Conspectus
In 1996, a snapshot of the field of synthesis was provided by many of its thought leaders in a Chemical Reviews thematic issue on “Frontiers in Organic Synthesis”. This Accounts of Chemical Research thematic issue on “Synthesis, Design, and Molecular Function” is intended to provide further perspective now from well into the 21st century. Much has happened in the past few decades. The targets, methods, strategies, reagents, procedures, goals, funding, practices, and practitioners of synthesis have changed, some in dramatic ways as documented in impressive contributions to this issue. However, a constant for most synthesis studies continues to be the goal of achieving function with synthetic economy. Whether in the form of new catalysts, reagents, therapeutic leads, diagnostics, drug delivery systems, imaging agents, sensors, materials, energy generation and storage systems, bioremediation strategies, or molecules that challenge old theories or test new ones, the function of a target has been and continues to be a major and compelling justification for its synthesis. While the targets of synthesis have historically been heavily represented by natural products, increasingly design, often inspired by natural structures, is providing a new source of target structures exhibiting new or natural functions and new or natural synthetic challenges. Complementing isolation and screening approaches to new target identification, design enables one to create targets de novo with an emphasis on sought-after function and synthetic innovation with step-economy. Design provides choice. It allows one to determine how close a synthesis will come to the ideal synthesis and how close a structure will come to the ideal function.
In this Account, we address studies in our laboratory on function-oriented synthesis (FOS), a strategy to achieve function by design and with synthetic economy. By starting with function rather than structure, FOS places an initial emphasis on target design, thereby harnessing the power of chemists and computers to create new structures with desired functions that could be prepared in a simple, safe, economical, and green, if not ideal, fashion. Reported herein are examples of FOS associated with (a) molecular recognition, leading to the first designed phorbol-inspired protein kinase C regulatory ligands, the first designed bryostatin analogs, the newest bryologs, and a new family of designed kinase inhibitors, (b) target modification, leading to highly simplified but functionally competent photonucleases—molecules that cleave DNA upon photoactivation, (c) drug delivery, leading to cell penetrating molecular transporters, molecules that ferry other attached or complexed molecules across biological barriers, and (d) new reactivity-regenerating reagents in the form of functional equivalents of butatrienes, reagents that allow for back-to-back three-component cycloaddition reactions, thus achieving structural complexity and value with step-economy. While retrosynthetic analysis seeks to identify the best way to make a target, retrofunction analysis seeks to identify the best targets to make. In essence, form (structure) follows function.
The extraordinary and to some extent inexplicable production of urea without the assistance of vital functions, for which we are indebted to Wöhler, must be considered one of the discoveries with which a new era in science has commenced.
Justus von Liebig (ca. 1837)1
That new era has since transformed science with consequences of immense societal benefit. From new medicines to materials and extraordinary research tools, our ability to make and manipulate molecules has profoundly advanced our understanding of molecular structure, reactivity, and function and with that contributed to the molecularization of science from molecular anthropology to zoology and most disciplines in between.2 Over a period of less than 200 years, the synthesis of natural products, once considered impossible, has become increasingly common, increasingly impressive, and increasingly consequential to research, our economy, our health care, and our standard of living.
In 1996, a snapshot of the field of synthesis was provided by many of its thought leaders.3 This Accounts of Chemical Research thematic issue on “Synthesis, Design, and Molecular Function” provides an inspiring sequel now with a 21st century perspective. Our own contribution addresses studies in our laboratory on function-oriented synthesis (FOS), a strategy for achieving function with synthetic economy, a goal of most “orientations” in synthesis.4 By starting with function rather than structure, FOS places an initial emphasis on target design, thereby harnessing the power of chemists and computers to create new structures with natural or new functions that could be prepared in a simple, safe, economical, and green if not ideal fashion.5 While retrosynthetic analysis seeks to identify the best way to make a target, retrofunction analysis seeks to identify the best targets to make. In essence, form (structure) follows function.
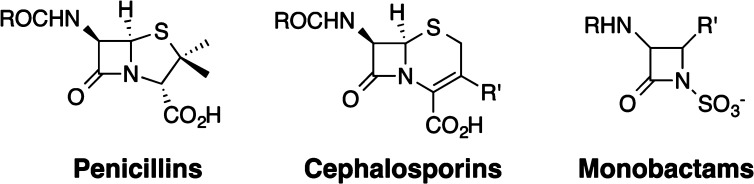
Whether natural or designed, the targets of organic synthesis have increased in number and diversity over the years from simple to complex molecules and even molecular systems. As impressively chronicled by Cragg, Grothaus, and Newman,6 the sources of new chemical entities (NCEs) over the last few decades have been diverse. Natural products, historically structures of great synthetic and medicinal interest, continue to figure as new therapeutic leads, accounting for 6% of the 1024 NCEs reported between January 1981 and October 2009. An additional 27% of the reported NCEs are derivatives of natural products. Significantly, 30% of the NCEs are synthetic compounds that share a functional or pharmacophoric relationship with natural products, while 37% are synthetic compounds with no natural product connection. These distributions are likely to fluctuate due to funding decisions and the realization that many natural products, while significant leads, are not optimized for their intended use as, for example, therapeutic agents. Thus, while natural products continue to inspire new synthetic strategies and methods based on their structures, they increasingly also inspire the design of new and more synthetically accessible structures based on their function (activity). Because a given function can be achieved with many different structures, design-for-function becomes a powerful strategy for creating totally new targets inspired by natural product leads or by abiological needs. β-Lactam antibiotic structures, as one example, changed over time from natural to designed and often from complex to less so, while their activity (function) was largely preserved or improved (Figure 1).7 A key to this success was knowledge of their mechanism of action and its use in designing simpler and thus more synthetically accessible targets with similar or improved function. Pertinent to current discussions about making molecules, fermentation, biosynthesis, semisynthesis, synthetic biology, synthetic methodology, and abiological synthesis all played prominent and often complementary roles in advancing this field.
Figure 1.
β-Lactam antibiotics: Similar function, different structures.
What’s next? The answer is complex and not driven only by scientific curiosity since funding also influences direction. It is however hard to imagine a time when natural products, representing 3.8 billion years of chemical experimentation and information, would not figure as prominent sources of inspiration and value. At the same time, given the structure generating and searching capabilities of computers, it is equally hard to imagine that virtual structures and libraries would not increasingly figure as sources of new structures and inspiration. Think about it. How many of us have drawn or doodled a new structure perhaps as a chiral ligand for asymmetric catalysis, a reagent for synthesis or a probe to test a theory. Computers can do this on a massive scale. The recent report of a 166 billion compound library exemplifies this potentially game-changing source of new structures, synthesis inspiration, and function.8 Driven by graph theory and guided by structural theory, this virtual approach to structure generation can identify all possible connections of selected atoms found, for example, in many known and as yet unknown drug leads. It deselects potentially problematic features (e.g., strained bonds), providing a virtual library that can be searched for structures of interest (Figure 2). While a work in progress, it is projected to reach a trillion compounds in the near future and should vastly exceed that as its potential is further explored.
Figure 2.
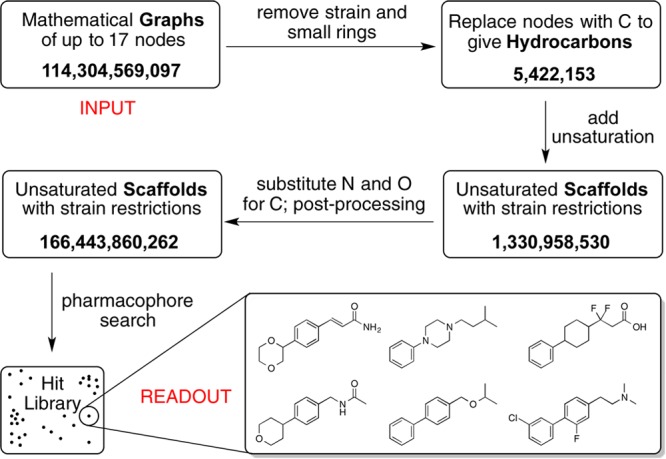
Generation of a 166 billion compound virtual library. Modified from Reymond et al.8
Virtual structure generation has great but largely unexploited value. It can be used to generate not only as yet undiscovered natural products but also structures that exhibit atom connectivities not encountered in biosynthetic pathways. While searching such a vast library, say for pharmacophoric features,9 is beyond human capabilities, it is as simple computationally as doing a word search on Google. In collaboration with the Reymond Group, for example, we have extracted a library of 90 000 hits from a 166 billion compound library based on a search for structures with a bryostatin- or phorbol-like pharmacophore (function). Many of these hits are as inspiring synthetically, and potentially functionally, as the natural products upon which the pharmacophores are based. Fundamentally, this is not new, since any lecture on constitutional isomers has addressed this approach “by hand” often in connection with identifying all the constitutional isomers of, say, pentane. What is new is the power of computers to do this exhaustively for larger collections of atoms and to identify and characterize the expected properties of the newly identified targets in terms of attributes of structural and functional interest.
Nature’s vast library and computer-derived virtual libraries can be expected to provide a continuing source of new structural challenges to test existing reactions and strategies and to inspire the design of new ones. If so, then what’s next?2 Are there other sources of structures? Or in the words of Dieter Seebach: “Organic Synthesis – Where Now?”10 Answers to these questions will vary from individual to individual. However, most would agree that the goals of synthesis are increasingly related to not just target structure but target function. It is indeed the function of a structure that is often used as justification and motivation for its synthesis. In 2014, for example, ∼134 total syntheses were reported in ACS journals alone. The vast majority of these efforts called attention, in varying degree, to the biological activity of the targets. With over 7000 diseases for which we have no effective therapy, it is likely that synthetic chemists will continue to rise to meet these, as yet unmet, challenges associated with function.
Structures with function can be discovered through screening of natural and virtual libraries. A third source is through design. This differs from natural and virtual approaches in that the structures are created de novo to achieve function and ease of synthesis. Such design-driven structure generation has lagged behind discovery-driven screening in producing functional targets partly because it requires an understanding of complex and dynamic molecular systems. Increasingly, however, with the emergence of powerful analytical and computational tools, such knowledge becomes more accessible and design becomes a more promising path to creating new structures with function. An attractive aspect of this approach is that design can also be used to create new synthetic challenges and ensure a more practical, step-economical, and “green” outcome.5,11 Here we describe the genesis and an overview of representative FOS studies in our laboratory with examples of FOS directed at molecular recognition, reactive intermediate generation, drug delivery, and reagent equivalency.
Our FOS studies evolved from an early interest in carcinogenesis.12 The basis for that interest was quite logical. Cancer is a major killer. There are three ways to intervene in addressing cancer or any disease: therapy, detection, and prevention. Of these, prevention is the most effective and least costly. Prevention in turn requires knowledge of how a disease starts. For cancer, it was clear then that carcinogens play a role. What was less clear then and still not fully understood now is the role of tumor promoters in carcinogenesis. Tumor promoters are noncarcinogenic agents that amplify the effect of a carcinogen. The most potent tumor promoters are the plant-derived phorbol esters (PEs).13 We thus set out to make these agents and to study their role in carcinogenesis.
Contemporaneously, Nishizuka reported a new family of proteins now collectively known as protein kinase C (PKC).14 It was found that PKC isoforms are regulated by the binding of endogenous diacylglycerol (DAG). In what for us was a remarkable confluence of events, it was subsequently found that mammalian DAG competes with plant-derived PEs in binding to PKC. Thus, simple DAG and the more complex PEs shared a similar function (binding to the same site of PKC) but not a similar structure. Through extensive computer analyses of the two structures, we identified pharmacophore candidates, among which the best spatial correlation was between the oxygens at C9, C20, and C4/C3 of PEs and a similar set of three hydrogen-bond donor and acceptor oxygens in DAG. This provided a blueprint for the design of PKC modulators and led to the first designed ligand (1) that bound to the regulatory domain of PKC competitively with the PEs and DAG and elicited function (phosphorylation) (Figure 3).12 While years later we successfully completed the total synthesis of phorbol,15 our designed and notably simpler agent was prepared in only 2 weeks, providing function with step-economy by design.
Figure 3.
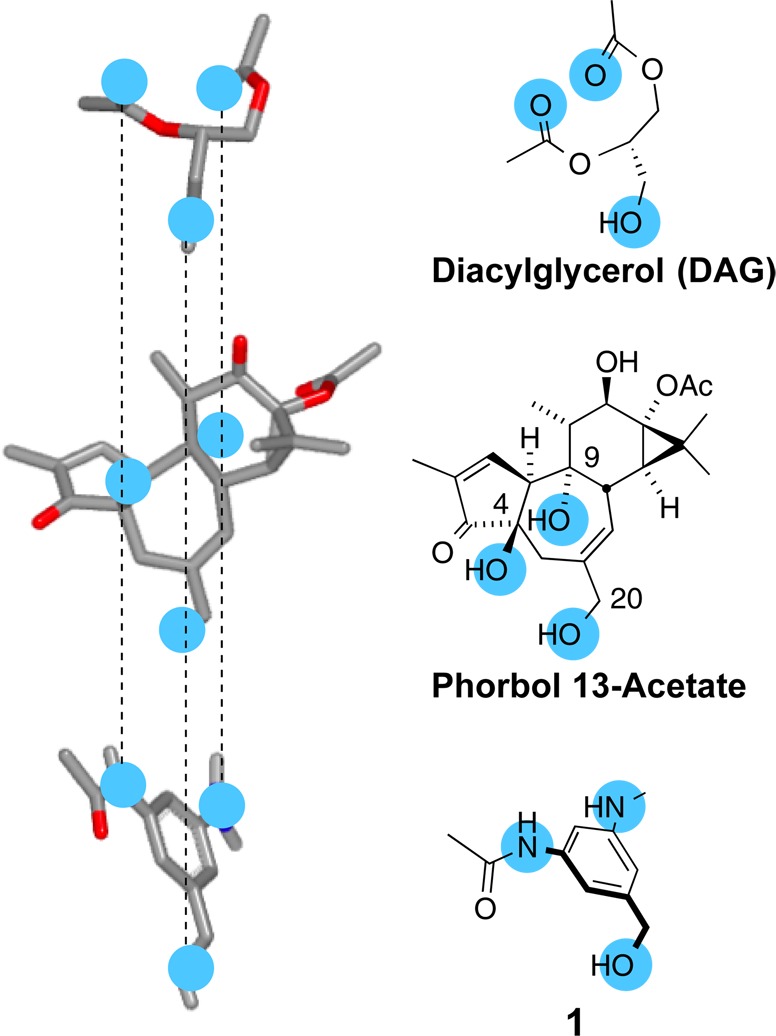
PKC activators (phorbol 13-acetate, PDB 1PTR):16 Different structures, similar pharmacophores and function.
There were several lessons learned from this early study. The function of a molecule, in this case binding to PKC, can be emulated by only a subset of its complex structure. Thus, the time required to design and synthesize simpler systems can often be greatly reduced relative to more complex systems, thereby favoring time-economy,5 a priority goal in research and especially that directed at clinical needs. Finally, the step-economy achieved through synthesis-informed design can dramatically lower the cost of accessing tool compounds or therapeutic leads. The success of this FOS strategy is based on using natural products to inspire design, thereby speeding access to their functions. It is noteworthy however that the designed product is not a derivative of the natural product, that is, a structural analog, but rather a simplified functional analog incorporating only its pharmacophoric features. This difference is sometimes lost in discussions about diverted total syntheses.17 There are no diversions in design. One starts with a functional goal and creates a structure to achieve it.
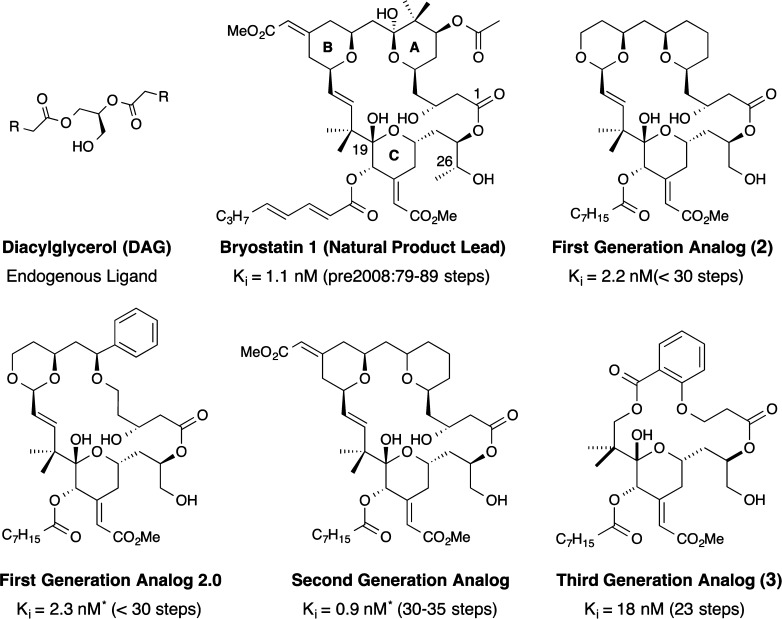
The impact of this study was heightened by the report of Pettit18 on another DAG competitive PKC binder, namely, marine-derived bryostatin. Significantly, bryostatin, unlike PEs, does not exhibit tumor promoter activity, exemplifying PKC’s complex role in cellular function. PKC has since been connected to many therapeutic indications including cancer,19 Alzheimer’s disease,20 and eradication of HIV/AIDS.21 As the story of bryostatin’s clinical potential took shape, concerns about its availability grew. Bryostatin is thought to be produced in a symbiotic relationship involving a bryozoan (Bugula neritina) and a bacterium (Endobugula sertula) in its gut. Fourteen tons of the source organism was needed to produce only 18 g of bryostatin for clinical trials. While the potency of bryostatin (dosed at ∼25–125 μg/m2) allowed for many clinical studies, its timely supply remained an issue and therefore became a superb test for FOS.
Following the protocol successfully used with DAG and PEs, computer generated structures of bryostatin and DAG were compared to identify shared pharmacophoric features.22 The best correlations suggested a hydrogen-bonding role for the C1, C26, and C19 oxygens of bryostatin in PKC binding. However, unlike conformationally restricted PEs, bryostatin can assume thousands of conformations, many energetically close to the global minimum. As such, we took a conservative approach to designing functional analogs. The entire lower half of the bryostatin structure (largely the C-ring and C1 lactone), which included the putative pharmacophoric features and thus the functionality contacting PKC, was kept intact while the A- and B-rings were dramatically simplified and modified (C14 replaced with an oxygen to enable a convergent synthesis). Synthesis of the resultant simplified structure 2 required only ∼30 steps, while at the time, syntheses of natural bryostatins required 69–89 steps. Significantly, this study led to the synthesis of the first designed bryostatin analog, which along with other “bryologs” exhibited affinities to PKC comparable to or better than the natural bryostatins (Figure 4).23 Importantly, the best analogs at the time, dubbed nanolog and picolog (2) based on their PKC affinities, performed significantly better than bryostatin in inhibiting the growth of human cancer cells as determined in a study by the National Cancer Institute (NIH).23 These and related functional analogs have since been advanced to animal studies and are in preclinical evaluation for the treatment of Alzheimer’s disease and for the eradication of HIV/AIDS. Some of this work and related FOS contributions on bryostatin analogs from others have been reviewed recently.23 For this analysis, the important take-home message from our work is that computer-assisted design provided simplified and thus more synthetically accessible functional analogs that perform as well as or better than the natural product.
Figure 4.
Function with step-economy through design: PKC potency with progressive simplification. The Ki values are for PKCδ. *Ki values are for PKC rat brain mix.
Are further structural simplifications possible? It follows from our pharmacophore hypothesis that the A- and B-rings of bryostatin contribute to PKC binding by preorganizing the pharmacophoric groups in contact with the protein and as such are subject to further simplification. Recently, we reported analogs in which these rings are replaced by commercially available salicylic acid, a substitution that further reduces step count (3, Figure 4).24 Computer comparisons of the designed analog with bryostatin showed an excellent spatial correlation of recognition domain atoms (RMSD = 0.05 Å). In line with this analysis, the lead salicylate-derived analog showed potent binding to PKC β and δ with Ki’s of 3.1 and 1.9 nM, respectively, comparable to bryostatin with Ki’s of 1.0 and 1.1 nM. Requiring only ∼23 steps to make relative to the ∼40 steps required for the best syntheses to date of natural bryostatins,23 this recent FOS study provides step-economical access to potent new PKC modulators.
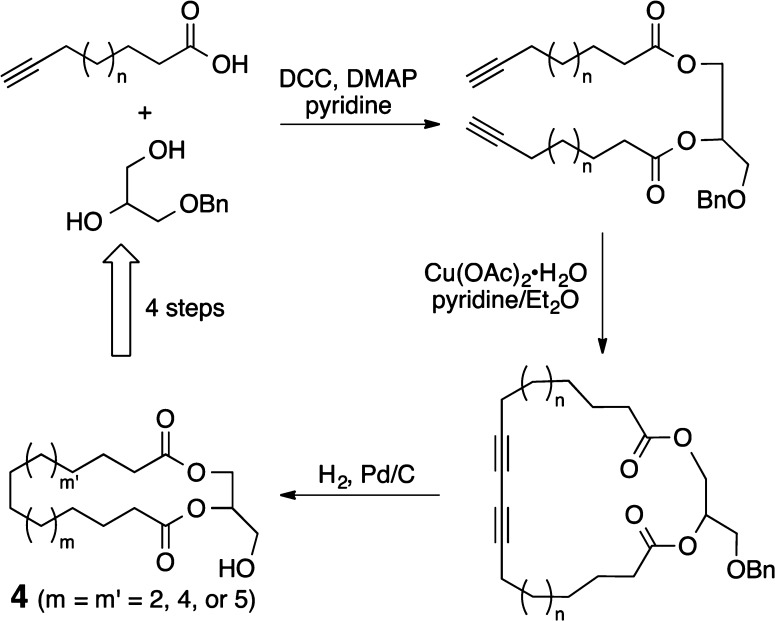
The aforementioned studies sought to understand how complex molecules like PEs and bryostatin mimic DAG in binding to PKC and from that how to design simplified PE and bryostatin analogs. A series of deletions and simplifying substitutions were employed. An unaddressed complementary question is whether DAG, an already simplified, natural PKC regulator, could be modified to create PE- or bryostatin-like activity. Interestingly, DAG is significantly less potent than plant-derived PEs and marine-derived bryostatin, suggesting that its greater flexibility (entropy) might contribute to its lower PKC affinity. The FOS challenge thus becomes one of designing a less flexible DAG. Early studies showed, however, that modifications of DAG, including heteroatom substitution, changes in the ester linkages, and insertion of additional atoms in the glycerol backbone, result in compounds with lower PKC affinities. Thus, one needs a way to restrict the DAG conformation without significant changes to its structure. Our approach to this challenge led to the creation of a new class of molecules, cyclic DAGs (cDAGs) (Figure 5).25 By connecting the lipid termini in DAG one creates a cDAG, a macrocyclic bis-lactone (4). Like other macrolides this changes the population of conformers without changing the functionality. Preliminary studies show that cDAGs bind to PKC isoforms with affinities that vary expectedly with ring size but approach values in the single digit nanomolar range and differ from acyclic DAGs in PKC selectivity. Significantly, synthetic access to cDAGs requires only 4 steps. Our cDAG analogs compete with the original triad of PKC modulators, providing new, more accessible, and potent leads for each scaffold. Our early and ongoing FOS work on bryostatin and related compounds has now been joined by others making noteworthy contributions to this field.26
Figure 5.
Four steps to cyclic diacylglycerols (4), a new family of PKC modulators.
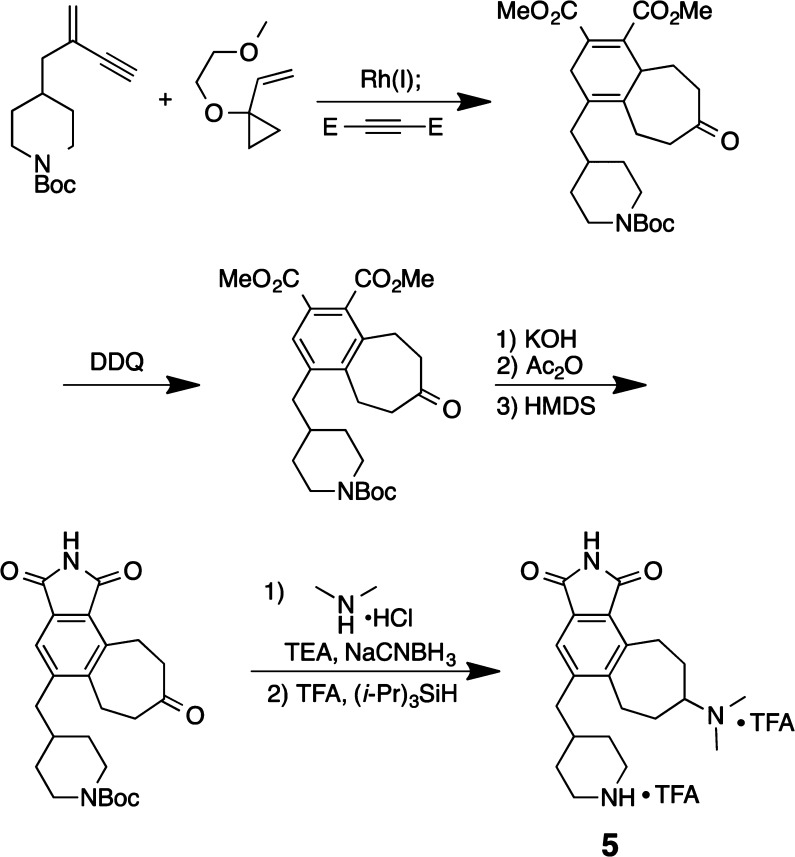
How general is this FOS approach? Given the role of kinases in human diseases, major research efforts have been directed at identifying kinase inhibitors by screening natural and non-natural chemical libraries. Several years ago, we launched a FOS program directed at designing kinase inhibitors that would bind to the kinase ATP-binding site and thus block function. Our starting point was, not unlike in the above studies, a natural product, staurosporine. Staurosporine is a promiscuous kinase inhibitor, affecting >253 kinases with Kd’s in the micro- to nanomolar range.27 Our goal here was to first achieve binding using readily accessible designed compounds and then to tune these compounds for function. We recently disclosed success with the first of these goals.28
Staurosporine inhibits kinase function through hydrogen-bonding and electrostatic interactions associated with its lactam and cationic amine subunits.29 Computer models suggest that these key elements are separated by ∼9.9 Å. The roles of the indolocarbazole and pyranyl subunits of staurosporine are less clear but certainly conformationally restrict its binding elements. These function-determining features are encoded in our first generation designed analogs (Figure 6). Our first strategy to step-economically access these analogs involved a three-component serial cycloaddition based on an initial [5 + 2] cycloaddition that we introduced in 1995.30 The diene product of that step would then be captured in a second (Diels–Alder) cycloaddition, producing most of the target analog structure in one operation. In fact, while the process proved less direct, this approach did provide analogs in only seven steps with the lead analog sufficiently potent (IC50 = 2.9 μM) to encourage further study.
Figure 6.
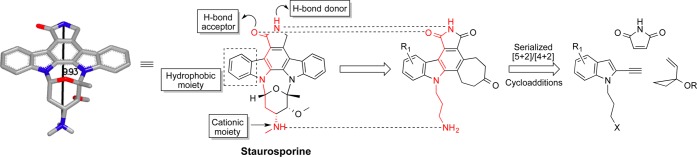
Design and synthesis of simplified staurosporine (PDB 1XJD) analogs16
Our next iteration involved further simplification of the analog structure while retaining the complexity-increasing and step-decreasing value of back-to-back cycloadditions (Figure 7). Gratifyingly, our newest analog 5 exhibits improved potency with an IC50 value of 13 nM and improved step-economy, being prepared in only five steps using a novel three-component, serial [5 + 2]/[4 + 2] cycloaddition strategy.
Figure 7.
Step-economical synthesis of designed, simplified staurosporine analog 5.
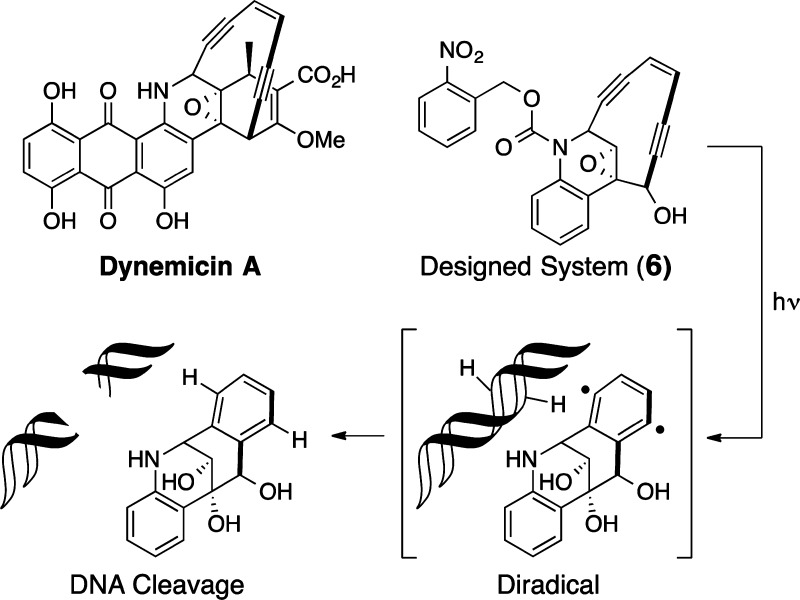
What other functions might FOS address? As exemplified above, function can involve binding to a target. Function can also involve target modification. For example, complex ene-diyne natural products are known to cleave DNA through the generation of an arene 1,4-diyl involving a Bergman cycloaromatization.31 Dynemicin A is an exquisite example and reminder of the wonders of natural products.32 Its ene-diyne subunit is encased in a 10-membered ring incorporating a trans-epoxide that prevents Bergman cycloaromatization. Cleavage of the epoxide, triggered by reduction of the anthraquinone subunit, induces cycloaromatization of the ene-diyne to an arene 1,4-diyl that abstracts hydrogens from proximate DNA nucleotides. Based on this analysis, FOS can be used to design simpler but effective mimics of mechanism. To mimic dynemicin A,33 we incorporated an epoxide-ene-diyne subunit into a hydroisoquinoline (Figure 8) and regulated electron density on its amine “trigger” by attaching it to a cleavable carbonyl group. By incorporating photocleavable or other cleavable groups on the nitrogen, one could thus control initiation.34 In the event, upon irradiation in the presence of DNA, ene-diyne 6 is readily converted to an arene 1,4-diyl, resulting in single and double strand cleavage of DNA. An especially attractive aspect of this approach, as with other FOS studies, is that function is achieved with step- and time-economy. The synthesis of “photonuclease” 6 required only eight steps.
Figure 8.
A designed, simplified dynemicin A analog that cleaves (function) DNA.
Are further simplifications possible? The basis for function is not always obvious. Upon further analysis, we came to realize that the key to DNA cleavage (function) was not a diyl but something simpler, an arene radical. In unrelated research, we showed that photolysis of certain triazoles proceeds with extrusion of nitrogen and formation of what is operationally an arene radical. When this photoextrusion is conducted in D8-THF, one isolates a deuterated product. In the presence of DNA, this process proceeds with DNA cleavage. To control cleavage selectivity, we next attached a DNA recognition element to the benzotriazole (7, Figure 9).35 The resulting photonuclease upon excitation caused single and double DNA strand breaks, with selective cleavage at predominantly one site in a 167 base pair sequence.36 While our first generation DNA cleavers required only eight steps to make, the functional part of this this simpler system proved to be a commercially available molecule.
Figure 9.
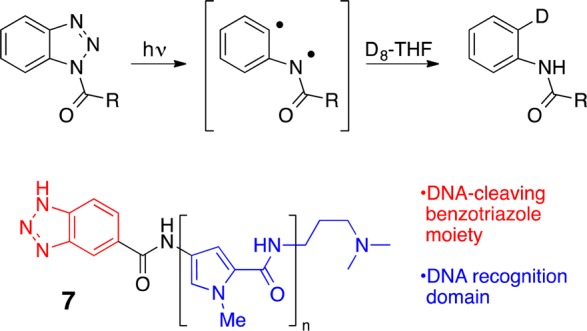
Designed simple photonucleases for selective DNA cleavage.
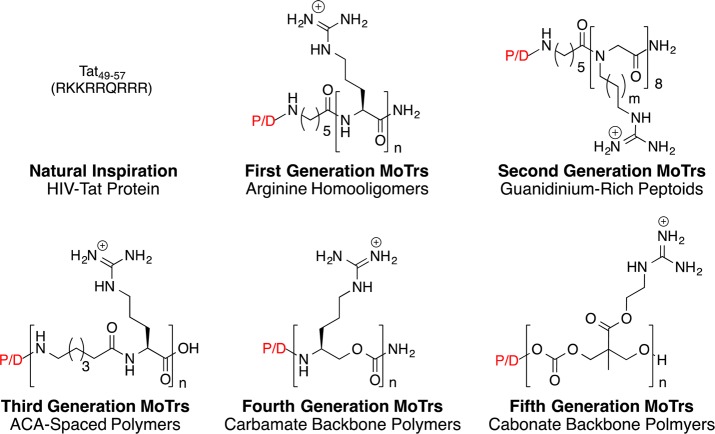
Beyond molecular recognition (PKC) and modification (DNA cleavage), another function of great significance is molecular transport, a process pertinent to drug delivery, imaging, diagnostics, and medicine. As illustrated previously, FOS design is often naturally inspired and hypothesis driven. Our studies on transport started in 1997 with an interest in drug delivery, that is, moving molecules across biological barriers, and were inspired by the “natural product” HIV-Tat, a protein that, unlike most, enters cells.37
At the time, HIV-Tat’s ability to enter cells was attributed to its polycationic domain (Tat nine-mer: RKKRRQRRR). Our interest was stimulated by the counterintuitive nature of this suggestion, specifically that a polycharged, water-soluble compound would cross a nonpolar membrane. Clearly, there were lessons to be learned. Reverse-engineering experiments were therefore conducted to generate the information needed to design superior, more synthetically accessible agents.38 An arginine nine-mer was found to outperform the Tat nine-mer. Charge type mattered: oligoarginines outperform oligolysines. Spacing mattered: greater spaces between arginines favor cellular uptake. Length mattered: various oligomers worked but octaarginine is the best compromise between function (uptake) and cost of goods. Stereochemistry mattered less: non-natural oligoarginine is better than its enantiomer.
The above reverse-engineering studies indicated that the number and spatial array of guanidinium groups were keys to cell entry. We proposed that entry was initiated by hydrogen-bonding and electrostatic association of cationic guanidinium groups with negatively charged cell surface carboxylates, sulfates, or phosphates.39 In one mechanistic scenario (adaptive translocation), the cell-surface bound oligoguanidinium transporter is driven inward by the inward polarization of the membrane potential. For larger cargos, endocytotic pathways participate or dominate. Single molecule experiments suggested that multiple mechanisms could operate concurrently.37,40
The special role that we first proposed for the guanidinium group (hydrogen-bonding to cell-surface anions), the number of guanidinium groups, their spatial array, and membrane potential, collectively provided the information for the first designed cell-penetrating guanidinium-rich transporters, peptoids with an arginine side chain attached to an amide backbone (Figure 10).38 We subsequently reported the first “spaced” cell-penetrating peptides showing that spacing between side chains influences uptake.37 Our expectation that more effective nonpeptidic agents could be designed led to our introduction of the more general term “molecular transporter” (MoTr). We have since shown that peptoids, spaced peptides, oligocarbamates, oligocarbonates, dendrimers, and other backbones are effective (Figure 10). Impressive contributions to this rapidly growing field from other groups have been reported in numerous reviews.37
Figure 10.
“Natural product” inspired design of cell-penetrating guanidinium-rich molecular transporters. P/D = probe/drug, n ≥ 5.
The beneficial impact of design on step-economy is again apparent. The Tat nine-mer requires ∼18 steps to make. Using a segment doubling strategy, one achieves synthesis of octaarginine in only 9 steps and on a practical GMP scale. Finally, by employing polymer concepts, the synthesis of guanidinium-rich oligocarbonates requires only two steps irrespective of length. Thus, an 8-mer or a 20-mer can be made in only two steps by controlling the initiator-to-monomer ratio.41Superior function (cell entry) is again achieved with step-economy through design.
Function extends beyond biological readouts. For example, in studies directed at new kinase inhibitors, we developed an interest in 1,2,3-butatriene as a reagent for one-flask back-to-back cycloadditions.30 Enophilic cycloaddition to its 2,3-double bond would generate a diene for a subsequent dienophilic cycloaddition. However, butatriene is difficult to make, is unsafe to handle, and reacts uncontrollably with most reactants. So we sought to design a reagent that would function like butatriene but without its liabilities. 4-(Trimethylsilyl)but-2-yn-1-ol (TMSBO, 8) fit our design criteria. Unlike butatriene, it is easily prepared and handled. Moreover, it would be expected to engage in ynophilic cycloadditions at its 2,3-π bond, producing, after a vinylogous Peterson elimination, a diene for a subsequent cycloaddition. This proved to be the case. Rhodium-catalyzed [5 + 2] cycloaddition of commercially available vinylcyclopropane 9 and TMSBO produced diene 10, which was captured in a second metal-catalyzed or Diels–Alder [4 + 2] cycloaddition to produce polycycle 11 (Figure 11). This and related reagents are now being developed in connection with cascade catalysis and multicomponent reactions.
Figure 11.
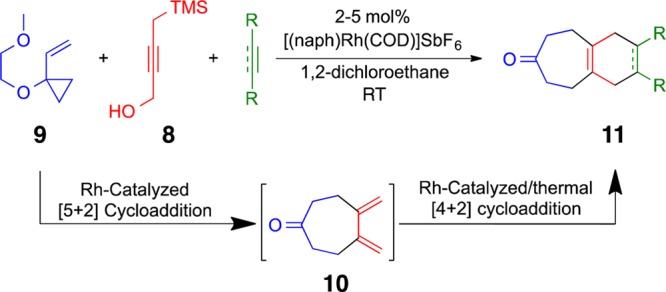
TMSBO, a reagent designed to function like butatriene in three-component serialized cycloadditions.
Form follows function. Design unifies the two. Screening of natural, non-natural, and virtual libraries allows one to uncover structures with known evolutionary or new functions. Known or new functions can also be created through knowledge-driven design. Design provides choice over form and thus whether a structure and its function would be synthetically accessible in a step- and time-economical fashion. Chemists are uniquely positioned to drive design utilizing knowledge of mechanism and synthesis to create function and form in ways limited only by imagination.
Acknowledgments
Grants NIH-CA031841 and NIH-CA031845 from the National Institutes of Health and fellowships from the National Science Foundation (R.V.Q., M.C.S.) are acknowledged, along with the exceptional contributions of referenced co-workers.
Biographies
Paul A. Wender is a product of Wilkes, Yale, and Columbia Universities, served on the Harvard faculty, is now Bergstrom Professor of Chemistry and Professor, by courtesy, of Chemical and Systems Biology at Stanford, and has the pleasure to be working with the coauthors and similarly gifted co-workers on major problems in synthesis, chemistry, biology, and medicine.
Ryan V. Quiroz received his B.S. degree in Biochemistry (2012) from UCLA and is currently an NSF Graduate Research Fellow at Stanford University. His research in Paul Wender’s laboratory focuses on new reaction methodology and the design and synthesis of small molecule kinase inhibitors.
Matthew C. Stevens received his B.S. degree in Chemistry (2012) from the University of Massachusetts Amherst and is currently an NSF Graduate Research Fellow at Stanford University. His research in Paul Wender’s laboratory focuses on new reaction methodology and the design and synthesis of small molecule therapeutics.
The authors declare no competing financial interest.
Special Issue
Published as part of the Accounts of Chemical Research special issue “Synthesis, Design, and Molecular Function”.
Funding Statement
National Institutes of Health, United States
References
- Cohen P. S.; Cohen S. M. Wöhler’s Synthesis of Urea: How Do the Textbooks Report It?. J. Chem. Educ. 1996, 73, 883–886. [Google Scholar]
- Wender P. A.; Miller B. L. Synthesis at the Molecular Frontier. Nature 2009, 460, 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender P. A. Introduction: Frontiers in Organic Synthesis. Chem. Rev. 1996, 96, 1–2and other articles in the special issue.. [DOI] [PubMed] [Google Scholar]
- Wender P. A.; Verma V. A.; Paxton T. J.; Pillow T. H. Function-Oriented Synthesis, Step Economy, and Drug Design. Acc. Chem. Res. 2007, 41, 40–49. [DOI] [PubMed] [Google Scholar]
- Wender P. A. Toward the Ideal Synthesis and Molecular Function through Synthesis-Informed Design. Nat. Prod. Rep. 2014, 31, 433–440. [DOI] [PubMed] [Google Scholar]
- a Cragg G. M.; Grothaus P. G.; Newman D. J. Impact of Natural Products on Developing New Anti-Cancer Agents. Chem. Rev. 2009, 109, 3012–3043. [DOI] [PubMed] [Google Scholar]; b Cragg G. M.; Grothaus P. G.; Newman D. J. New Horizons for Old Drugs and Drug Leads. J. Nat. Prod. 2014, 77, 703–723. [DOI] [PubMed] [Google Scholar]
- Konaklieva M. I. Molecular Targets of β-Lactam-Based Antimicrobials: Beyond the Usual Suspects. Antibiotics 2014, 3, 128–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddigkeit L.; van Deursen R.; Blum L. C.; Reymond J.-L. Enumeration of 166 Billion Organic Small Molecules in the Chemical Universe Database GDB-17. J. Chem. Inf. Model. 2012, 52, 2864–2875. [DOI] [PubMed] [Google Scholar]
- Awale M.; Reymond J.-L. Atom Pair 2D-Fingerprints Perceive 3D-Molecular Shape and Pharmacophores for Very Fast Virtual Screening of ZINC and GDB-17. J. Chem. Inf. Model. 2014, 54, 1892–1907. [DOI] [PubMed] [Google Scholar]
- Seebach D. Organic Synthesis—Where Now?. Angew. Chem., Int. Ed. Engl. 1990, 29, 1320–1367. [Google Scholar]
- a Sheldon R.Introduction to Green Chemistry, Organic Synthesis and Pharmaceuticals. In Green Chemistry in the Pharmaceutical Industry; Dunn P. J., Wells A. S., Williams M. T., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2010; pp 1–20. [Google Scholar]; b Trost B. M. The Atom Economy--a Search for Synthetic Efficiency. Science 1991, 254, 1471–1477. [DOI] [PubMed] [Google Scholar]; c Bryan M. C.; Dillon B.; Hamann L. G.; Hughes G. J.; Kopach M. E.; Peterson E. A.; Pourashraf M.; Raheem I.; Richardson P.; Richter D.; Sneddon H. F. Sustainable Practices in Medicinal Chemistry: Current State and Future Directions. J. Med. Chem. 2013, 56, 6007–6021. [DOI] [PubMed] [Google Scholar]
- Wender P. A.; Koehler K. F.; Sharkey N. A.; Dell’Aquila M. L.; Blumberg P. M. Analysis of the Phorbol Ester Pharmacophore on Protein Kinase C as a Guide to the Rational Design of New Classes of Analogs. Proc. Natl. Acad. Sci. U. S. A. 1986, 83, 4214–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker E. In Carcinogenesis – A Comprehensive Survey; Slaga T. J., Sivak A., Boutwell R. K., Eds.; Raven Press: New York, 1978; Vol. 2, p 11. [Google Scholar]
- For PKC reviews, see:; a Steinberg S. F. Structural Basis of Protein Kinase C Isoform Function. Physiol. Rev. 2008, 88, 1341–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Newton A. C. Protein Kinase C: Poised to Signal. Am. J. Physiol.: Endocrinol. Metab. 2010, 298, E395–E402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender P. A.; Rice K. D.; Schnute M. E. The First Formal Asymmetric Synthesis of Phorbol. J. Am. Chem. Soc. 1997, 119, 7897–7898and references cited therein.. [Google Scholar]
- a Hawkins P. C. D.; Skillman A. G.; Warren G. L.; Ellingson B. A.; Stahl M. T. OMEGA 2.5.1.4: OpenEye Scientific Software, Santa Fe, NM, http://www.eyesopen.com.; b Molecular structures visualized using OpenEye VIDA software.
- Wender P. A.; Loy B. A.; Schrier A. J. Translating Nature’s Library: The Bryostatins and Function-Oriented Synthesis. Isr. J. Chem. 2011, 51, 453–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit G. R.; Herald C. L.; Doubek D. L.; Herald D. L.; Arnold E.; Clardy J. Isolation and Structure of Bryostatin 1. J. Am. Chem. Soc. 1982, 104, 6846–6848. [Google Scholar]
- a DeChristopher B. A.; Fan A. C.; Felsher D. W.; Wender P. A. “Picolog,” a Synthetically-Available Bryostatin Analog, Inhibits Growth of MYC-Induced Lymphoma In Vivo. Oncotarget 2012, 3, 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Barr P. M.; Lazarus H. M.; Cooper B. W.; Schluchter M. D.; Panneerselvam A.; Jacobberger J. W.; Hsu J. W.; Janakiraman N.; Simic A.; Dowlati A.; Remick S. C. Phase II Study of Bryostatin 1 and Vincristine for Aggressive Non-Hodgkin Lymphoma Relapsing after an Autologous Stem Cell Transplant. Am. J. Hematol. 2009, 84, 484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan T. K.; Nelson T. J.; Verma V. A.; Wender P. A.; Alkon D. L. A Cellular Model of Alzheimer’s Disease Therapeutic Efficacy: PKC Activation Reverses Abeta-Induced Biomarker Abnormality on Cultured Fibroblasts. Neurobiol. Dis. 2009, 34, 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Wender P. A.; Kee J.-M.; Warrington J. M. Practical Synthesis of Prostratin, DPP, and Their Analogs, Adjuvant Leads Against Latent HIV. Science 2008, 320, 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Bullen C. K.; Laird G. M.; Durand C. M.; Siliciano J. D.; Siliciano R. F. New Ex Vivo Approaches Distinguish Effective and Ineffective Single Agents for Reversing HIV-1 Latency in Vivo. Nat. Med. 2014, 20, 425–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender P. A.; Cribbs C. M.; Koehler K. F.; Sharkey N. A.; Herald C. L.; Kamano Y.; Pettit G. R.; Blumberg P. M. Modeling of the Bryostatins to the Phorbol Ester Pharmacophore on Protein Kinase C. Proc. Natl. Acad. Sci. U. S. A. 1988, 85, 7197–7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender P. A.; Donnelly A. C.; Loy B. A.; Near K. E.; Staveness D.. Rethinking the Role of Natural Products: Function-Oriented Synthesis, Bryostatin and Bryologs. In Natural Products in Medicinal Chemistry; Hanessian S., Ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2014; Chapter 14, pp 475–544. [Google Scholar]
- a Wender P. A.; Nakagawa Y.; Near K. E.; Staveness D. Computer-Guided Design, Synthesis, and Protein Kinase C Affinity of a New Salicylate-Based Class of Bryostatin Analogs. Org. Lett. 2014, 16, 5136–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wender P. A.; Staveness D. Improved Protein Kinase C Affinity through Final Step Diversification of Simplified Salicylate-Derived Bryostatin Analog Scaffold. Org. Lett. 2014, 16, 5140–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender P. A.; Cribbs C. M.. Computer Assisted Molecular Design Related to the Protein Kinase C Receptor. In Advances in Medicinal Chemistry; Maryanoff C. A., Maryanoff B. E., Eds.; JAI: Greenwich, CT, 1992; Vol. 1, pp 1–53. [Google Scholar]
- a Kikumori M.; Yanagita R. C.; Tokuda H.; Suzuki N.; Nagai H.; Suenaga K.; Irie K. Structure–Activity Studies on the Spiroketal Moiety of a Simplified Analogue of Debromoaplysiatoxin with Antiproliferative Activity. J. Med. Chem. 2012, 55, 5614–5626. [DOI] [PubMed] [Google Scholar]
- Karaman M. W.; Herrgard S.; Treiber D. K.; Gallant P.; Atteridge C. E.; Campbell B. T.; Chan K. W.; Ciceri P.; Davis M. I.; Edeen P. T.; Faraoni R.; Floyd M.; Hunt J. P.; Lockhart D. J.; Milanov Z. V.; Morrison M. J.; Pallares G.; Patel H. K.; Pritchard S.; Wodicka L. M.; Zarrinkar P. P. A Quantitative Analysis of Kinase Inhibitor Selectivity. Nat. Biotechnol. 2008, 26, 127–132. [DOI] [PubMed] [Google Scholar]
- Wender P. A.; Axtman A. D.; Golden J. E.; Kee J.-M.; Sirois L. E.; Quiroz R. V.; Stevens M. C. Function through Bio-Inspired, Synthesis-Informed Design: Step-Economical Syntheses of Designed Kinase Inhibitors. Org. Chem. Front. 2014, 1, 1166–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gani O. A. B. S. M.; Engh R. A. Protein Kinase Inhibition of Clinically Important Staurosporine Analogues. Nat. Prod. Rep. 2010, 27, 489–498. [DOI] [PubMed] [Google Scholar]
- For a recent example:Wender P. A.; Fournogerakis D. N.; Jeffreys M. S.; Quiroz R. V.; Inagaki F.; Pfaffenbach M. Structural Complexity Through Multicomponent Cycloaddition Cascades Enabled by Dual-Purpose, Reactivity Regenerating 1,2,3-Triene Equivalents. Nat. Chem. 2014, 6, 448–452and references therein.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman R. G. Reactive 1,4-dehydroaromatics. Acc. Chem. Res. 1973, 6, 25–31. [Google Scholar]
- Konishi M.; Ohkuma H.; Tsuno T.; Oki T.; VanDuyne G. D.; Clardy J. Crystal and Molecular Structure of Dynemicin A: A Novel 1,5-diyn-3-ene Antitumor Antibiotic. J. Am. Chem. Soc. 1990, 112, 3715–3716. [Google Scholar]
- Wender P. A.; Zercher C. K. Studies on DNA-cleaving Agents: Synthesis of a Functional Dynemicin Analogue. J. Am. Chem. Soc. 1991, 113, 2311–2313. [Google Scholar]
- Wender P. A.; Beckham S.; O’Leary J. G. A Second Generation Photochemically Activatable Dynemicin Analog: A Concise Synthesis and DNA Cleavage Studies. Synthesis 1994, 1994, 1278–1282and references therein.. [Google Scholar]
- Wender P. A.; Touami S. M.; Alayrac C.; Philipp U. C. Triazole Photonucleases: A New Family of Light Activatable DNA Cleaving Agents. J. Am. Chem. Soc. 1996, 118, 6522–6523. [Google Scholar]
- Touami S. M.; Poon C. C.; Wender P. A. Sequence Specific DNA Cleavage by Conjugates of Benzotriazoles and Minor Groove Binders. J. Am. Chem. Soc. 1997, 119, 7611–7612. [Google Scholar]
- Stanzl E. G.; Trantow B. M.; Vargas J. R.; Wender P. A. Fifteen Years of Cell-Penetrating, Guanidinium-Rich Molecular Transporters: Basic Science, Research Tools, and Clinical Applications. Acc. Chem. Res. 2013, 46, 2944–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wender P. A.; Mitchell D. J.; Pattabiraman K.; Pelkey E. T.; Steinman L.; Rothbard J. B. The Design, Synthesis, and Evaluation of Molecules That Enable or Enhance Cellular Uptake: Peptoid Molecular Transporters. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 13003–13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbard J. B.; Jessop T. C.; Lewis R. S.; Murray B. A.; Wender P. A. Role of Membrane Potential and Hydrogen Bonding in the Mechanism of Translocation of Guanidinium-Rich Peptides into Cells. J. Am. Chem. Soc. 2004, 126, 9506–9507. [DOI] [PubMed] [Google Scholar]
- Wender P. A.; Galliher W. C.; Goun E. A.; Jones L. R.; Pillow T. H. The Design of Guanidinium-rich Transporters and Their Internalization Mechanisms. Adv. Drug Delivery Rev. 2008, 60, 452–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geihe E. I.; Cooley C. B.; Simon J. R.; Kiesewetter M. K.; Edward J. A.; Hickerson R. P.; Kaspar R. L.; Hedrick J. L.; Waymouth R. M.; Wender P. A. Designed Guanidinium-rich Amphipathic Oligocarbonate Molecular Transporters Complex, Deliver and Release siRNA in Cells. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 13171–13176and references therein.. [DOI] [PMC free article] [PubMed] [Google Scholar]