1002/(SICI)1097-0169(1997)38:4<385::AID-CM8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE. Myosin-V is a processive actin-based motor. Nature. 1999;400(6744):590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- Nakano K, Mabuchi I. Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol Biol Cell. 2006;17(4):1933–1945. doi: 10.1091/mbc.E05-09-0900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguchi Y, Ishizuka J, Hitchcock-DeGregori SE, Ishiwata S, Kawai M. The role of tropomyosin domains in cooperative activation of the actin-myosin interaction. J Mol Biol. 2011;414(5):667–680. doi: 10.1016/j.jmb.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156(6):1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard LW, Onishi M, Pringle JR, Lord M. Fission yeast Cyk3p is a transglutaminase-like protein that participates in cytokinesis and cell morphogenesis. Mol Biol Cell. 2012;23:2433–2444. doi: 10.1091/mbc.E11-07-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne DW, Schott DH, Bretscher A. Tropomyosin-containing actin cables direct the Myo2p–dependent polarized delivery of secretory vesicles in budding yeast. J Cell Biol. 1998;143(7):1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- Siemankowski RF, Wiseman MO, White HD. ADP dissociation from actomyosin subfragment 1 is sufficiently slow to limit the unloaded shortening velocity in vertebrate muscle. Proc Natl Acad Sci U S A. 1985;82(3):658–662. doi: 10.1073/pnas.82.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Hitchcock-DeGregori SE. Tropomyosin's periods are quasi-equivalent for actin binding but have specific regulatory functions. Biochemistry. 2007;46(51):14917–14927. doi: 10.1021/bi701570b. [DOI] [PubMed] [Google Scholar]
- Skau CT, Kovar DR. Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Curr Biol. 2010;20(16):1415–1422. doi: 10.1016/j.cub.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau CT, Neidt EM, Kovar DR. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell. 2009;20(8):2160–2173. doi: 10.1091/mbc.E08-12-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoumpla K, Coulton AT, Lehman W, Geeves MA, Mulvihill DP. Acetylation regulates tropomyosin function in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 2007;120(Pt 9):1635–1645. doi: 10.1242/jcs.001115. [DOI] [PubMed] [Google Scholar]
- Snaith HA, Anders A, Samejima I, Sawin KE. New and old reagents for fluorescent protein tagging of microtubules in fission yeast; experimental and critical evaluation. Methods Cell Biol. 2010;97:147–172. doi: 10.1016/S0091-679X(10)97009-X. [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246(15):4866–4871. [PubMed] [Google Scholar]
- Stark BC, Sladewski TE, Pollard LW, Lord M. Tropomyosin and myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Mol Biol Cell. 2010;21(6):989–1000. doi: 10.1091/mbc.E09-10-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehn JR, Haass NK, Bonello T, Desouza M, Kottyan G, Treutlein H, Zeng J, Nascimento PR, Sequeira VB, Butler TL, et al. A novel class of anticancer compounds targets the actin cytoskeleton in tumor cells. Cancer Res. 2013;73(16):5169–5182. doi: 10.1158/0008-5472.CAN-12-4501. [DOI] [PubMed] [Google Scholar]
- Stehn JR, Schevzov G, O'Neill GM, Gunning PW. Specialisation of the tropomyosin composition of actin filaments provides new potential targets for chemotherapy. Curr Cancer Drug Targets. 2006;6(3):245–256. doi: 10.2174/156800906776842948. [DOI] [PubMed] [Google Scholar]
- Takagi Y, Yang Y, Fujiwara I, Jacobs D, Cheney RE, Sellers JR, Kovacs M. Human myosin Vc is a low duty ratio, nonprocessive molecular motor. J Biol Chem. 2008;283(13):8527–8537. doi: 10.1074/jbc.M709150200. [DOI] [PubMed] [Google Scholar]
- Tang N, Ostap EM. Motor domain-dependent localization of myo1b (myr-1) Curr Biol. 2001;11(14):1131–1135. doi: 10.1016/s0960-9822(01)00320-7. [DOI] [PubMed] [Google Scholar]
- Tobacman LS. Cooperative binding of tropomyosin to actin. Adv Exp Med Biol. 2008;644:85–94. doi: 10.1007/978-0-387-85766-4_7. [DOI] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Schevzov G, Hotulainen P, Naumanen P, Martin C, Gunning PW, Lappalainen P. A molecular pathway for myosin-II recruitment to stress fibers. Curr Biol. 2011;21(7):539–550. doi: 10.1016/j.cub.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Toth J, Kovacs M, Wang F, Nyitray L, Sellers JR. Myosin V from Drosophila reveals diversity of motor mechanisms within the myosin V family. J Biol Chem. 2005;280(34):30594–30603. doi: 10.1074/jbc.M505209200. [DOI] [PubMed] [Google Scholar]
- Trybus KM. Myosin V from head to tail. Cell Mol Life Sci. 2008;65(9):1378–1389. doi: 10.1007/s00018-008-7507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbancikova M, Hitchcock-DeGregori SE. Requirement of amino-terminal modification for striated muscle alpha-tropomyosin function. J Biol Chem. 1994;269(39):24310–24315. [PubMed] [Google Scholar]
- Watanabe S, Watanabe TM, Sato O, Awata J, Homma K, Umeki N, Higuchi H, Ikebe R, Ikebe M. Human myosin Vc is a low duty ratio nonprocessive motor. J Biol Chem. 2008;283(16):10581–10592. doi: 10.1074/jbc.M707657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawro B, Greenfield NJ, Wear MA, Cooper JA, Higgs HN, Hitchcock-DeGregori SE. Tropomyosin regulates elongation by formin at the fast-growing end of the actin filament. Biochemistry. 2007;46(27):8146–8155. doi: 10.1021/bi700686p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Pennise CR, Babcock GG, Fowler VM. Tropomodulin caps the pointed ends of actin filaments. J Cell Biol. 1994;127(6 Pt 1):1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.