Abstract
Meta-BASIC (http://basic.bioinfo.pl) is a novel sensitive approach for recognition of distant similarity between proteins based on consensus alignments of meta profiles. Specifically, Meta-BASIC compares sequence profiles combined with predicted secondary structure by utilizing several scoring systems and alignment algorithms. In our benchmarking tests, Meta-BASIC outperforms many individual servers, including fold recognition servers, and it can compete with meta predictors that base their strength on the structural comparison of models. In addition, Meta-BASIC, which enables detection of very distant relationships even if the tertiary structure for the reference protein is not known, has a high-throughput capability. This new method is applied to 860 PfamA protein families with unknown function (DUF) and provides many novel structure–functional assignments available on-line at http://basic.bioinfo.pl/duf.pl. Detailed discussion is provided for two of the most interesting assignments. DUF271 and DUF431 are predicted to be a nucleotide-diphospho-sugar transferase and an α/β-knot SAM-dependent RNA methyltransferase, respectively.
INTRODUCTION
The fastest approach to annotating a novel protein is to infer its function from an experimentally studied homologue. However, simple and reliable sequence similarity search tools such as Fasta (1) or Blast (2) are frequently not powerful enough to detect homology unambiguously. In the 1990s threading methods (3–6) emerged, which were able to go further in predicting similarities between proteins, but only when one of the proteins to be aligned had a known three-dimensional (3D) structure. Threading methods were developed with the hope of detecting analogues, i.e. structurally similar proteins with no evolutionary relationship. However, for most predictions found by threading, homology (evolutionary relationship) was later supported by new and advanced sequence comparison methods, such as PSI-Blast (7). The competition and partnership between sequence-based and structure-based prediction strategies has led to considerable improvements and changes in recent years. The fold recognition field is now dominated by the meta predictors [3D-Jury (8), Pcons (9), Shotgun (10), Libullela (11)] exploiting many different prediction methods of both types to generate consensus models. The usage of profiles is now generalized to the alignment of two sequence profiles (12–18) or two meta profiles [ORFeus (19)]. Here we present a method, Meta-BASIC (Bilaterally Amplified Sequence Information Comparison), that combines the achievements in sequence profile-based strategies with secondary structure predictions to generate fast and reliable predictions using meta profile alignment algorithms. We apply Meta-BASIC to obtain large-scale structure–functional predictions for catalogued protein families of unknown function in the Pfam database (20) and find many surprising functional connections. This shows the general applicability of the method, which, similar to structural genomic initiatives, is aimed at extracting functional information from poorly studied protein sequences.
METHODS
Overview
Our method, Meta-BASIC, derives its strength from four sources. First, it is a novel sequence profile-based method. Profile methods, including PSI-Blast, set the standard in the field as accurate predictors of remote links between proteins. High-scoring PSI-Blast hits are essentially correct and biologically meaningful. In addition, a skilful PSI-Blast user is able to pick a few non-trivial homologues by careful analysis of hits in the twilight zone. However, many interesting but very remote homologues still remain undetected at the sequence level. Second, Meta-BASIC uses predicted local structure (secondary structure) information. Adding structural information to a sequence profile often helps to find homologues that diverge beyond recognition sequence-wise but remain structurally similar. In contrast to Meta-BASIC, many conventional threading algorithms utilize experimental global 3D structure to score similarity. Therefore, as a pre-requisite for correct prediction, a protein of interest must have the structure of its homologue determined. Using predicted secondary structure, we are free from that requirement and can find links between proteins of unknown structure. In addition, parting with the global threading allows for a faster algorithm and higher throughput. Third, Meta-BASIC not only combines sequence profile with secondary structure profile to form what we call meta profile, but also utilizes several scoring systems and alignment algorithms. Averaging between the results obtained by slightly different approaches helps to boost the accuracy. Fourth, Meta-BASIC has a high-throughput capability since it is a stand-alone program in contrast to most meta servers, which collect predictions from several remotely located prediction services.
Algorithm
In brief, the current version of Meta-BASIC (BasD) uses two versions of highly sensitive similarity detection algorithms. Both algorithms are based on dynamic programming and gapped alignment of meta profiles. Specifically, meta profiles combine the conventional positional variability of amino acids (sequence profiles) with secondary structure predictions. Our recent studies demonstrated that alignment of meta profiles as implemented in ORFeus is more sensitive than the alignment of pure sequence profiles (19). As a further improvement of this approach, Meta-BASIC uses the combination of two main components that differ in the way the score for aligning two positions of meta profiles is calculated. The first method (zdotc) computes a dot product of sequence variability vectors for the two positions, while the second method (zmatc) multiplies one vector by the BLOSUM62 matrix and the result by the other vector. The predicted preferences for the three secondary structure states (α-helix, β-strand and loop) are compared using the city block metric (the sum of the absolute differences of propensities). Alignment scores obtained with both versions are normalized to enable direct comparison. To ensure standardization, each profile is aligned to a set of 300 unrelated profiles and the collected scores are used to estimate the parameters of the distribution of random scores. Each alignment score is then converted to a Z-score by subtracting the mean of the distribution and dividing the result by the standard deviation. The final score reported by Meta-BASIC is equal to the average of the two Z-scores obtained with both methods, and the alignment is selected from the version which reported higher Z-score. A more detailed description of Meta-BASIC is available on-line at http://basic.bioinfo.pl/about.pl.
RESULTS AND DISCUSSION
Comparison with other methods
Meta-BASIC has been extensively evaluated and compared with other protein structure prediction methods in the framework of the LiveBench program (http://bioinfo.pl/LiveBench/) (21). Methods that are being continuously evaluated by LiveBench cover pure sequence algorithms, threading approaches, and various meta predictors that combine sequence-based and structure-based methods. Several meta servers obtain many models generated for the target protein from diverse prediction servers that may be located in several countries around the world. These meta servers compare the models with each other to find a consensus. All benchmarks, including the recent CASP5 experiment (22), confirm that consensus-based methods that use many prediction servers are more powerful than individual servers and thus represent the best of what researchers can explore today. However, these meta servers are slow since they need to collect models from many sources.
The main asset of Meta-BASIC is the high specificity of the reported confidence score, which means that very few high-scoring hits represent incorrect predictions. Importantly, 12 different components and 3 different versions of Meta-BASIC were tested in the LiveBench-8 experiment to finally select BasD as having the highest specificity (Table 1). Our benchmarking of Meta-BASIC reveals that it outperforms many individual servers, including fold recognition servers, and it can compete with meta predictors basing their strength on the structural comparison of models. Specifically, Meta-BASIC (BasD) has achieved rank 2 in the specificity assessment in LiveBench-8. Only one version of Shotgun meta predictor, Shotgun-on-3 (3DS3), has obtained a higher specificity. While competitive in accuracy with other meta predictors, Meta-BASIC has clear advantages. It is local, relatively fast and can be used for high-throughput annotation of genomes (1000 predictions per day are feasible on the current server at http://basic.bioinfo.pl), while the meta predictors coupled to the Meta Server (23) can handle only about 50 predictions per day. Another crucial advantage is that Meta-BASIC does not require the structure of the template to be known. This makes it possible to compare the target protein not only with proteins of known structure, such as those extracted from the PDB (24), but also with protein families of unknown structure, such as those from Pfam database, various genomic resources or any other sequence databases.
Table 1. Comparison of 3 Meta-BASIC versions and 12 components in LiveBench-8.
| EASY | Score | Hits | HARD | Score | Hits | ROC | Score |
|---|---|---|---|---|---|---|---|
| rdotb | 6002 | 67 | mBAS | 2306 | 34 | BasD | 92.0 |
| zdotb | 5971 | 68 | BasD | 2261 | 33 | rorfc | 92.0 |
| mBAS | 5964 | 68 | rorfc | 2233 | 32 | zmatc | 90.9 |
| BasP | 5958 | 67 | zdotb | 2225 | 32 | mBAS | 90.1 |
| BasD | 5951 | 68 | zdotc | 2213 | 32 | zorfc | 89.4 |
| rdotc | 5932 | 68 | BasP | 2157 | 31 | zdotc | 88.7 |
| zmatb | 5909 | 68 | zorfc | 2150 | 31 | zdotb | 87.9 |
| zdotc | 5908 | 68 | rorfb | 2144 | 31 | zmatb | 87.9 |
| zmatc | 5886 | 68 | rdotb | 2143 | 30 | rorfb | 87.7 |
| rorfc | 5880 | 68 | rdotc | 2125 | 31 | BasP | 87.6 |
| rorfb | 5875 | 66 | zmatc | 2063 | 30 | rdotc | 85.6 |
| zorfb | 5869 | 66 | zorfb | 2030 | 29 | zorfb | 83.5 |
| zorfc | 5861 | 68 | zmatb | 1996 | 30 | rmatc | 81.8 |
| rmatb | 5843 | 67 | rmatb | 1973 | 28 | rdotb | 79.5 |
| rmatc | 5822 | 68 | rmatc | 1902 | 27 | rmatb | 76.7 |
The targets are divided into EASY and HARD. The ‘Score’ columns for both categories show the total score obtained with the 3D-eval evaluation method. The ‘Hits’ columns indicate the number of correct hits produced by the methods. The specificity of the methods is evaluated in the ROC column. A higher ROC score indicates a higher reliability of the confidence score generated by each method. The details of the evaluation procedure are described on the LiveBench pages (http://bioinfo.pl/LiveBench/) and in related publications. Three tested Meta-BASIC versions are shown in boldface. There are three basic component methods dot, orf and mat. orf and dot use dot product calculation when comparing two vectors of the aligned profiles. mat uses vector times matrix times vector multiplication to compare the two vectors. Methods with names starting with the letter r return the raw score of the alignment while methods starting with the letter z return the Z-score transformation of the raw score. Methods with names ending with letter b use three PSI-Blast iterations to calculate the profile for each family while methods with names ending with the letter c use six PSI-Blast iterations. mBAS calculates the score by averaging the results obtained with all six methods which return Z-scores (zorfb, zorfc, zdotb, zdotc, zmatb, zmatc). The underlying raw scores cannot be easily compared with each other. BasP uses only zdotb and zmatb (three PSI-Blast iterations). BasD uses zdotc and zmatc (six PSI-Blast iterations) and it was selected as the current version of Meta-BASIC.
Structure–functional annotation for uncharacterized protein families
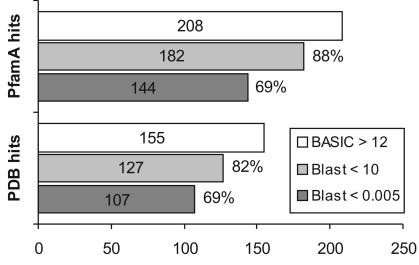
Exploiting this positive aspect of the new method, we searched for putative homologues for 860 PfamA protein families without functional annotation (DUF, catalogued by Pfam developers) to generate hypotheses about the functions of these proteins. Each target family was compared with all 6249 PfamA families and with 7225 proteins (representatives at 90% of sequence identity) extracted from PDB. For each PDB entry and PfamA/DUF family (represented as consensus sequence), sequence alignments utilized in the profile building were created with PSI-Blast, while secondary structure predictions were obtained using the PSIPRED program (25). The same comparison was also conducted using RPS-Blast and PSI-Blast (combined with Meta-Blast). Meta-BASIC was able to find significant hits to PfamA for 208 families and significant hits to PDB for 155 families, when the conservative Z-score of 12 was used as a threshold. Predictions with Z-score above 12 have <5% probability of being incorrect (using rigorous structural criteria). Of those hits, 70% can be confirmed with significant Meta-Blast E-value of <0.005, and 85% achieved E-value of <10 (Figure 1). As a necessary disclaimer, these statistics may change, since all DUF families are recalculated periodically to keep the database of assignments up to date with regard to the currently available sequential and structural information. When the Meta-BASIC threshold is relaxed to lower values, many more potential links can be found, but in such cases additional evidence is necessary to confirm the validity of the predictions. All predictions are available on-line at http://basic.bioinfo.pl/duf.pl and in our opinion represent a goldmine for undiscovered homologies. Detection of unexpected but relatively reliable relationships enables researchers to assign function, and frequently also a structure, to a few completely uncharacterized families of hypothetical proteins. Table 2 shows selected examples of such unexpected assignments obtained (in October 2003) with Meta-BASIC score above 12 that could not be confirmed by PSI-Blast or RPS-Blast in our setting. Table 2 includes also the highest scoring Meta-BASIC matches to proteins of known structure. All of the five hits have confident fold assignments as confirmed by detailed manual analysis and all five provide unexpected functional predictions to the best of our knowledge not reported before. Two of the five predictions are discussed below. It should also be stressed that in several cases where no reliable structural assignments were obtained, Meta-BASIC (but not Meta-Blast) confidently mapped analysed DUF families to other PfamA families of unknown structure. For instance, DUF820 was linked by Meta-BASIC with Z-score of 21.74 to the Competence protein CoiA-like family (PF06054). This further emphasizes the applicability of the new method to detecting distant relationship between proteins even if the tertiary structure for the reference protein is not known.
Figure 1.
Statistics of putative annotations obtained with Meta-BASIC for 860 PfamA families of unknown function. The figure shows the number of similarities found with Meta-BASIC between the target families with unknown function (DUF) and other PfamA families or PDB proteins with a conservative Meta-BASIC score cut-off of 12 (BASIC). The bars labelled ‘Blast’ indicate the number of similarities found with Meta-BASIC and supported by Meta-Blast with an E-value below 10 or 0.005, respectively. The Meta-Blast E-values correspond to the lowest E-values reported by PSI-Blast or RPS-Blast.
Table 2. Selected examples of unexpected but reliable Meta-BASIC predictions.
| DUF | PfamA | Name | Score | PDB | Name | Score |
|---|---|---|---|---|---|---|
| DUF271 | PF01501 | Glycosyl transferase family 8 | 23.55 | 1ll0A | Glycogenin-1 | 23.70 |
| DUF431 | PF04243 | tRNA m(1)G methyltransferase | 17.06 | 1ipaA | RNA 2′-O-Ribose Methyltransferase | 10.81 |
| DUF393 | PF00462 | Glutaredoxin | 13.29 | 1fovA | Glutaredoxin 3 | 14.16 |
| DUF920 | PF00797 | N-acetyltransferase | 13.02 | 1e2tA | N-Hydroxyarylamine O-Acetyltransferase | 15.57 |
| DUF833 | PF03577 | Peptidase family U34 | 12.73 | 3pvaA | Penicillin V Acylase | 9.24 |
Five out of twenty-six significant similarities between PfamA families with unknown function (DUF) and other PfamA families detected by Meta-BASIC but not with Meta-Blast E-value below 10 are displayed in left part of the table. The structural assignment for the five families based on detected similarities to proteins of known structure is reported in the right part. In all cases the name of the related family or protein and the Meta-BASIC score is shown. Only first, strongest PfamA and PDB hits are listed. Our literature search indicated that these similarities for the selected DUF families have not been reported before.
Prediction highlights
DUF271 belongs to the superfamily of nucleotide-diphospho-sugar transferases
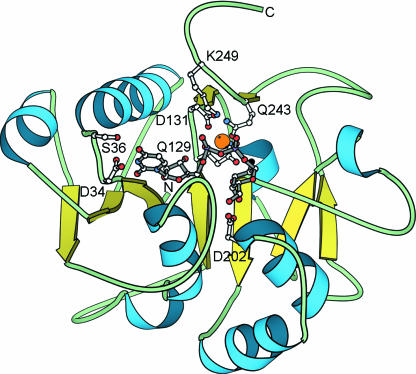
PfamA family DUF271 encompasses several hypothetical proteins from Caernorhabditis elegans. While Meta-Blast was unable to find any reliable matches to other PfamA families or to proteins of known structure, Meta-BASIC predicted the nucleotide-diphospho-sugar transferases fold for this family. Meta-BASIC assigned reliable scores to Glycosyl transferase family 8 (GT8) (PF01501) and, in particular, to the structures of galactosyl transferase LgtC (26) and Glycogenin-1 (27). Apart from LgtC and Glycogenin, PF01501 contains about 200 different proteins involved in the biosynthesis of disaccharides, oligosaccharides and polysaccharides. The GT8 enzymes belong to the EC 2.4.1 group that catalyse the transfer of sugar moieties from activated donor molecules (UDP-sugar) to specific acceptor molecules, forming glycosidic bonds (28,29). The consensus sequence of DUF271 contains four known motifs that are necessary for UDP-sugar binding (Figure 2). The first conserved motif (33YDSSN37) is responsible for the interaction between the enzyme and the nucleotide. Conserved Asp34 forms a hydrogen bond with the nitrogen of the uracil base (27). The DUF271 also contains the DXD-like motif (129QQD131) that is conserved in most prokaryotic and eukaryotic glycosyltransferases. This motif appears to function primarily in the coordination of the divalent cation, most commonly Mn2+, essential for the binding of the nucleotide sugar-donor substrate (26). Asp202, is the next key residue that is highly conserved among all DUF271 sequences. The 4′ and 6′ sugar oxygens make hydrogen bonds to the side-chain carboxylate of Asp202, indicating that this residue has an important role in binding and most likely in catalysis as well. The fourth sequence motif (243QLDGEKK249) forms hydrogen bonds with Mn2+ and both phosphates. Gln243 possibly coordinates the cation while the backbone nitrogen of conserved Gly246 and the side-chain of conserved Lys249 directly interact with the phosphates. Therefore, based on the conservation of residues characteristic for the uridine 5′-diphosphate-sugar (UDP-sugar) binding site, the glycosyltransferase function can be confidently inferred for DUF271.
Figure 2.
DUF271 belongs to the superfamily of nucleotide-diphospho-sugar transferases. The 3D model of the DUF271 consensus sequence was based on the structure of the galactosyl transferase LgtC (26) (PDB code: 1ga8) from Neisseria meningitides. The Mn2+ cation (orange), UDP-sugar (gray) and the side-chains (white) of the key residues (Asp34, Ser36, Gln129, Asp131, Asp202, Gln243, Lys249) essential for UDP-sugar binding are shown.
DUF431 belongs to the superfamily of α/β-knot SAM-dependent RNA methyltransferases
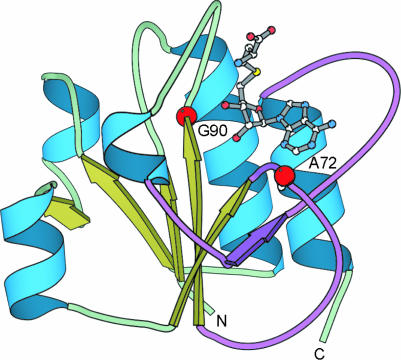
DUF431, encompassing several hypothetical proteins from Saccharomyces cerevisiae, Plasmodium falciparum, Plasmodium yoelii, Neurospora crassa and a few archeal organisms, is another PfamA family for which no reliable structure–functional assignment can be obtained with PSI-Blast or RPS-Blast. Meta-BASIC assigned an above-threshold score to the tRNA m(1)G methyltransferase family (PF04243), which includes biochemically characterized S.cerevisiae protein Trm10p responsible for the methylation of G residues to m(1)G in tRNAGly at position 9 (30). In addition, Meta-BASIC mapped the consensus sequence of DUF431 to the structures of the cofactor-binding domain of RrmH (31), RlmB (32), YibK (33), YggJ (34) and YbeA, which are members of the SPOUT (35) superfamily of known or predicted S-adenosylmethionine(SAM)-dependent tRNA and rRNA methyltransferases. This domain adopts an α/β-knot fold with topology differing from that of the classical methyltransferases. Importantly, unique pseudoknot structure provides the binding site for SAM, which interacts mainly with the main chain amide and carbonyl groups as well as with the surrounding side-chains of hydrophobic residues that are also conserved in the DUF431 family. DUF431 and SPOUT methyltransferases share the conserved Gly90 in the SAM-binding loop (35), Ala72 at the position occupied by tiny side-chains, important for the pseudoknot formation, as well as several conservatively replaced residues at the dimer interface. Despite very weak sequence similarity, conservation of these unique features as well as reasonable mapping of predicted and observed secondary structure elements are additional indicators of the correct structure–functional assignment (Figure 3). All these findings demonstrate that DUF431 is yet another family of α/β-knot SAM-dependent RNA methyltransferases. In addition, we propose that the last 60 residues not mapped on the cofactor-binding domain may form a substrate-binding domain. Its C-terminal localization indicates that DUF431 proteins possibly use tRNA as substrates (33).
Figure 3.
DUF431 belongs to the superfamily of α/β-knot SAM-dependent RNA methyltransfeases. The 3D model of the cofactor-binding domain of the DUF431 consensus sequence was based on the structure of the YibK, hypothetical tRNA/rRNA methyltransferase HI0766 (33) (PDB code: 1mxi) from Haemophilus influenzae. The pseudoknot is coloured in violet. The S-adenosylhomocysteine (SAH) (grey) and positions of highly conserved residues (red dots) essential for the pseudoknot formation (Ala72) and SAH binding (Gly90) are shown.
CONCLUSIONS
The database of potential structure–functional annotations generated by Meta-BASIC for PfamA families of unknown function contains many more examples of non-trivial and potentially useful assignments that can be studied and verified by researchers. Methods such as Meta-BASIC that push homology inference further and allow for large-scale annotations may represent a cheaper alternative to, and clearly complement, many experimental efforts in the diverse field of genomics-oriented research. The growing amounts of data obtained in large-scale sequencing projects, structural genomics efforts, DNA-chip experiments, two-hybrid interaction mapping and many others provide the foundation for, and a strong boost to, the systematic approach to investigating cells and organisms [systems biology (36)]. To fully understand the large networks of interactions, it is crucial to know as much as possible about every individual member of the system. Yet, functional annotation of proteins with standard methods leaves a prohibitively large gap of more than 30% of the proteome. The goal of Meta-BASIC is to reduce this gap and we hope that scientists annotating genomes will take advantage of the recent progress in the field of protein structure–functional prediction.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by 5th Framework Project Grants and KBN grants ELM (QLRI-CT2000-00127 E-617/SPB/5.PR UE/DZ 438/2003-2004) and MiFriend (QLR3-CT2000-00170 E-617/SPB/5.PR UE/DZ 439/2003), and an NIH grant GM67165 to N.V.G.
REFERENCES
- 1.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci., USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 3.Narayana S.V. and Argos,P. (1984) Residue contacts in protein structures and implications for protein folding. Int. J. Pept. Protein Res., 24, 25–39. [DOI] [PubMed] [Google Scholar]
- 4.Bowie J.U., Luthy,R. and Eisenberg,D. (1991) A method to identify protein sequences that fold into a known three-dimensional structure. Science, 253, 164–170. [DOI] [PubMed] [Google Scholar]
- 5.Blundell T.L. (1991) Comparative analysis of protein three-dimensional structures and an approach to the inverse folding problem. Ciba Found. Symp., 161, 28–36; discussion 37–51. [DOI] [PubMed] [Google Scholar]
- 6.Godzik A., Kolinski,A. and Skolnick,J. (1992) Topology fingerprint approach to the inverse protein folding problem. J. Mol. Biol., 227, 227–238. [DOI] [PubMed] [Google Scholar]
- 7.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ginalski K., Elofsson,A., Fischer,D. and Rychlewski,L. (2003) 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics, 19, 1015–1018. [DOI] [PubMed] [Google Scholar]
- 9.Lundstrom J., Rychlewski,L., Bujnicki,J. and Elofsson,A. (2001) Pcons: a neural-network-based consensus predictor that improves fold recognition. Protein Sci., 10, 2354–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer D. (2003) 3D-SHOTGUN: a novel, cooperative, fold-recognition meta-predictor. Proteins, 51, 434–441. [DOI] [PubMed] [Google Scholar]
- 11.Juan D., Grana,O., Pazos,F., Fariselli,P., Casadio,R. and Valencia,A. (2003) A neural network approach to evaluate fold recognition results. Proteins, 50, 600–608. [DOI] [PubMed] [Google Scholar]
- 12.von Ohsen N., Sommer,I. and Zimmer,R. (2003) Profile-profile alignment: a powerful tool for protein structure prediction. Pac. Symp. Biocomput., 252–263. [PubMed] [Google Scholar]
- 13.Sadreyev R. and Grishin,N. (2003) COMPASS: a tool for comparison of multiple protein alignments with assessment of statistical significance. J. Mol. Biol., 326, 317–336. [DOI] [PubMed] [Google Scholar]
- 14.Panchenko A.R. (2003) Finding weak similarities between proteins by sequence profile comparison. Nucleic Acids Res., 31, 683–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yona G. and Levitt,M. (2002) Within the twilight zone: a sensitive profile-profile comparison tool based on information theory. J. Mol. Biol., 315, 1257–1275. [DOI] [PubMed] [Google Scholar]
- 16.Heger A. and Holm,L. (2001) Picasso: generating a covering set of protein family profiles. Bioinformatics, 17, 272–279. [DOI] [PubMed] [Google Scholar]
- 17.Rychlewski L., Jaroszewski,L., Li,W. and Godzik,A. (2000) Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci., 9, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rychlewski L., Zhang,B. and Godzik,A. (1998) Fold and function predictions for Mycoplasma genitalium proteins. Fold Des., 3, 229–238. [DOI] [PubMed] [Google Scholar]
- 19.Ginalski K., Pas,J., Wyrwicz,L.S., von Grotthuss,M., Bujnicki,J.M. and Rychlewski,L. (2003) ORFeus: Detection of distant homology using sequence profiles and predicted secondary structure. Nucleic Acids Res., 31, 3804–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rychlewski L., Fischer,D. and Elofsson,A. (2003) LiveBench-6: large-scale automated evaluation of protein structure prediction servers. Proteins, 53(Suppl. 6), 542–547. [DOI] [PubMed] [Google Scholar]
- 22.Kinch L.N., Wrabl,J.O., Krishna,S.S., Majumdar,I., Sadreyev,R.I., Qi,Y., Pei,J., Cheng,H. and Grishin,N.V. (2003) CASP5 assessment of fold recognition target predictions. Proteins, 53(Suppl. 6), 395–409. [DOI] [PubMed] [Google Scholar]
- 23.Bujnicki J.M., Elofsson,A., Fischer,D. and Rychlewski,L. (2001) Structure prediction meta server. Bioinformatics, 17, 750–751. [DOI] [PubMed] [Google Scholar]
- 24.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones D.T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol., 292, 195–202. [DOI] [PubMed] [Google Scholar]
- 26.Persson K., Ly,H.D., Dieckelmann,M., Wakarchuk,W.W., Withers,S.G. and Strynadka,N.C. (2001) Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. Nat. Struct. Biol., 8, 166–175. [DOI] [PubMed] [Google Scholar]
- 27.Gibbons B.J., Roach,P.J. and Hurley,T.D. (2002) Crystal structure of the autocatalytic initiator of glycogen biosynthesis, glycogenin. J. Mol. Biol., 319, 463–477. [DOI] [PubMed] [Google Scholar]
- 28.Campbell J.A., Davies,G.J., Bulone,V. and Henrissat,B. (1997) A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J., 326, 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell J.A., Davies,G.J., Bulone,V.V. and Henrissat,B. (1998) A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J., 329, 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackman J.E., Montange,R.K., Malik,H.S. and Phizicky,E.M. (2003) Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA, 9, 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nureki O., Shirouzu,M., Hashimoto,K., Ishitani,R., Terada,T., Tamakoshi,M., Oshima,T., Chijimatsu,M., Takio,K., Vassylyev,D.G. et al. (2002) An enzyme with a deep trefoil knot for the active-site architecture. Acta Crystallogr. D Biol. Crystallogr., 58, 1129–1137. [DOI] [PubMed] [Google Scholar]
- 32.Michel G., Sauve,V., Larocque,R., Li,Y., Matte,A. and Cygler,M. (2002) The structure of the RlmB 23S rRNA methyltransferase reveals a new methyltransferase fold with a unique knot. Structure (Camb), 10, 1303–1315. [DOI] [PubMed] [Google Scholar]
- 33.Lim K., Zhang,H., Tempczyk,A., Krajewski,W., Bonander,N., Toedt,J., Howard,A., Eisenstein,E. and Herzberg,O. (2003) Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): a cofactor bound at a site formed by a knot. Proteins, 51, 56–67. [DOI] [PubMed] [Google Scholar]
- 34.Forouhar F., Shen,J., Xiao,R., Acton,T.B., Montelione,G.T. and Tong,L. (2003) Functional assignment based on structural analysis: Crystal structure of the yggJ protein (HI0303) of Haemophilus influenzae reveals an RNA methyltransferase with a deep trefoil knot. Proteins, 53, 329–332. [DOI] [PubMed] [Google Scholar]
- 35.Anantharaman V., Koonin,E.V. and Aravind,L. (2002) SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol., 4, 71–75. [PubMed] [Google Scholar]
- 36.Hood L. and Galas,D. (2003) The digital code of DNA. Nature, 421, 444–448. [DOI] [PubMed] [Google Scholar]