Abstract
Purpose
CRISPR-Cas systems are prokaryotic immune systems against invading nucleic acids that adapt as new environmental threats arise. There are emerging examples of CRISPR-Cas functions in bacterial physiology beyond their role in adaptive immunity. This highlights the poorly understood, but potentially common, moonlighting functions of these abundant systems. We propose that these non-canonical CRISPR-Cas activities have evolved to respond to stresses at the cell envelope.
Recent findings
Here, we discuss recent literature describing the impact of the extracellular environment on the regulation of CRISPR-Cas systems, and the influence of CRISPR-Cas activity on bacterial physiology. The described non-canonical CRISPR-Cas functions allow the bacterial cell to respond to the extracellular environment, primarily through changes in envelope physiology.
Summary
This review discusses the expanding non-canonical functions of CRISPR-Cas systems, including their roles in virulence, focusing mainly on their relationship to the cell envelope. We first examine the effects of the extracellular environment on regulation of CRISPR-Cas components, and then discuss the impact of CRISPR-Cas systems on bacterial physiology, focusing on their roles in influencing interactions with the environment including host organisms.
Keywords: CRISPR-Cas, envelope stress, membrane composition, bacterial pathogenesis
Introduction
Prokaryotic organisms have evolved unique, adaptive, nucleic acid restriction machineries to prevent the uptake of mobile genetic elements, such as those derived from bacteriophages and plasmids(1). Termed CRISPR (clustered, regularly interspaced, short, palindromic repeats) - Cas (CRISPR-associated) systems, these RNA-guided endonuclease machineries canonically act in a sequence-specific fashion to cleave foreign DNA or RNA targets(2-5). This protects cells from exposure to potentially harmful genetic elements(2-4). Beyond this well-established function, CRISPR-Cas systems have been observed to play alternative roles in physiology. These moonlighting functions of CRISPR-Cas systems include roles in oxidative stress tolerance, antibiotic resistance, extracellular structure formation, DNA repair, and host-microbe interactions.
The molecular mechanism of many alternative CRISPR-Cas functions has not yet been fully elucidated, but may utilize a similar activity to that used in canonical targeting of foreign nucleic acids (6, 7). The signature component of CRISPR-Cas systems is the CRISPR array, composed of short, repetitive, and often palindromic sequences(8). These repeats are interspaced by short, unique, spacer sequences that are complementary to different nucleic acid targets (2, 9, 10). In most systems, the CRISPR array is transcribed as a single transcript (the pre-crRNA array) and is cleaved into small targeting RNAs (crRNAs) (11-14). These crRNAs form complexes with Cas proteins, which are encoded in adjacent, conserved operons (4). The complexes are capable of sequence-specific interaction with foreign nucleic acids (6). Upon hybridization of the crRNA to its target sequence, endonuclease activity of the associated Cas protein(s) is triggered, resulting in target cleavage (6). CRISPR-Cas systems are diverse and can be grouped into three main subtypes (types I,II, and III) defined by the unique Cas proteins used in crRNA processing and targeting/cleavage(1). While the type I and III systems use multimeric protein complexes for these processes, the type II system requires a single Cas protein, Cas9, as well as a unique accessory RNA, the trans-activating CRISPR RNA (tracrRNA) (1, 13, 15, 16). Uniquely, CRISPR-Cas systems can also acquire new spacer sequences within the CRISPR array as the nucleic acid threats (such as bacteriophages) in the environment change (2, 17).
Interestingly, many of the alternative activities (not involving the targeted degradation of foreign nucleic acid) of CRISPR-Cas systems are linked to processes occurring at the bacterial envelope. Herein, we present a CRISPR view of how CRISPR-Cas systems monitor and respond to stresses at the cell envelope, allowing bacteria to counteract not only bacteriophage infection, but also diverse insults such as antibiotics and host defenses. First, we discuss the transcriptional regulation of CRISPR-Cas systems in response to environmental changes signaled by the status of the bacterial envelope. We then describe the current understanding of how CRISPR-Cas systems regulate bacterial physiology, largely through changes at the cell surface, to promote resistance to environmental stresses. Finally, we highlight unanswered questions in the field of CRISPR-Cas biology, the exploration of which will provide insight into the evolution of CRISPR-Cas systems and the origins of their increasingly broad functions in bacterial physiology.
Activation and function of CRISPR-Cas systems in response to envelope stress
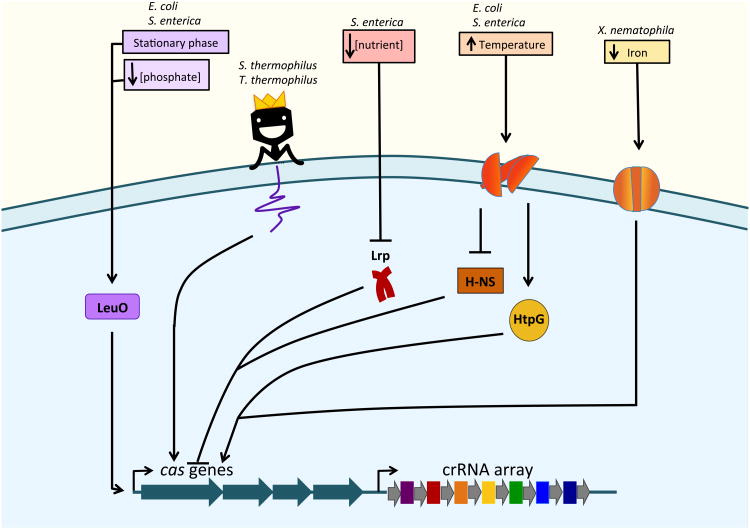
Since CRISPR-Cas systems target nucleic acids that have entered the cell through the envelope, it is interesting to note that their transcriptional activation often occurs directly, and indirectly, in response to envelope stresses (Figure 1). The most explicit example of this occurs during bacteriophage infection. It is logical to think that upon bacteriophage adsorption and DNA injection the envelope is disrupted, resulting in an envelope stress response(18-20). Concomitantly, activation of CRISPR-Cas transcription has been observed, suggesting that the cell actively senses the status of the envelope in order to respond to invading threats(21, 22). Furthermore, it has been observed that membrane protein dysregulation is capable of inducing the increased expression of CRISPR-Cas systems. For instance, in Escherichia coli, the BaeSR extracytoplasmic stress response regulator acts to activate its CRISPR-Cas system when the bacterial envelope is perturbed (23). Furthermore, the transcriptional regulator H-NS is an inhibitor of CRISPR-Cas expression. Upon an envelope stress response, H-NS is inhibited, leading to an upregulation of a CRISPR-Cas system in Salmonella enterica and E. coli (24, 25). Additionally, high temperatures result in misfolding of membrane proteins and an envelope stress response leading to activation of heat shock protein G (HtpG) (26, 27). HtpG has subsequently been shown to activate transcription of CRISPR-Cas systems in E. coli (27). Thus, CRISPR-Cas systems can be primed by stress at the envelope, likely at least in part to counter actin coming foreign nucleic acids.
Figure 1. Activation of CRISPR-Cas systems in response to environmental changes.
CRISPR-Cas systems can be activated in response to the broader environmental stressors of nutrient starvation, stationary phase growth, and iron limitation. Likewise, CRISPR-Cas systems can be activated directly in response to envelope stressors, such as phage infection and high temperature. These examples highlight the influence of the extracellular environment on the regulation of CRISPR-Cas systems.
In line with this idea, a recent study of Streptococcus mutans, a cause of tooth decay, revealed that expression of the Type II-A CRISPR-Cas system was negatively affected by the stress response regulator VicK/R two-component system, which also positively regulated the expression of its Type I-C system (28-30). Additionally, it was observed that both of these CRISPR-Cas systems play a role in temperature stress tolerance. CRISPR-Cas locus deletion mutants exhibited reduced survival after heat exposure, and surprisingly, double mutants in both loci had a greater sensitivity to high temperature than mutants from either locus alone, suggesting independent activity of each system (30). Furthermore, CRISPR-Cas mutants in the type II-A system, but not the Type I-C system, displayed reduced growth upon exposure to membrane stress (detergents) as well as oxidative stress (paraquat and hydrogen peroxide) (30). Together, these data directly link CRISPR-Cas function to envelope stresses, and further suggest that VicK/R may differentially regulate each CRISPR-Cas system under specific conditions. This raises the questions of whether these systems work together in nucleic acid defense as well, if they have distinct defense activities beyond adaptive immunity, or if they diverged in function to fulfill distinct regulatory roles, perhaps by altering the envelope. Exactly how these CRISPR-Cas systems regulate stress tolerance remains to be elucidated, and continued study of this phenomenon in diverse bacteria will be necessary to identify common themes. It is reasonable to postulate that this occurs through physiological changes at the envelope, which acts as the frontline to counteract environmental stressors.
CRISPR-Cas control of population behaviors
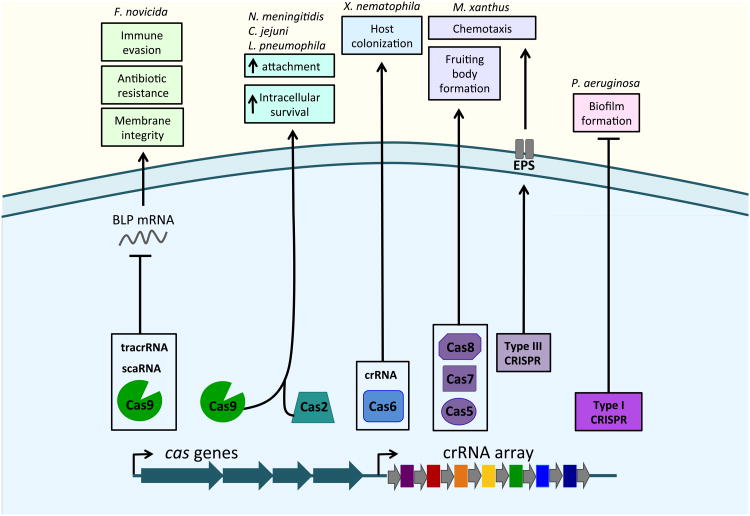
In addition to roles in the envelope stress response, CRISPR-Cas systems have been implicated in complex population behaviors that involve extensive envelope alterations, such as biofilm formation and fruiting body development (Figure 2). Before CRISPR-Cas systems were identified, three genes encoded by the Gram-negative saprophytic bacterium Myxococcusxanthus, were found to be necessary for sporulation and fruiting body development (31-33). Interestingly, the three genes, devT, devR, and devS, respectively correspond to cas8, cas7, and cas5from a type I CRISPR-Cas system. In the absence of devT (cas8), M. xanthus displayed delayed cellular aggregation, sporulation, and chemotaxis, as well as decreased transcript levels for a fruiting body transcriptional activator(31). While the mechanism of regulation has not been fully elucidated, the M. xanthus CRISPR array encodes two spacers that have identity to endogenous sequences on the bacterial chromosome. One has identity to an integrase of a Myxococcus bacteriophage, while the other has identity to a casgene in a different CRISPR-Cas locus, raising the intriguing possibility that the CRISPR-Cas system regulates endogenous targets (33). However, whether the CRISPR array itself is required for control of the aforementioned processes remains unknown.
Figure 2. CRISPR-Cas mediated physiological changes.
CRISPR-Cas systems influence bacterial physiology, altering population behavior and host-microbe interactions through events that are centered at the envelope. In Francisellanovicida, Cas9, tracrRNA and scaRNA form a complex that represses a bacterial lipoprotein mRNA (BLP). Repression of the BLP increases membrane integrity, conferring resistance to membrane targeting antibiotics and enabling evasion of the host immune system, increasing virulence. Cas9 from Neisseria meningitidis and Cas2 from Legionella pneumophila type II systems increase host-cell attachment and intracellular survival. In Xenorhabdus nematophila, Cas6 and a CRISPRRNA (crRNA) of the type I-E system are required for host colonization. In Myxococcusxanthus, the type III CRISPR-Cas system regulates exopolysacchride production (EPS) to enable chemotaxis, while negatively effecting fruiting body formation. Conversely, Cas5, Cas7, and Cas8 of its type III CRISPR-Cas system are necessary for fruiting body formation and sporulation. Finally, in Pseudomonas aeruginosa, all interference components of the Type I CRISPR system are required for biofilm formation and swarming motility. These examples provide a framework for understanding the alternative functions of CRISPR-Cas systems from interactions at the prokaryotic envelope.
M. xanthus regulation of fruiting body formation is further influenced by a type III-B CRISPR-Cas locus, which also regulates exopolysaccharide (EPS) production and type IV pili mediated chemotaxis (34). Not only is crRNA processing required for this regulatory activity, but the associated cas genes are as well (34). Further studies are needed to determine if and how the type I and III systems in M. xanthus interact to regulate fruiting body formation, as well as the mechanism of CRISPR-Cas mediated EPS regulation. It will be interesting to determine whether these functions evolved due to pressures to restrict mobile genetic elements, broader stresses at the envelope, or from entirely different environmental pressures.
Another population behavior involving extensive envelope changes, biofilm formation, is regulated by the type I CRISPR-Cas system in the opportunistic pathogen Pseudomonas aeruginosa (35, 36). A spacer within the P. aeruginosa CRISPR array has sequence similarity to a gene within a chromosomally integrated prophage (36). The CRISPR-Cas system interaction with this chromosomal element is necessary to represss warming motility and biofilm formation (35, 36). While it is not known how repression occurs, it is established as a sequence-specific activity requiring all interference components of this CRISPR-Cas system (36, 37). Given the importance of biofilm formation to antibiotic resistance and pathogenesis in P. aeruginosa, it is likely that this CRISPR-Cas system plays an important role in mediating infection of eukaryotic hosts.
CRISPR-Cas mediated regulation of host-pathogen interactions
While all bacteria encounter numerous environmental stresses, those bacteria that interact with eukaryotes, particularly mammalian hosts, are subjected to a variety of microenvironments and stressors as they traffic through the host and encounter the immune system (Figure 2). It is therefore an exciting proposition that CRISPR-Cas systems may be utilized in response to these host-derived stresses and ultimately mediate host-microbe interactions.
Recently, it has been observed that CRISPR-Cas systems can modulate host immune evasion. The intracellular pathogen Francisellanovicida upregulates its type II-B CRISPR-Cas system in the phagosome of host macrophages, a stressful environment containing a plethora of host defenses that attack the bacterial envelope (38). Components of this system (Cas9, tracrRNA, and a small CRISPR-Cas associated RNA [scaRNA]) regulate the production of an endogenous bacterial lipoprotein (BLP), a process necessary for strengthening the bacterial envelope (38, 39). Loss of these components results in increased envelope permeability and subsequently increases susceptibility to membrane damaging compounds, such as those found in the macrophage phagosome (39). Furthermore, regulation of the BLP dramatically alters how F. novicida survives within its mammalian host. In fact, cas9 mutants are attenuated in a mouse model by 103-104 fold compared to wild-type bacteria (38). Cas9 and its associated RNAs enable evasion of the host innate immune response through two distinct pathways, both of which originate due to changes at the membrane. In the absence of Cas9, the BLP transcript is de-repressed, and the bacteria are detected by the host pattern recognition receptor (PRR) Toll-like receptor 2 (TLR2), which initiates a proinflammatory response upon recognition of BLP(38). Additionally, repression of the BLP increases envelope integrity and reduces activation of the AIM2/ASC inflammasome, a protein complex involved in a programmed host cell death pathway that results in loss of Francisella's replicative niche (39). This CRISPR-Cas mediated evasion of both TLR2 and the AIM2/ASC inflammasome is critical for the ability of F. novicida to cause disease.
Consistent with the idea that CRISPR-Cas systems have evolved functions to mediate interactions with eukaryotic hosts, Neisseria meningitidis Cas9 is necessary for intracellular survival in human epithelial cells (38). Further, N. meningitidis Cas9 is also required for attachment and entry into these cells, processes dependent on surface components, suggesting that it may regulate envelope structures in this bacterium (38). Cas9 is likewise necessary for attachment and intracellular survival of Campylobacterjejuni, a cause of diarrheal disease and Guillain-Barré syndrome, in epithelial cells (40). Furthermore, C. jejunicas9 mutants displayed increased surface antibody binding, as well as increased envelope permeability and antibiotic susceptibility, all potentially linking Cas9 to the regulation of envelope components (40). Finally, it was observed bioinformatically that the presence of envelope sialylation correlates with a loss of the type II CRISPR-Cas system in multiple bacteria (including N. meningitidis, C. jejuni, and Haemophilus parainfluenzae) (40). Taken together, these data provide additional evidence for alternative functions of CRISPR-Cas systems in regulating envelope functions in response to environmental pressures.
Another example of a CRISPR-Cas system promoting host-microbe interactions is observed in the Gram-negative bacterium Xenorhabdus nematophila. Here, an orphan CRISPR RNA, termed NilD, is necessary for X. nematophila to colonize Steinemema spp. nematodes, a symbiotic relationship that facilitates the pathogenesis of these nematodes for their insect hosts (41). This is the first example of a CRISPR-Cas system modulating a mutualistic and tripartite interaction, and sheds light on the under explored complexity of CRISPR-Cas functions in broader ecological niches. Interestingly, this CRISPR-Cas system is expressed at a higher level in iron limiting conditions, furthering the concept that these machineries respond to extracellular changes and to events that are tightly regulated at the bacterial envelope (41). Additionally, the role of the crRNA from this system in colonization is independent of the effector protein Cas3, suggesting that the NilD CRISPR RNA has a unique function not involving canonical CRISPR-Cas activity (41). Further studies to elucidate the molecular mechanism of NilD-mediated nematode colonization will shed light not only on envelope changes that facilitate colonization, but also on how orphan crRNAs can potentially function as regulatory elements.
Similar to NilD, it was observed that the cas2 gene of the type II-B CRISPR-Cas system of Legionella pneumophila was required for intracellular survival within amoebae, and that cas2 was upregulated during intra-amoeba growth (42). Interestingly, no other cas gene was required, and cas2 was not required for growth in broth culture or intracellular infection of macrophages (42). Furthermore, expression of cas2 in a L. pneumophila strain that lacks a CRISPR-Cas system increased the strain's ability to replicate within amoebae, further indicating that Cas2 can act independently of canonical CRISPR-Cas function (43). Cas2 orthologs have RNase and/or DNase activity, depending on the organism, and are involved in spacer acquisition (17, 44-47). Cas2 nuclease activity is dependent on a single catalytic residue, which is also required for L. pneumophila intra-ameobal survival (43). In. L. pneumophila, not only is Cas2 RNase activity more efficient than DNase activity, but each requires a different divalent ion (Mg2+ or Mn2+, respectively)(43). Thus, preferred nuclease activity may change with shifts in the bacterial environment. It is unclear which nuclease activity promotes survival in amoebae, and a comparison of the ion concentrations in different growth environments may shed light on this difference. Likewise, the precise role of Cas2 in promoting intracellular survival is still unknown; it is tempting to consider that Cas2 has functions in mRNA regulation, particularly given that residues in its nuclease motif are essential for its role in intra-amoeba survival. Studies to observe which nucleic acids associate with Cas2 in different stages and contexts of Legionella growth, as well as determining the environmental cues governing the independent regulation of this Cas protein, will significantly enhance the understanding of CRISPR-Cas function as a regulator of intracellular survival.
Are CRISPR-Cas systems more broadly involved in stress responses?
Intriguingly, CRISPR-Cas systems are also regulated by a broad range of environmental conditions not necessarily linked to envelope stress (Figure 1). For instance, in nutrient rich conditions, the leucine-responsive protein (Lrp) represses CRISPR-Cas expression in Salmonella enterica serovar Typhi (24). However, upon starvation, Lrp is inactivated and may de-repress CRISPR-Cas transcription (24). Additionally, the regulator LeuO is an activator of CRISPR-Cas expression in S. enterica and Escherichia coli (24, 25, 48). LeuO is active under low phosphate and stationary phase conditions, further suggesting that starvation responses can increase CRISPR-Cas expression (49, 50). It is interesting to speculate that expression of CRISPR-Cas systems may also be tied to nutrient conditions since prokaryotic organisms may actively seek out nucleic acids as a nutrient source (51). While starvation is a stress in itself, it can indirectly result in dysregulation of membrane composition, as well as serve as a signal for prophages to become lytic (52, 53). The same is true for oxidative and osmotic stress, which have been shown to activate CRISPR-Cas systems and cause broad stress to the cell, including at the membrane (54, 55). Therefore, it is unclear whether there is a universal link between induction of CRISPR-Cas systems and envelope stress, or if these machineries may more broadly be induced by diverse stresses. In total, these examples provide further links between CRISPR-Cas activation and the response to environmental cues, which may occur through either their canonical or alternative functions.
In addition, CRISPR-Cas systems may act to regulate the cell's response against other diverse environmental stresses(38, 41, 54-57). For example, in E. coli, both the CRISPR array and Cas1 can participate in mediating DNA repair, while in Thermoproteustenax, a CRISPR-Cas system is activated in response to DNA damaging UV light (55, 56). Therefore, CRISPR-Cas systems may be responsible for alleviating the effects of stresses that damage the chromosome. In another example, the orphan CRISPR locus in Listeriamono cytogenes, rliB, acts to upregulate the production of the iron transport system feoAB, further demonstrating that CRISPR-Cas systems mediate physiological changes that are likely in response to environmental stress (57). Overall, these observations demonstrate that CRISPR-Cas systems may have evolved multiple functions to not only be activated in response to diverse environmental stress, but also to play active roles in preventing stress-promoted damage.
Conclusion
CRISPR-Cas systems are complex machineries that act to protect the cell against potentially harmful mobile genetic elements. As such, it would be efficient to regulate expression of these systems to times when the threat of such elements is imminent. Accordingly, there are now multiple examples of increased activation of CRISPR-Cas systems in response to envelope stress, such as bacteriophage binding and envelope disruption, ultimately enabling cells to activate defenses against potential genetic threats.
We have summarized numerous examples of CRISPR-Cas systems having functions beyond defense against foreign nucleic acids, many of which involve regulation of envelope physiology and how the cell interacts with its host and environment. It is interesting to consider how these non-canonical functions may have arisen. These observed roles could have appeared due to independent pressures, or stochastically due to accidental acquisition of spacers targeting self. Furthermore, the relationships between CRISPR-Cas system subtype and their non-canonical functions are poorly understood. Since some bacterial species encode multiple CRISPR-Cas subtypes within the same genome, each unique system may represent a fine-tuning of nucleic acid defense, perhaps based on niche and environmental cues. Alternatively, the presence of multiple systems may be linked to non-canonical functions, whereby some systems are preferentially used for nucleic acid defense and others to regulate bacterial physiology, or multiple systems facilitate different non-canonical functions. We hypothesize that clues to these interactions lie at the envelope, and that by studying the non-canonical functions of CRISPR-Cas systems from this perspective, we will gain insight into the evolution of both commensal and pathogenic bacteria to defend against their own pathogens and survive within their diverse replicative niches.
Key points.
CRISPR-Cas systems play roles in bacterial gene regulation.
Regulatory roles of CRISPR-Cas systems are linked to processes occurring at the bacterial envelope.
The ability to respond to envelope stress may have driven the involvement of CRISPR-Cas systems in gene regulation
Acknowledgments
Due to the rapidly expanding field, we have undoubtedly omitted some relevant studies. We apologize in advance to those authors whose work we did not cite.
Financial support and sponsorship: DSW is supported by NIH grant R01-AI110701 and a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease award.
Footnotes
Conflicts of interest: TRS and DSW have filed provisional patents based on CRISPR-Cas technological applications. HKR has no conflict of interest.
References
- 1.Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR–Cas systems. Nat Rev Micro. 2011;9(6):467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science. 2007;315(5819):1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 3.Marraffini LA, Sontheimer EJ. CRISPR Interference Limits Horizontal Gene Transfer in Staphylococci by Targeting DNA. Science. 2008;322(5909):1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–8. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 5.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139(5):945–56. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *6.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Micro. 2014;12(7):479–92. doi: 10.1038/nrmicro3279. A complete resource describing the molecular action of CRISPR-Cas systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westra ER, Buckling A, Fineran PC. CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Micro. 2014;12(5):317–26. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 8.Makarova KS, Wolf YI, Koonin EV. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013;41(8):4360–77. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. Journal of molecular evolution. 2005;60(2):174–82. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 10.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (Reading, England) 2005;151(Pt 8):2551–61. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 11.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pougach K, Semenova E, Bogdanova E, Datsenko KA, Djordjevic M, Wanner BL, et al. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol Microbiol. 2010;77(6):1367–79. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–7. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *14.Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, et al. Processing-Independent CRISPR RNAs Limit Natural Transformation in Neisseria meningitidis. Molecular cell. 2013;50(4):488–503. doi: 10.1016/j.molcel.2013.05.001. An elegant demonstration of a unique CRISPR-Cas system, whereby individual crRNAs contain their own promoter, rather than being processed from a single transcript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *15.Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42(10):6091–105. doi: 10.1093/nar/gku241. A comprehensive resource and forward thinking analysis of type II CRISPR-Cas systems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *16.Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biology. 2013;10(5):726–37. doi: 10.4161/rna.24321. A complete analysis of known type II CRISPR-Cas systems and their molecular functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *17.Heler R, Marraffini LA, Bikard D. Adapting to new threats: the generation of memory by CRISPR-Cas immune systems. Molecular Microbiology. 2014;93(1):1–9. doi: 10.1111/mmi.12640. An inclusive resource for the molecular mechanism of CRISPR-Cas adaptation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darwin AJ. Stress Relief during Host Infection: The Phage Shock Protein Response Supports Bacterial Virulence in Various Ways. PLoS Pathog. 2013;9(7):e1003388. doi: 10.1371/journal.ppat.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fallico V, Ross RP, Fitzgerald GF, McAuliffe O. Genetic response to bacteriophage infection in Lactococcus lactis reveals a four-strand approach involving induction of membrane stress proteins, D-alanylation of the cell wall, maintenance of proton motive force, and energy conservation. Journal of virology. 2011;85(22):12032–42. doi: 10.1128/JVI.00275-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raivio T. Identifying your enemies – could envelope stress trigger microbial immunity? Molecular Microbiology. 2011;79(3):557–61. doi: 10.1111/j.1365-2958.2010.07485.x. [DOI] [PubMed] [Google Scholar]
- 21.Young JC, Dill BD, Pan C, Hettich RL, Banfield JF, Shah M, et al. Phage-induced expression of CRISPR-associated proteins is revealed by shotgun proteomics in Streptococcus thermophilus. PloS one. 2012;7(5):e38077. doi: 10.1371/journal.pone.0038077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. Journal of molecular biology. 2010;395(2):270–81. doi: 10.1016/j.jmb.2009.10.057. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Rodriguez R, Haitjema C, Huang Q, Nam KH, Bernardis S, Ke A, et al. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol Microbiol. 2011;79(3):584–99. doi: 10.1111/j.1365-2958.2010.07482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina-Aparicio L, Rebollar-Flores JE, Gallego-Hernandez AL, Vazquez A, Olvera L, Gutierrez-Rios RM, et al. The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J Bacteriol. 2011;193(10):2396–407. doi: 10.1128/JB.01480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westra ER, Pul U, Heidrich N, Jore MM, Lundgren M, Stratmann T, et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol Microbiol. 2010;77(6):1380–93. doi: 10.1111/j.1365-2958.2010.07315.x. [DOI] [PubMed] [Google Scholar]
- 26.Bardwell JC, Craig EA. Eukaryotic Mr 83,000 heat shock protein has a homologue in Escherichia coli. Proceedings of the National Academy of Sciences. 1987;84(15):5177–81. doi: 10.1073/pnas.84.15.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yosef I, Goren MG, Kiro R, Edgar R, Qimron U. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proceedings of the National Academy of Sciences. 2011;108(50):20136–41. doi: 10.1073/pnas.1113519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiological Reviews. 1986;50(4):353–80. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senadheera D, Krastel K, Mair R, Persadmehr A, Abranches J, Burne RA, et al. Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J Bacteriol. 2009;191(20):6415–24. doi: 10.1128/JB.00793-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Serbanescu MA, Cordova M, Krastel K, Flick R, Beloglazova N, Latos A, et al. Role of the Streptococcus mutans CRISPR/Cas systems in immunity and cell physiology. J Bacteriol. 2014 doi: 10.1128/JB.02333-14. A new study revealing unique roles for the S mutans CRISPR-Cas systems in tolerance to temperature, oxidative, and envelope stress. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boysen A, Ellehauge E, Julien B, Sogaard-Andersen L. The DevT protein stimulates synthesis of FruA, a signal transduction protein required for fruiting body morphogenesis in Myxococcus xanthus. J Bacteriol. 2002;184(6):1540–6. doi: 10.1128/JB.184.6.1540-1546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thony-Meyer L, Kaiser D. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J Bacteriol. 1993;175(22):7450–62. doi: 10.1128/jb.175.22.7450-7462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viswanathan P, Murphy K, Julien B, Garza AG, Kroos L. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J Bacteriol. 2007;189(10):3738–50. doi: 10.1128/JB.00187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *34.Wallace RA, Black WP, Yang X, Yang Z. A CRISPR with roles in Myxococcus xanthus development and exopolysaccharide production. J Bacteriol. 2014;196(23):4036–43. doi: 10.1128/JB.02035-14. A recent study demonstrating a regulatory function of the M. xanthus CRISPR-Cas systems in the regulation of exopolysaccharide production, fruiting body formation, and motility. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O'Toole GA. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol. 2009;191(1):210–9. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cady KC, O'Toole GA. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol. 2011;193(14):3433–45. doi: 10.1128/JB.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O'Toole GA. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol. 2012;194(21):5728–38. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *38.Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497(7448):254–7. doi: 10.1038/nature12048. The first mechanistic observation of CRISPR-Cas mediated endogenous gene regulation with effects on the virulence of a bacterial pathogen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sampson TR, Napier BA, Schroeder MR, Louwen R, Zhao J, Chin CY, et al. A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(30):11163–8. doi: 10.1073/pnas.1323025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *40.Louwen R, Horst-Kreft D, de Boer AG, van der Graaf L, de Knegt G, Hamersma M, et al. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barre syndrome. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2013;32(2):207–26. doi: 10.1007/s10096-012-1733-4. The first demonstration that components of a CRISPR-Cas system are important for traits linked in virulence. [DOI] [PubMed] [Google Scholar]
- *41.Veesenmeyer JL, Andersen AW, Lu X, Hussa EA, Murfin KE, Chaston JM, et al. NilD CRISPR RNA contributes to Xenorhabdus nematophila colonization of symbiotic host nematodes. Mol Microbiol. 2014;93(5):1026–42. doi: 10.1111/mmi.12715. The first demonstration that components of a CRISPR-Cas system are involved in colonization of a symbiotic bacterium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *42.Gunderson FF, Cianciotto NP. The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. mBio. 2013;4(2):e00074–13. doi: 10.1128/mBio.00074-13. An interesting observation that a component of the adaptive machinery of CRISPR-Cas systems is required for intracellular survival. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *43.Gunderson FF, Mallama CA, Fairbairn SG, Cianciotto NP. The Nuclease Activity of Legionella pneumophila Cas2 Promotes Intracellular Infection of Amoebal Host Cells. Infection and immunity. 2014 doi: 10.1128/IAI.03102-14. A study describing the molceular action that Cas2 may require to mediate Legionella survival in amoeba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. The Journal of biological chemistry. 2008;283(29):20361–71. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nam KH, Ding F, Haitjema C, Huang Q, DeLisa MP, Ke A. Double-stranded endonuclease activity in Bacillus halodurans clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas2 protein. The Journal of biological chemistry. 2012;287(43):35943–52. doi: 10.1074/jbc.M112.382598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ka D, Kim D, Baek G, Bae E. Structural and functional characterization of Streptococcus pyogenes Cas2 protein under different pH conditions. Biochemical and biophysical research communications. 2014;451(1):152–7. doi: 10.1016/j.bbrc.2014.07.087. [DOI] [PubMed] [Google Scholar]
- 47.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40(12):5569–76. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hernandez-Lucas I, Gallego-Hernandez AL, Encarnacion S, Fernandez-Mora M, Martinez-Batallar AG, Salgado H, et al. The LysR-type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar Typhi. J Bacteriol. 2008;190(5):1658–70. doi: 10.1128/JB.01649-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.VanBogelen RA, Olson ER, Wanner BL, Neidhardt FC. Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli. J Bacteriol. 1996;178(15):4344–66. doi: 10.1128/jb.178.15.4344-4366.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stratmann T, Pul U, Wurm R, Wagner R, Schnetz K. RcsB-BglJ activates the Escherichia coli leuO gene, encoding an H-NS antagonist and pleiotropic regulator of virulence determinants. Mol Microbiol. 2012;83(6):1109–23. doi: 10.1111/j.1365-2958.2012.07993.x. [DOI] [PubMed] [Google Scholar]
- 51.Finkel SE, Kolter R. DNA as a Nutrient: Novel Role for Bacterial Competence Gene Homologs. Journal of Bacteriology. 2001;183(21):6288–93. doi: 10.1128/JB.183.21.6288-6293.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Death A, Notley L, Ferenci T. Derepression of LamB protein facilitates outer membrane permeation of carbohydrates into Escherichia coli under conditions of nutrient stress. J Bacteriol. 1993;175(5):1475–83. doi: 10.1128/jb.175.5.1475-1483.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen AM, Gu Y, Li M, Andrykovitch M, Waugh DS, Jin DJ, et al. Structural basis for the function of stringent starvation protein a as a transcription factor. The Journal of biological chemistry. 2005;280(17):17380–91. doi: 10.1074/jbc.M501444200. [DOI] [PubMed] [Google Scholar]
- 54.Strand K, Sun C, Li T, Jenney F, Jr, Schut G, Adams MW. Oxidative stress protection and the repair response to hydrogen peroxide in the hyperthermophilic archaeon Pyrococcus furiosus and in related species. Arch Microbiol. 2010;192(6):447–59. doi: 10.1007/s00203-010-0570-z. [DOI] [PubMed] [Google Scholar]
- 55.Plagens A, Tjaden B, Hagemann A, Randau L, Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J Bacteriol. 2012;194(10):2491–500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, et al. A dual function of the CRISPR–Cas system in bacterial antivirus immunity and DNA repair. Molecular Microbiology. 2011;79(2):484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 2007;35(3):962–74. doi: 10.1093/nar/gkl1096. [DOI] [PMC free article] [PubMed] [Google Scholar]