Abstract
ConSite is a user-friendly, web-based tool for finding cis-regulatory elements in genomic sequences. Predictions are based on the integration of binding site prediction generated with high-quality transcription factor models and cross-species comparison filtering (phylogenetic footprinting). By incorporating evolutionary constraints, selectivity is increased by an order of magnitude as compared to single-sequence analysis. ConSite offers several unique features, including an interactive expert system for retrieving orthologous regulatory sequences. Programming modules and biological databases that form the foundation of the ConSite service are freely available to the research community. ConSite is available at http:/www.phylofoot.org/consite.
INTRODUCTION
Understanding the mechanisms of coordinated regulation of gene activity is one of the primary goals of the post-genomic era of biology. Gene regulation at the transcriptional level is an ancient and central control mechanism present in all forms of life. RNA-polymerase II-mediated transcription is activated or repressed by sequence-specific DNA binding proteins called transcription factors (TFs). Transcription factor binding sites (TFBS) are typically short (∼5–12 bp), and considerable sequence variation between functional binding sites is tolerated by most TFs. While the laboratory elucidation of TFBS within genes is feasible, the process is arduous and time-consuming if no prior information is available. As regulatory elements are often scattered over regions spanning thousands of base pairs around the targeted gene in multi-cellular eukaryotes, additional methodology is required. Computational predictions have been successfully utilized for suggesting potential regulatory regions for further experimental analysis; in effect enabling researchers to determine key regulatory elements more efficiently (1–4).
Transcriptional regulation, in particular modeling and prediction of TFBS, is one of the most studied problems in computational biology (5,6). Reliable profile-based methods and model frameworks have been developed over the years which accurately describe the DNA-binding specificity of a TF (6). While these models can accurately identify sites bound in vitro (7), they are insufficiently selective for finding functional elements in vivo (8): the information contained in the interface between TFBS and TF is in itself not enough to discriminate between functional and non-functional sites in the complex cellular environment. The in vivo specificity of a factor depends on other, additional properties; for instance interacting proteins, DNA accessibility and effective concentrations. With rare exceptions, our understanding of these properties is insufficient to enable the creation of effective computational methods.
Phylogenetic footprinting
Cross-species sequence conservation can be used as an effective filter for improving selectivity of detection of functional elements in DNA sequences. This approach is known as phylogenetic footprinting: due to selective pressure, functional regulatory regions in non-coding sequence should be preferentially conserved compared to regions with no sequence-specific function (9,10). Two assumptions are implicit: the analyzed regions must be orthologous, and the selective pressure on the gene from each respective organism must be similar. A change of gene function will alter the functional constraints imposed on the regulation of the gene.
A further consideration is the evolutionary distance that is ideal for relevant filtering—i.e. what pair of organisms should be analyzed. The sequence difference between closely related species, for instance human and chimpanzee, is generally insufficient to confer any meaningful filtering in pairwise analysis. Conversely, relatively long evolutionary distances, such as between human and fish, often render similarities in promoters all but undetectable with current methodology (11).
Here we describe a web resource for TFBS prediction in genomic sequence using phylogenetic footprinting, available at www.phylofoot.org/consite. The web service is a user-friendly tool with a high degree of user interactivity and optional customization.
IMPLEMENTATION
General schema
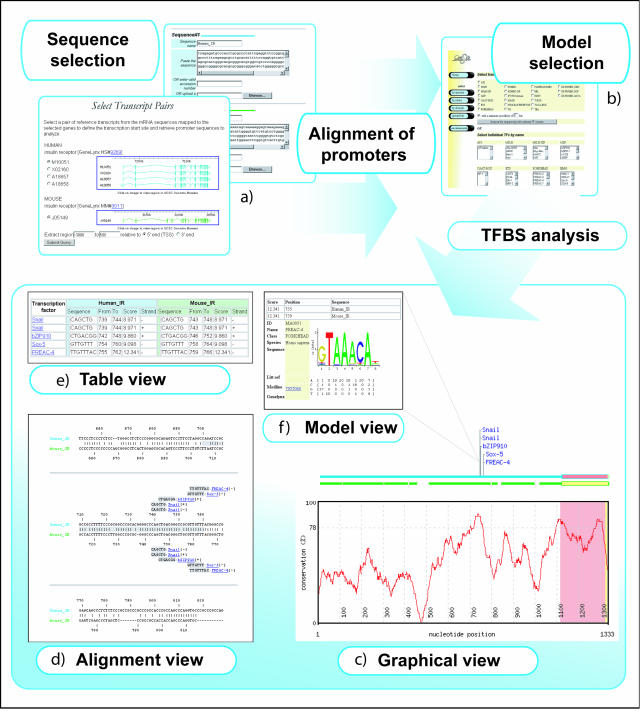
The ConSite program executes a number of analysis steps, each in interaction with the user (Figure 1). In brief, the program (i) aligns input promoter sequences, (ii) calculates the degree of conservation in the alignment, (iii) scans the sequences of a set of TF binding profile models, (iv) performs filtering on the initial sets of sites using phylogenetic footprinting and (v) presents the results in user-selected output formats (Figure 1).
Figure 1.
ConSite analysis schema and output formats. Pairs of promoter sequences can either be submitted directly or (for human/mouse) be retrieved by an interactive search (a). Sequences are aligned and analyzed for TFBS using a set of TF models chosen by the user (b). Results can be visualized as a graphical conservation plot (c), as an annotated alignment (d) or in tabular form (e). Individual TFBS and corresponding TF model can be displayed in pop-up windows using sequence logos (f).
Selection of regulatory sequences
The success of phylogenetic footprinting methods is critically dependent on the selection of regulatory sequences. As discussed above, only the corresponding regulatory regions of orthologous pairs of genes are appropriate. In ConSite, the user has the choice between manually locating promoter pairs of interest [e.g. by using genome browsers such as UCSC (12) and EnsEMBL (13)] and semi-automatically retrieving target mouse : human genomic sequences based on an accession number, keyword or sequence. In this process, users are aided by an intuitive graphical interface. This ortholog finder interface is unique among phylogenetic footprinting services. Its search engine is powered by GeneLynx (14)—a gene index and catalog. As GeneLynx expands to include additional species, the automated orthologous gene selection service will have increasing utility for promoter analysis of model organisms.
Sequence alignment
Once submitted or selected, sequences are aligned preferentially using the ORCA aligner (Arenillas and Wasserman, unpublished), a progressive global alignment program optimized for non-coding genomic sequences. For pairs of particularly long sequences, ConSite accepts pre-computed alignments in a variety of standard formats. For convenience, users can specify a cDNA sequence that will be used to identify and highlight coding regions and/or exons in the output.
Conservation calculation
The ‘degree of conservation’ is calculated by sliding a window of a user-defined width over the alignment. In each window location, the percentage of identical nucleotides is calculated. A potential problem with this approach is that short, highly-conserved regions adjacent to large gaps or insertions will be assigned low identity-scores. For each input sequence, we chose to collapse the gaps in the alignment for the purpose of the calculation of nucleotide identity. Thus, the analysis results are displayed as a pair of conservation plots, where the first (second) input sequence is continuous in the first (second) plot. For each analysis, we obtain a set of window identity scores Wi, corresponding to the percentage of identical nucleotides in the window starting at position i. For filtering purposes, only those windows with Wi exceeding an identity threshold I (typically 70–80%) are retained for further TFBS analysis.
TF binding profile collection and scoring
The mathematical background of profile models used for describing TF binding properties has been extensively reviewed elsewhere (5,6). In brief, a profile consists of a matrix tabulating observed nucleotides in each position of the protein–DNA interface, typically counted from an alignment of known sites. The profile collection in ConSite is drawn from the JASPAR database, an open-access, non-redundant collection of curated profiles (15). Profiles are converted to log-scaled position weight matrices (PWMs) in order to evaluate possible binding sites in an input sequence, as reviewed elsewhere (6). As score ranges are unique for each model, scores are normalized according to
![]()
In ConSite, a set of profiles chosen by the user are used to analyze each input sequence. In the final phylogenetic footprinting step, we retain only those sites that
have a site score S ≥ c (a user-adjustable TFBS detection threshold),
and are found in window where Wi ≥ I (as defined above),
and have a predicted site in the other input sequence in corresponding position, subjected to constraints (i) and (ii).
Graphical interpretation of analysis results
For the evaluation of results, researchers are given a choice of several distinct output formats (Figure 1), including:
Graphical view: showing an alignment overview and conservation plots. Conserved TFBS are shown as intuitive flags with mouse-over functionality.
Alignment view: detailed alignments labeled with detected conserved sites;
Table view: a tabular view of all detected sites with supplementary data.
All predicted sites are hyperlinked to pop-up summary pages describing TF binding models, including a sequence logo for graphical representation of the TF's binding specificity.
PERFORMANCE
For many individual genes, phylogenetic footprinting has been shown to be a highly useful method. However, until recently, no confirmation that the concept holds in regard to larger gene sets has been available. In a recent study (11), we sought to test the concept with two separate test sets: sites resulting from detailed literature analysis (40 sites in 14 promoters) and sites from the TRANSFAC database mapped onto genome assemblies (110 sites in 40 promoters). The latter set is the largest reference set to date for phylogenetic footprinting tests, available at http://www.phylofoot.org/consite/testset. In brief, the ConSite set of methods could, in both test cases, reduce the noise level by ∼85% while retaining high sensitivity compared to single sequence analysis (11).
ACCESS TO UNDERLYING SOFTWARE
The ConSite integration of TFBS prediction and cross-species comparison is based on the open-access TFBS Perl modules (15,16) (http://forkhead.cgb.ki.se/TFBS), which support a wide variety of analysis modes, including pattern-finding and pattern similarity analysis. TFBS enables automated analysis on genome-scale data sets for power users.
CONCLUSION
We have presented a graphical, web-based interface for computer-assisted prediction of regulatory regions in higher eukaryotes, powered by cross-species comparison. Besides the intuitive interface design, ConSite integrates several features not found in other TFBS prediction services such as TESS (17) or rVista (18). The features include a curated model dataset, a computer-assisted input sequence selection and a powerful underlying set of programming modules for genome-scale analysis.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Bill Wilson for useful comments on the manuscript. This work was supported in part by funding from Pharmacia Corporation (now Pfizer).
REFERENCES
- 1.Aparicio S., Morrison,A., Gould,A., Gilthorpe,J., Chaudhuri,C., Rigby,P., Krumlauf,R. and Brenner,S. (1995) Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc. Natl Acad. Sci. USA, 92, 1684–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagheri-Fam S., Ferraz,C., Demaille,J., Scherer,G. and Pfeifer,D. (2001) Comparative genomics of the SOX9 region in human and Fugu rubripes: conservation of short regulatory sequence elements within large intergenic regions. Genomics, 78, 73–82. [DOI] [PubMed] [Google Scholar]
- 3.Christensen T.H., Prentice,H., Gahlmann,R. and Kedes,L. (1993) Regulation of the human cardiac/slow-twitch troponin C gene by multiple, cooperative, cell-type-specific, and MyoD-responsive elements. Mol. Cell Biol., 13, 6752–6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gumucio D.L., Heilstedt-Williamson,H., Gray,T.A., Tarle,S.A., Shelton,D.A., Tagle,D.A., Slightom,J.L., Goodman,M. and Collins,F.S. (1992) Phylogenetic footprinting reveals a nuclear protein which binds to silencer sequences in the human gamma and epsilon globin genes. Mol. Cell Biol., 12, 4919–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wasserman W.W. and Krivan,W. (2003) In silico identification of metazoan transcriptional regulatory regions. Naturwissenschaften, 90, 156–166. [DOI] [PubMed] [Google Scholar]
- 6.Stormo G.D. (2000) DNA binding sites: representation and discovery. Bioinformatics, 16, 16–23. [DOI] [PubMed] [Google Scholar]
- 7.Tronche F., Ringeisen,F., Blumenfeld,M., Yaniv,M. and Pontoglio,M. (1997) Analysis of the distribution of binding sites for a tissue-specific transcription factor in the vertebrate genome. J. Mol. Biol., 266, 231–245. [DOI] [PubMed] [Google Scholar]
- 8.Fickett J.W. (1996) Quantitative discrimination of MEF2 sites. Mol. Cell Biol., 16, 437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tagle D.A., Koop,B.F., Goodman,M., Slightom,J.L., Hess,D.L. and Jones,R.T. (1988) Embryonic epsilon and gamma globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol., 203, 439–455. [DOI] [PubMed] [Google Scholar]
- 10.Wasserman W.W., Palumbo,M., Thompson,W., Fickett,J.W. and Lawrence,C.E. (2000) Human–mouse genome comparisons to locate regulatory sites. Nat. Genet., 26, 225–228. [DOI] [PubMed] [Google Scholar]
- 11.Lenhard B., Sandelin,A., Mendoza,L., Engstrom,P., Jareborg,N. and Wasserman,W.W. (2003) Identification of conserved regulatory elements by comparative genome analysis. J. Biol., 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karolchik D., Baertsch,R., Diekhans,M., Furey,T.S., Hinrichs,A., Lu,Y.T., Roskin,K.M., Schwartz,M., Sugnet,C.W., Thomas,D.J. et al. (2003) The UCSC Genome Browser Database. Nucleic Acids Res., 31, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clamp M., Andrews,D., Barker,D., Bevan,P., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V. et al. (2003) EnsEMBL 2002: accommodating comparative genomics. Nucleic Acids Res., 31, 38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenhard B., Hayes,W.S. and Wasserman,W.W. (2001) GeneLynx: a gene-centric portal to the human genome. Genome Res., 11, 2151–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandelin A., Alkema,W., Engstrom,P., Wasserman,W.W. and Lenhard,B. (2004) JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res., 32, D91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenhard B. and Wasserman,W.W. (2002) TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics, 18, 1135–1136. [DOI] [PubMed] [Google Scholar]
- 17.Schug J. and Overton,G.C. (1997) TESS: Transcription Element Search Software on the WWW (report CBIL-TR-1997-1001-v0.0). Computational Biology and Informatics Laboratory, School of Medicine, University of Pennsylvania.
- 18.Loots G.G., Ovcharenko,I., Pachter,L., Dubchak,I. and Rubin,E.M. (2002) rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res., 12, 832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]