Abstract
The DICHROWEB web server enables on-line analyses of circular dichroism (CD) spectroscopic data, providing calculated secondary structure content and graphical analyses comparing calculated structures and experimental data. The server is located at http://www.cryst.bbk.ac.uk/cdweb and may be accessed via a password-limited user ID, available upon completion of a registration form. The server facilitates analyses using five popular algorithms and (currently) seven different reference databases by accepting data in a user-friendly manner in a wide range of formats, including those output by both commercial CD instruments and synchrotron radiation-based circular dichroism beamlines, as well as those produced by spectral processing software packages. It produces as output calculated secondary structures, a goodness-of-fit parameter for the analyses, and tabular and graphical displays of experimental, calculated and difference spectra. The web pages associated with the server provide information on CD spectroscopic methods and terms, literature references and aids for interpreting the analysis results.
INTRODUCTION
Circular dichroism (CD) is a spectroscopic technique that can be used to determine the secondary structural content of proteins. This is because the electronic transitions of polypeptide backbone peptide bonds in different conformations produce differential absorption spectra for left- and right-handed circularly polarized light in the far UV and vacuum UV wavelength ranges, which are both distinct and linearly-independent. Therefore the data can be used to deconvolute the secondary structural types present in the net spectrum measured for a protein. Typically, conventional CD (cCD) data collected on laboratory-based instruments are in the wavelength range between ∼190 and 300 nm. In recent years, the use of synchrotron radiation as an intense light source has led to the development of the technique of synchrotron radiation circular dichroism (SRCD) spectroscopy, which can measure spectra into the even lower VUV wavelength range, which also includes data between 160 and 190 nm (1).
The information contained in CD spectra can be treated as a sum of the characteristic individual spectra arising from each type of secondary structure present in a protein sample. Typically, empirical analysis methods utilize a reference database comprised of spectra of proteins whose crystal structures (and therefore their secondary structures) are known. Using singular value deconvolutions, principal component analyses, variable selection procedures or neural networks, they calculate the fraction that each component structure contributes to the net experimental spectrum. The procedures are necessarily iterative due to their empirical nature, and the output generally consists of a list of secondary structural fractions and a calculated spectrum, which can be compared to the experimental spectrum in order to evaluate the quality of the data analysis. The number of different types of secondary structure that can be analysed for depends both on the available wavelength range of the spectra [spectra at lower wavelengths have a higher information content, and hence can be deconvoluted into more component types (2)] and on the range of secondary structural types present in the reference database proteins.
Several algorithms developed to perform the deconvolution calculations (3–9) are publicly available for downloading to be installed locally by users. However, considerable technical effort may be required to compile the source codes and to execute programs from the command line; this is often time-consuming, especially for casual users. The different programs tend to require different input data formats or units; in addition, different CD instruments output different formats, which do not tend to coincide with the required input formats of the analysis programs. The format differences include headers and footers, ordering of data (from high to low wavelength or vice versa), number of significant figures, format of data strings, wavelength range, wavelength interval and spacings/punctuation in the file formats. Likewise the data output formats of the programs may not be compatible with external graphics software, and the programs do not calculate a goodness-of-fit parameter in a consistent manner, so direct comparisons of the outputs are not facile.
To address some of these issues, a different approach is needed, either by crafting a pre-compiled executable for each program, such as CDPro (10), or by making an algorithm available on a web server as a delocalized service such as the K2d web server (http://www.embl-heidelberg.de/~andrade/k2d/). Both these approaches aid the usability of CD analysis software; but in isolation still leave some pre-processing and computational work for the spectroscopist to do, and make direct-comparisons between different algorithms difficult and time consuming. As an alternative, DICHROWEB provides a user-friendly interface to the existing programs and databases, which enables a wide range of input formats and limits the need for pre-analysis processing and conversion programs. An additional advantage is that it calculates a single type of goodness-of-fit parameter as well as providing an easily interpretable graphical comparison of the experimental and calculated data. The purpose of DICHROWEB, therefore, is to facilitate access to CD deconvolution algorithms in a manner which is independent of where the data were collected, what units the data were collected in, and what file structure the data were recorded in. DICHROWEB is run via a web server in order that the latest version is always available and that the analysis package is easily accessible, even to the casual user.
WEB SERVER DESCRIPTION
The primary function of the DICHROWEB server is to provide a user-friendly interface and calculation platform for a range of popular secondary structure calculation algorithms and reference databases, thereby facilitating the analysis of CD spectroscopic data. The server interface supports all of the major file formats and data units produced both by conventional CD and SRCD instruments. Currently it is being operated in the ‘supported’ mode, which enables users to email the administrator (cdweb@mail.cryst.bbk.ac.uk) if problems arise in their analyses, or if they have questions regarding the suitability of their data for analysis, or for advice on interpretation of the results. Feedback from users regarding website features is also welcomed via this email address.
ALGORITHMS AVAILABLE
DICHROWEB currently supports five popular and freely available analysis algorithms: SELCON3 (3,4), CONTINLL (5,6), CDSSTR (7), VARSLC (8) and K2d (9). The first three of these algorithms may be used in conjunction with any of the seven publicly available reference datasets (11), whilst VARSLC and K2d have built-in protein reference data.
As part of the password registration process, the user is required to agree to cite the original algorithms and databases accessed through DICHROWEB, in order that those authors receive the appropriate credit for their work.
INPUT
The web page with the input form requires the user to select a file to upload for analysis and to provide some information about the data contained within the file, i.e. the format type, data units, initial and final wavelengths (files written with wavelengths ordered either from high to low or from low to high are acceptable in ‘free format’) and wavelength interval.
The following data formats are currently accepted: data files from several different versions of Aviv, Jasco or Applied Photophysics CD instrument software, Daresbury SRCD data format (DRS), the Brookhaven SRCD data format (SDS/2000), the BP and YY output formats of the Super3 CD data processing program (12) and a ‘free’ format. The latter is an intuitive general file-reading routine provided to interpret other, less common file formats. In most cases, a ‘preview’ is available which plots the data and allows the user to check that they have been read properly prior to analysis.
Data in units of either delta epsilon, mean residue ellipticity, theta (machine units) or SRCD counts are acceptable. Input data at wavelength intervals of 0.1, 0.2, 0.5 and 1.0 nm are accommodated.
A further input option allows the user to select the lowest wavelength data to use in the analysis. This feature can be used to exclude portions of the data that have poor signal-to-noise ratios, such as are typically encountered at low wavelengths where the diode or HT (total absorption) level is high.
The user must then select an analysis method and, where appropriate, a reference dataset. At present seven different reference datasets are available, which include different proteins with different structural characteristics, and which require a minimum amount of data in different wavelength ranges. Reference datasets 1, 2 and 5 require data at least between 178 and 260 nm (usually only achievable with SRCD), whilst datasets 3 and 6 require data at least between 185 and 240 nm. Datasets 4 and 7 are the most flexible and require a minimum range of 190–240 nm, as does the VARSLC algorithm with default parameters. The neural-network-based algorithm K2d is based on an internal set database and is usable with a minimum range of data between 200 and 241 nm.
An optional scaling factor may be applied to spectra when it is anticipated that there may be small errors in the determination of protein concentration or pathlength, or in instrument calibration (A. J. Miles, L. Whitmore and B. A. Wallace, in preparation). The minimum and maximum scaling values allowed are 0.5 and 1.5, respectively, but the recommended values are in the range of 0.95–1.05, assuming proper concentration determinations have been done.
Finally, if the user is unclear about the parameter requested in a given blank on the input form, they can click on the ‘help’ link next to each blank to obtain suggestions on the types of information needed, the units, and the acceptable limits (and expected magnitudes) for the requested parameter.
ERROR MESSAGES
Error messages are generated when the data files do not match the input parameters or, rarely, if the experimental spectrum is so different from the reference databases that analyses are not possible, or if the data quality is very poor. The latter situation can arise if the signal-to-noise level is too low, the data have not been properly zeroed in the near UV region or the wavelength range of the data is not sufficient to permit accurate analyses (i.e. it does not extend to at least 190 nm at the low wavelength end—or to the lower wavelength limits if reference databases 1, 2, 3, 5 or 6 are used). If the error messages persist and none of the above conditions is obvious, it is recommended that the user contact the web administrator for advice.
OUTPUT
Each of the deconvolution algorithms aims to provide information about a range of structural elements. The techniques rely on empirical, comparative methods and thus are restricted by the structure assignments and features present in the component proteins in the reference datasets available.
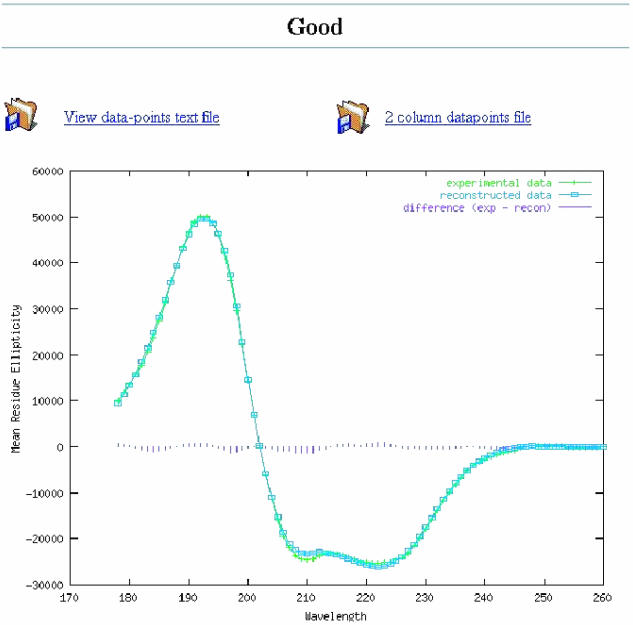
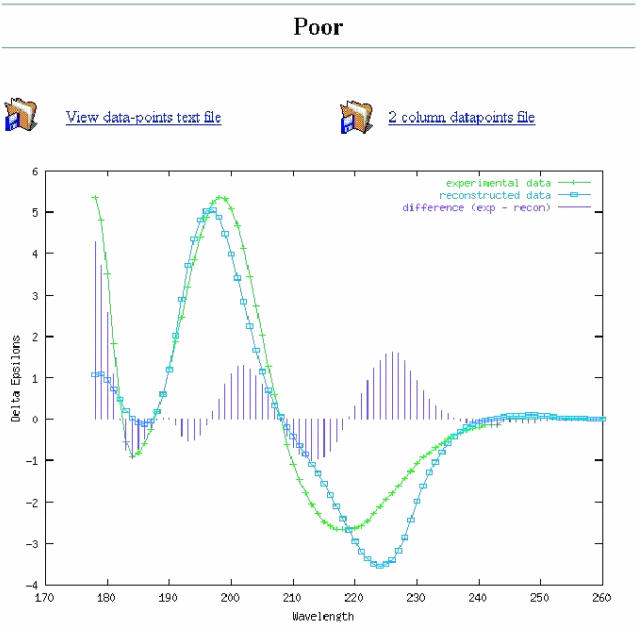
The results page presents the output at various levels of detail: a short summary of the best deconvolution (listing only the secondary structure composition and total content), a longer report including the deconvolution details and the normalized root mean square deviation (NRMSD) (and where applicable, R-value) fit parameters, and a graph of the experimental and calculated spectra, with the difference graph superimposed (Figure 1). From the latter, it is possible to view (and download) an ASCII file containing the wavelength, experimental and calculated data in the output units specified on the input page. This file can be easily ported into commercial software plotting packages. Each of the results options is displayed in a separate window. The graphical output and NRMSD values are not available when using the VARSLC algorithm as it does not provide the calculated spectrum required to generate the graphs and values. All of the results produced by DICHROWEB are presented in tables that may be copied and pasted into popular spreadsheet programs.
Figure 1.
Example of the graphical output of DICHROWEB. Experimental (input) data are plotted with crosses in green, the calculated spectrum derived from the calculated output secondary structure is plotted with open boxes in blue, and the difference spectra is depicted in vertical lines in purple, (a) for a ‘good’ fit, and (b) for a ‘poor’ fit.
The NRMSD is a goodness-of-fit parameter defined as ![]() (13), summed over all wavelengths, where θexp and θcal are, respectively, the experimental ellipticities and the ellipticities of the back-calculated spectra for the derived structure. It is produced for all calculated spectra. This parameter is an important measure of the correspondence between the experimental and calculated spectra and can be used to judge the quality of the results (14). A low value for the NRMSD is a necessary but not sufficient condition for concluding that an analysis has produced a good result (15). That is, if the NRMSD is high (>0.1) the correspondence of the calculated secondary structure with the actual one is unlikely to be ‘correct’, but a low NRMSD alone does not mean that an analysis is accurate. Since DICHROWEB defines this parameter in the same way for all analyses, it does, however, provide the user with one way of comparing the results obtained by using the different databases and algorithms, and a means of assessing which may be the most appropriate method and reference dataset for their protein. Because the different reference datasets consist of different proteins, one may be more appropriate than the others for the analysis of a given ‘unknown’ protein if it contains the types of structural elements that are present in the protein under study. For example, if the unknown protein contains a significant amount of polyproline II structure, it may be better analysed with the databases that contain this component (4,16).
(13), summed over all wavelengths, where θexp and θcal are, respectively, the experimental ellipticities and the ellipticities of the back-calculated spectra for the derived structure. It is produced for all calculated spectra. This parameter is an important measure of the correspondence between the experimental and calculated spectra and can be used to judge the quality of the results (14). A low value for the NRMSD is a necessary but not sufficient condition for concluding that an analysis has produced a good result (15). That is, if the NRMSD is high (>0.1) the correspondence of the calculated secondary structure with the actual one is unlikely to be ‘correct’, but a low NRMSD alone does not mean that an analysis is accurate. Since DICHROWEB defines this parameter in the same way for all analyses, it does, however, provide the user with one way of comparing the results obtained by using the different databases and algorithms, and a means of assessing which may be the most appropriate method and reference dataset for their protein. Because the different reference datasets consist of different proteins, one may be more appropriate than the others for the analysis of a given ‘unknown’ protein if it contains the types of structural elements that are present in the protein under study. For example, if the unknown protein contains a significant amount of polyproline II structure, it may be better analysed with the databases that contain this component (4,16).
In the course of creating this server software, we have tested the algorithms and databases for a large number of different proteins. Whilst we make no general conclusions regarding which produce the ‘best results’, several general observations can be noted: (i) membrane proteins are not well analysed by the existing reference databases (14), a finding which has led us to create a new membrane-protein specific database (B. A. Wallace et al., unpublished) and (ii) CDSSTR tends (but not always) to give the lowest NRMSD values for a given reference database, although this does not necessarily mean that those results are the most correct.
Finally, the output can be produced in any of the common CD units. For conversion to some units, additional information is needed, so the server will prompt for the mean residue weight, sample concentration and sample cell path length. As a consequence, this server can also be used as a simple conversion program, changing input formats and units to alternative output formats and units.
INFORMATION PAGES
There are a number of web pages associated with the server which provide information on CD spectroscopic methods (including literature references), definitions of units and terms, FAQs for use of the server and links to the original algorithms and other relevant links. There is also an extensive user guide.
NEW FEATURES
The first version of DICHROWEB was implemented in-house in 2000, and released in test format in 2001 (17). The current version contains many enhancements of and improvements on the earlier version described in Lobley et al. (18). These include additional input formats (additional instrument types and free format), the ability to select a low wavelength cut-off, scaling options, graphical outputs including difference plots and expanded information pages. It is under continuous development. An important recent addition is the presence of FAQ pages, where (anonymous) questions of general interest raised by users are answered, and where suggestions for good practice are noted.
PRECAUTIONS
The home page includes a list of precautionary notes for undertaking and interpreting the CD analyses; it is suggested that these be heeded in order to produce valid results using the algorithms (regardless of whether this server is used to access them or not). These include warnings about the importance of precise determinations of cell pathlength and protein concentration, the requirement to include data down to at least 190 nm for accurate analyses, the necessity that the data has been zeroed properly prior to submission to the server, and the observations (noted above) that the best NRMSD is not always the correct solution and that the reference databases do not work well for peptides or proteins in non-aqueous solutions (i.e. membrane proteins).
AVAILABILITY
DICHROWEB can be accessed at http://www.cryst.bbk.ac.uk/cdweb. The calculation server is freely available to academic and non-profit organizations but requires a server password/userID, which may be obtained by emailing cdweb@mail.cryst.bbk.ac.uk and completing and signing an application form. To date more than 24 000 deconvolutions have been performed on the server. The information pages, FAQs and ‘helps’ can be accessed without a password or user ID.
FUTURE DEVELOPMENTS
Future developments will include the membrane-protein CD reference database (B. A. Wallace, F. Wien and J. G. Lees, in preparation), a new bioinformatics-defined soluble protein reference database suitable for both CD and SRCD data (R. W. Janes and A. Cuff, unpublished results), and the ability to select specialty user-defined reference databases. Ultimately the reference databases may be extended to include analyses for fold motifs and supersecondary structures (F. Wien, J. G. Lees, A. J. Miles and B. A. Wallace, unpublished results) for use with the low-wavelength SRCD data (19). It is also planned that other algorithms such as themagnitude-independent normalized least squares SUPER3 method (12) may be incorporated in the future. This method has the advantage that, unlike any of the algorithms currently included in DICHROWEB, it does not require precise knowledge of the sample concentration. In addition, calibration curves for various CD and SRCD instruments will be provided, along with software to enable cross-spectrometer calibration, as described in Miles et al. (20). Finally, it is expected that in the future servers may be available at other sites, specifically at a number of SRCD facilities, including Brookhaven National Laboratory (21).
CONCLUSIONS
The DICHROWEB web server provides access to a number of circular dichroism secondary structure calculation algorithms and reference databases in a user-friendly manner. It accepts a wide range of data formats and units and provides a goodness-of-fit parameter for assessing the quality of the analyses and a range of tabular and graphical output formats of experimental, calculated and difference spectra.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Anna Lobley for creating the original implementation of DICHROWEB, the authors of the algorithms for providing access to their programs, as well as the many researchers (especially those in the Wallace group) who tested early versions of the program. DICHROWEB development and curation has been supported by BBSRC grant B13586. This project is part of the BBSRC Centre for Protein and Membrane Structure and Dynamics (CPMSD).
REFERENCES
- 1.Wallace B.A. (2000) Synchrotron radiation circular dichroism spectroscopy as a tool for investigating protein structures. J. Synchrotron Radiat., 7, 289–295. [DOI] [PubMed] [Google Scholar]
- 2.Toumadje A., Alcorn,S.W. and Johnson,W.C., Jr (1992) Extending CD spectra of proteins to 168 nm improves the analysis for secondary structures. Anal. Biochem., 200, 321–331. [DOI] [PubMed] [Google Scholar]
- 3.Sreerama N. and Woody,R.W. (1993) A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem., 209, 32–44. [DOI] [PubMed] [Google Scholar]
- 4.Sreerama N., Venyaminov,S.Y. and Woody,R.W. (1999) Estimation of the number of helical and strand segments in proteins using circular dichroism spectroscopy. Protein Sci., 8, 370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Provencher S.W. and Glockner,J. (1981) Estimation of globular protein secondary structure from circular dichroism. Biochemistry, 20, 33–37. [DOI] [PubMed] [Google Scholar]
- 6.Van Stokkum I.H.M., Spoelder,H.J.W., Bloemendal,M., Van Grondelle,R. and Groen,F.C.A. (1990) Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal. Biochem., 191, 110–118. [DOI] [PubMed] [Google Scholar]
- 7.Compton L.A. and Johnson,W.C., Jr (1986) Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal. Biochem., 155, 155–167. [DOI] [PubMed] [Google Scholar]
- 8.Manavalan P. and Johnson,W.C., Jr (1987) Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal. Biochem., 167, 76–85. [DOI] [PubMed] [Google Scholar]
- 9.Andrade M.A., Chacón,P., Merelo,J.J. and Morán,F. (1993) Evaluation of secondary structure of proteins from UV circular dichroism using an unsupervised learning neural network. Protein Eng., 6, 383–390. [DOI] [PubMed] [Google Scholar]
- 10.Sreerama N. and Woody,R.W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON and CDSSTR methods with an expanded reference set. Anal. Biochem., 287, 252–260. [DOI] [PubMed] [Google Scholar]
- 11.Sreerama N., Venyaminov,S.Y. and Woody,R.W. (2000) Estimation of protein secondary structure from circular dichroism spectra: inclusion of denatured proteins with native protein in the analysis. Anal. Biochem., 287, 243–251. [DOI] [PubMed] [Google Scholar]
- 12.Wallace B.A. and Teeters,C.L. (1987) Differential absorption flattening optical effects are significant in the circular dichroism spectra of large membrane fragments. Biochemistry, 26, 65–70. [DOI] [PubMed] [Google Scholar]
- 13.Mao D., Wachter,E. and Wallace,B.A. (1982) Folding of the H+-ATPase proteolipid in phospholipid vesicles. Biochemistry, 21, 4960–4968. [DOI] [PubMed] [Google Scholar]
- 14.Wallace B.A., Lees,J., Orry,A.J.W., Lobley,A. and Janes,R.W. (2003) Analyses of circular dichroism spectra of membrane proteins. Protein Sci., 12, 875–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brahms S. and Brahms,J. (1980) Determination of protein secondary structure in solution by vacuum ultraviolet circular dichroism. J. Mol. Biol., 138, 149–178. [DOI] [PubMed] [Google Scholar]
- 16.Cascio M., Gogol,E. and Wallace,B.A. (1990) The secondary structure of gap junctions: influence of isolation methods and proteolysis. J. Biol. Chem., 265, 2358–2364. [PubMed] [Google Scholar]
- 17.Lobley A. and Wallace,B.A. (2001) DICHROWEB: a website for the analysis of protein secondary structure from circular dichroism spectra. Biophys. J., 80, 373a. [DOI] [PubMed] [Google Scholar]
- 18.Lobley A., Whitmore,L. and Wallace,B.A. (2002) DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics, 18, 211–212. [DOI] [PubMed] [Google Scholar]
- 19.Wallace B.A. and Janes,R.W. (2001) Synchrotron radiation circular dichroism spectrosocpy of proteins: secondary structure, fold recognition and structural genomics. Curr. Opin. Chem. Biol., 5, 567–571. [DOI] [PubMed] [Google Scholar]
- 20.Miles A.J., Wien,F., Lees,J.G., Rodger,A., Janes,R.W. and Wallace,B.A. (2003) Calibration and standardisation of synchrotron radiation circular dichroism amplitudes and conventional circular dichroism spectrophotometers. Spectroscopy, 17, 653–661. [Google Scholar]
- 21.Wallace B.A. (2002) First international workshop on SRCD spectroscopy. Synchrotron Radiat. News, 15, 20–22. [Google Scholar]