Abstract
We have developed a simple but powerful method and web server to quickly locate charged and hydrophobic clusters in proteins (http://www.netasa.org/qgrid/index.html). For the charged clusters, each atom in the protein is first assigned a charge according to a standard force field. Then a box is created with dimensions corresponding to the range of atomic coordinates. This box is then divided into cubic grids of selected size, which now have one or more charged atoms in them. This leaves each grid with a certain amount of charge. Cubic grids with more than a cutoff charge are then clustered using a hierarchical clustering method based on Euclidean distance. A tree diagram made from the resulting clusters indicates the distribution of charged and hydrophobic regions of the protein. Hydrophobic clusters are developed by grouping the positions of Cα atoms of such residues. We propose that such a tree representation will be helpful in detecting protein–protein interfaces, structure similarity and motif detection.
INTRODUCTION
Positive and negative charge clusters in proteins have been implicated in different biologically important functions and their importance has been realized for among other things, protein–protein interactions, DNA-binding, Ca+ and Na+ channeling and gating, electron transport and domain swapping (1–8). Similarly, hydrophobic clusters have been found to be of central importance in determining the stability, folding pattern, guanosine diphosphate (GDP) dissociation and similar properties of proteins (9–13). Despite enormous need to detect such charged and hydrophobic clusters in proteins, there is no web server to allow molecular biologists to detect such regions in proteins quickly. Molecular structure visualization programs such as Rasmol (now Protein Explorer) (14), Chime (http://www.mdli.com/) and VMD (15) may at best be used to locate surface distribution of residues and generate Connolly surfaces. Apart from their inability to locate the clusters in the interior of the protein, their very three-dimensional rendering makes it impossible to visualize the overall distribution without a need to rotate the structure and explore all possible orientations. Here we present a web server which can give the distribution of charged and hydrophobic regions and allow their quick visual location in a nutshell in two-dimensional cluster-tree diagrams. These two-dimensional diagrams allow us to inspect clusters in every part of the protein and present the results in a more concise manner which is free from the protein orientation and overall symmetry properties such as molecular chirality.
MATERIALS AND METHODS
The principle of Qgrid is quite straightforward. The atomic co-ordinates are read from a PDB-formatted file (16). Charges are assigned to every atom according to a standard force field [the current implementation uses parameters from Cornell et al. (1995) (17)]. Using these potentials, the box parameters are calculated by choosing the extremities and forming a cube along those dimensions. This box is then divided into cubic grids of a selected dimension. The center of each grid now identifies it uniquely and (for charge clustering) charges are calculated on each grid (which may be due to one or more atoms falling inside the grid). Using these values of charges the cubic grids are clustered using a simple criterion of Euclidian distance and hierarchical clustering. For generating the postscript tree structures, cluster diagrams and the distance tables, we use the open source free software provided by P. Kleiweg (http://odur.let.rug.nl/~kleiweg/indexs.html). For hydrophobic clusters, charges are not assigned to all atoms. Only the Cα atoms of hydrophobic residues are assumed to have a pseudo-charge of 1.0 each. The rest of the clustering proceeds in the same way as that of the charged grids. Chain breaks are implemented whenever clustering of only one chain is desired.
Clustering is started by first calculating the pairwise (Euclidean) distance between the grid centers. Once the first distance matrix is available, the first branches of the tree are constructed based on these distances. These first-level clusters (or pairs) are then joined by calculating distance between pairs. The distance between the clusters (or pairs) here is defined in two ways. In the group average or average linkage method each cluster location is the geometric center of all the points (grid centers) in the cluster. In the single linkage method, this distance refers to the distance between the nearest two points (grid centers) in the two clusters. The group average clustering makes more sense if the cluster geometry is spherical or closer to spherical. The single linkage may be useful if the cluster members in the original structure are less spherical in nature. Furthermore, single linkage will be preferable when a cluster of residues may be caused by successive attachment of residues to a region of interest, e.g. hydrophobic residues in a transmembrane helix. On the other hand group average or average linkage will reflect a more realistic situation if members of a cluster are all important to each other as is the case of hydrophobic cores in proteins and charged patches in DNA binding proteins. This process of joining pairs of grids and sub-clusters is continued until all grids have been joined and is termed ‘hierarchical clustering’.
QGRID QUERY INTERFACE
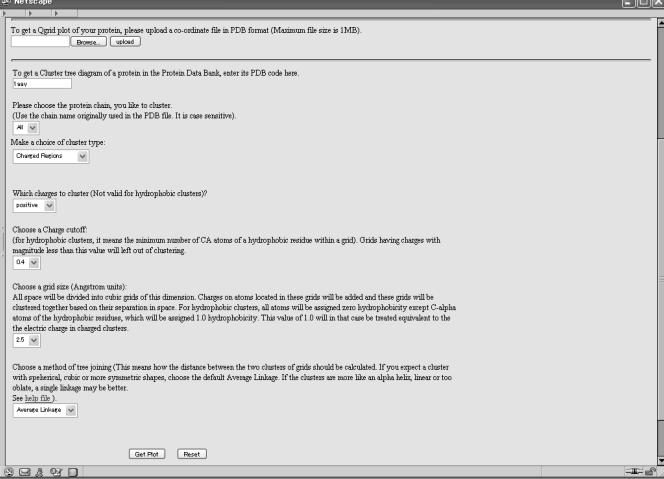
Qgrid has a simple HTML query interface, which takes the following inputs as the options (Figure 1):
Figure 1.
Query interface of Qgrid.
File upload or PDB code: This is simply the four-letter PDB code of the protein for which clusters are desired. We have a local mirror of PDB from which this data will be subsequently retrieved for calculations.
Cluster type: Here the users can decide if they want to generate a cluster of hydrophobic or charged regions. This field input is implemented by way of <select> and <option> keywords in HTML. Only two options are provided in the current implementation.
Chain name: This field is also provided by way of <select> and <option> tags. All possible chain names are provided as an option and users can select their chain by a pull-down menu. This field is case sensitive.
Charge cutoffs: Once the grids are formed according to the dimensions of the protein, there is no need to include all grids in the final clusters. This option will remove all those grids which have a charge less than the selected cutoff. In order to avoid nonsensical user inputs in this field, we do not allow text input and provide a choice of numbers which can be selected by users. In hydrophobic clusters, it will eliminate those grids which have less than this number of Cα atoms from any hydrophobic residue (only integer values will make sense).
Grid size: This option will be needed if the protein size is too big or too small, in order to make the graphical outputs manageable/visible. Large grid size will give smaller tree diagrams and vice versa. Users can select from a list of options provided here.
Tree joining and clustering method: Here users can select how the distance between two clusters is calculated. Description of these methods is already provided above.
QUERY RESULTS
The main output format of this server is postscript. Once the query is submitted a tree diagram in postscript format will be placed in a temporary HTML folder from where it can be downloaded almost immediately. The query result just provides the link to the page where results are expected. The generated postscript file may either be saved on a local system to visualize later or be seen within the browser. Results of postscript data are best viewed using ghostview (http://www.ghostscript.com; http://www.gnu.org/software/ghostview/ghostview.html) and we recommend use of this software for viewing these files, especially because it is free and open source software. For further convenience of online viewing, these postscript files are also converted to GIF and PDF formats and the same can be viewed from the link on the query results.
INTERPRETING THE CLUSTER TREE
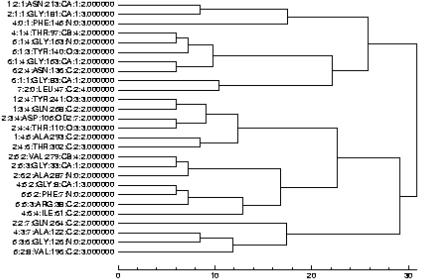
The simple nature of the tree diagram leaves little need for explanation as the distances in the tree branches are self-explanatory. In summary, each node or grid of the cluster is identified by a notation which provides useful information about the location of that node (Figure 2). The first three numbers on the nodes represent the indices of the grids along the x-, y- and z-axes of the box. The fourth index is the name of the residue which falls in this grid (only one residue name will appear for brevity). The next is the residue number in the original PDB data. Then the name of the atom from that residue, located within this grid, is presented (again, only one atom name will be shown for one grid). This is followed by the index of that atom within the atoms of the residue mentioned on the left. The last entry in this line refers to the total charge on that grid. Lengths of the tree nodes refer to the actual distances and hence the location of clusters in space can be easily understood. At the bottom of the graphics a scale is made to estimate the locations for these nodes and clusters. A complete map of cubic grids and how each atom of the protein was assigned to the grids is also available through a link at the bottom of the query result page. This may be helpful in examining each grid for its atomic contents in a more detailed manner.
Figure 2.
An example output of Qgrid (a hydrophobic cluster tree for a transmembrane protein PDB code 1e54; grid size = 6.0 Å and pseudo-charge cutoff of 2.0, i.e. the minimum number of Cα atoms of a hydrophobic residue in a grid is two; clustering is based on average linkage).
APPLICATIONS
Detection of charged and hydrophobic clusters in proteins based on the above method can be used for the following purposes:
Detection of the number and size of the charged and hydrophobic clusters in proteins: By simply looking at the tree diagram, the compactness of these clusters and their separations can be immediately estimated.
Comparison of structures: It has been noticed by looking at various charged and hydrophobic clusters that domains and chains in proteins that have similar structures form similar types of tree. Hence, this property can be used for comparing two protein structures. In the case when one molecule is a stereoisomer of the other, the root mean square distance between the superimposed structures will fail to detect the structural similarity. However, because the distance tree diagrams are directionless, two mirror images of molecules will form identical tree structures. This property can be used for comparing such structures.
Detecting new structure motifs: This is an interesting potential application of such cluster trees. Currently, structure motifs are represented in terms of their secondary structure elements (e.g. HTH for helix–turn–helix). Conserved charged and hydrophobic regions can also be represented using this method, such that similar structure patterns will have similar substructure in their cluster tree diagram. One advantage here is that we can introduce the information about charge/hydrophobicity naturally into such representations. We are working on compiling a database of such representations of motifs.
Complementary structures for protein–protein interactions: A positively charged domain structure should be a structural complement of a similar negatively charged domain structure. Such complementary structures can be detected by visual inspections of their charge clusters, made by Qgrid.
Detection of transmembrane segments in membrane proteins: One application of Qgrid may be to locate the hydrophobic clusters in a protein in order to hypothesize transmembrane regions of such proteins. Transmembrane, helical, beta barrel and other proteins are observed to form hydrophobic clusters and proximity of hydrophobic grids will be a good measure of the spatial distribution of such segments.
CONCLUSION
We developed an online web server by which charged and hydrophobic clusters can be located in proteins. Inputs of the server are the PDB code and chain name, and several user options are available to detect clusters in a two-dimensional tree diagram. These charged and hydrophobic clusters can be used to identify functionally and structurally important regions in proteins. In addition, such representations have a promising application in structure comparison and detection of motifs and complementary structures.
REFERENCES
- 1.Groome J.R., Fujimoto,E. and Ruben,P.C. (2003) Negative charges in DIII-DIV linker human skeletal muscle Na+ channels regulate deactivation gating. J. Physiol., 548, 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watanabe J., Beck,C., Kuner,T., Premkumar,L.S. and Wollmuth,L.P. (2002) DRPEER: a motif in extra-cellular vestibule conferring high Ca2+ flux rates in NMDA receptor channels. J. Neurosci., 22, 10209–10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vanhooren A., Vanhee,K., Noyelle,K., Majer,Z., Joniau,M. and Hanssens,I. (2002) Structural basis for differences in heat capacity increments for Ca2+ binding to two alpha-lactalbumins. Biophys. J., 82, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duin E.C., Bauer,C., Jaun,B. and Hedderich,R. (2003) Coenzyme M binds to a 4Fe-4S cluster in the active site of heterodisulfide reductase as deduced from EPR studies with the [33S] coenzyme M-treated enzyme. FEBS Lett., 538, 81–84. [DOI] [PubMed] [Google Scholar]
- 5.Sendak R.A., Berryman,D.E., Gellman,G., Melford,K. and Bensadoun,A. (2000) Binding of hepatic lipase to heparin. Identification of specific heparin-binding residues in two distinct positive charge clusters. J. Lipid Res., 41, 260–268. [PubMed] [Google Scholar]
- 6.Stawiski E.W., Gregoret,L.Mm. and Mandel-Gutfreund,Y. (2003) Annotating nucleic acid binding function based on protein structure. J. Mol. Biol., 326, 1065–1079. [DOI] [PubMed] [Google Scholar]
- 7.Nadassy K., Wodak,S.J. and Janin,J. (1999) Structural features of protein–nucleic acid recognition sites. Biochemistry, 38, 1999–2017. [DOI] [PubMed] [Google Scholar]
- 8.Shiba T., Takatsu,H., Nogi,T., Matsugaki,N., Kawasaki,M., Igarashi,N., Suzuki,M., Kato,R., Earnest,T., Nakayama,K. et al. (2002) Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature, 415, 937–941. [DOI] [PubMed] [Google Scholar]
- 9.Hakansson M., Svensson,A., Fast,J. and Linse,S. (2001) An extended hydrophobic core induces EF-hand swapping. Protein Sci., 10, 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espinosa J.F., Munoz,V. and Gellman,S.H. (2001) Interplay between hydrophobic cluster and loop propensity in beta-hairpin formation. J. Mol. Biol., 306, 397–402. [DOI] [PubMed] [Google Scholar]
- 11.Kuai J. and Kahn,R.A. (2000) Residues forming a hydrophobic pocket in ARF3 are determinants of GDP dissociation and effector interactions. FEBS Lett., 487, 252–256. [DOI] [PubMed] [Google Scholar]
- 12.Desrumaux C., Labeur,C., Verhee,A., Tavernier,J., Vandekerchhove,J., Rosseneu,M. and Peelman,F. (2001) A hydrophobic cluster at the surface of the human plasma phospholipid transfer protein is critical for activity on high density lipoproteins. J. Biol. Chem., 276, 5908–5915. [DOI] [PubMed] [Google Scholar]
- 13.Kwok S.C. and Hidges,R.S. (2003) Clustering of large hydrophobes in the hydrophobic core of two-stranded alpha-helical coiled-coils controls protein-folding and stability. J. Biol. Chem., 278, 35248–35254. [DOI] [PubMed] [Google Scholar]
- 14.Martz E. (2002) Protein Explorer: easy yet powerful macromolecular visualization. Trends Biochem. Sci., 27, 107–109. [DOI] [PubMed] [Google Scholar]
- 15.Humphrey W., Dalke,A. and Schulten,K. (1996) VMD—Visual Molecular Dynamics. J. Mol. Graphics, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- 16.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cornell D.C., Cieplak,P., Bayly,C.I., Gould,I.R., Merz,K.M., Ferguson,D.M., Spellmeyer,D.C., Fox,T., Caldwell,J.W. and Kollman,P.A. (1995) A second generation force field for the simulation of proteins, nucleic acids and organic molecules. JACS, 117, 5179–5197. [Google Scholar]