Abstract
DIALIGN is a widely used software tool for multiple DNA and protein sequence alignment. The program combines local and global alignment features and can therefore be applied to sequence data that cannot be correctly aligned by more traditional approaches. DIALIGN is available online through Bielefeld Bioinformatics Server (BiBiServ). The downloadable version of the program offers several new program features. To compare the output of different alignment programs, we developed the program AltAVisT. Our software is available at http://bibiserv.TechFak.Uni-Bielefeld.DE/dialign/.
Multiple sequence alignment is of fundamental importance in all aspects of DNA and protein sequence analysis. It is used as a first and critical step in protein structure prediction and classification, phylogenetic reconstruction, analysis of protein domains and identification of functional sites in genomic sequences, to mention just a few important applications. Development and improvement of multi-alignment methodology is therefore crucial for all branches of molecular biology and genomics, see (1) for an overview on existing multiple-alignment approaches. A widely used software tool for DNA and protein multiple alignment is DIALIGN (2,3). This program differs in various aspects from more traditional multi-alignment algorithms. Those traditional approaches are generally classified as either global or local methods. This is not appropriate for distantly related sequences where multiple conserved domains may be separated by non-related parts of the sequences. In such situations neither purely global nor purely local methods can produce meaningful alignments.
THE DIALIGN APPROACH
In contrast to standard methods, DIALIGN combines both global and local alignment features. It assembles pair-wise and multiple alignments from local fragment alignments or fragments. More precisely, alignments are composed of equal-length segment pairs exhibiting some statistically significant similarity. Note that these segment pairs are not directly visible to the user; they are used internally by the program to construct alignments from given input data sets. Gaps are not penalized, and the program does not try to align parts of the sequences that do not show significant similarity to other input sequences. Consequently, for sequence sets with local homology only, alignments are restricted to those homologies and the program ignores non-related parts of the sequences. In such a situation, DIALIGN returns a local alignment. For globally related sequences, however, the program finds segment pairs (fragments) covering the entire length of the sequences, so it returns a full global alignment. A mixture of global and local alignments is created for sequence families where islands of local similarity are separated by unrelated sequence. Here, the program aligns these similarities and leaves non-related sequence parts unaligned. As a result, DIALIGN is far more versatile than traditional alignment methods; it can be applied to a large range of sequence data that cannot be correctly aligned using standard approaches.
During the last few years, several independent studies have been carried out to compare and evaluate the performance of multiple protein alignment software. Thompson et al. used the BAliBASE database of benchmark alignments (4) to evaluate the commonly used software tools. BAliBASE mainly contains globally related proteins. In this study, DIALIGN was found to be the best program on sequences with large insertions and deletions, while standard programs such as CLUSTAL W (5) were superior on globally related protein families (6). A more recent program comparison confirmed these findings (7). This study also included the newly developed program T-COFFEE (7), which was found to be superior on all five sequence categories in BAliBASE. Like DIALIGN, T-COFFEE combines global and local alignment features. The most comprehensive evaluation of protein multi-alignment programs so far has been carried out by Lassmann and Sonnhammer (8). These authors used not only globally related sequences but also artificial sequences where conserved motifs are separated by non-related random sequences, corresponding to the domain organization of proteins. They concluded that there are currently three methods that perform best for multiple protein alignment, DIALIGN, T-COFFEE and POA (9); their paper summarizes: ‘Overall, DIALIGN was the most accurate in cases with low sequence identity, while T-COFFEE won in cases with high sequence identity. The fast POA algorithm was almost as accurate’ (8).
DIALIGN AT BiBiServ
From the beginning, DIALIGN has been developed as informal cooperation between non-commercial research groups. Researchers from different institutes contributed in various ways to the development of the program (10–14) and explored new algorithmical solutions to the segment-based alignment problem (15,16). To advance the further development of the program and to make it available without restriction to the scientific community, future versions will be released as open source code under the GNU general public licence.
Much of the program development work has been carried out at the University of Bielefeld, Germany. The DIALIGN homepage is therefore located at Bielefeld Bioinformatics Server (BiBiServ), where the latest program versions are available, either as executable files for UNIX/LINUX platforms or as source code on request. To make DIALIGN easily available to non-computer experts, we set up a WWW server at BiBiServ. The server accepts multiple sequence sets of up to 100 DNA or protein sequences. Input sequences are entered in FASTA format, either through a window or by uploading a single multi-sequence file. A web page is created that contains links to the output alignment in three different formats, DIALIGN, FASTA and MSF. For larger data sets, this page can be book-marked to retrieve the results once the program run has terminated. A threshold parameter is available that controls the minimum similarity score of the fragments (segment pairs) from which the alignment is assembled. For DNA sequences, the similarity of segment pairs can be calculated either at the nucleotide level—by comparing segments nucleotide-by-nucleotide—or at the protein level, by first translating them according to the genetic code and then comparing the implied peptide segments.
NEW PROGRAM FEATURES
The downloadable program comes with several new options for modified alignment strategies and alternative output formats.
• The most important addition to the program is the possibility to use alignment anchors (13). This means that the user can specify arbitrary sequence positions and force the program to align these positions to each other. These local alignments are then used as anchor points imposing certain constraints on the subsequent automatic alignment procedure. The anchoring option can be applied to use expert knowledge for improved alignment quality. Alternatively, anchor points can be used in order to reduce the alignment search space and running time. In this case, anchor points can be created using a fast local alignment search tool such as CHAOS (14). DIALIGN checks the list of pre-defined anchor points for consistency and rejects inconsistent anchors where necessary.
• Conversely, the user can specify segment pairs that are to be excluded from the alignment without enforcing alternative alignments. This option is useful in situations where the standard program output is known to be biologically wrong but an alternative, correct alignment is not obvious.
• Several new alignment features are available for alignment of genomic sequences. These options are mainly concerned with different levels of sequence similarity between genomic sequences; details are explained in (13).
• Various heuristics are available to speed up the program. For example, it is possible to reduce the maximum length of segment pairs (fragments), and a threshold can be imposed on the initial similarity values of fragments in order to reduce the total number of fragments considered for alignment.
• In addition to the default DIALIGN output format, alignments can be returned in FASTA, CLUSTAL or MSF format.
• A list of all fragments considered for alignment can be returned in a separate output file. Alternatively, lists of fragments contained in the respective optimal pair-wise alignments or in the final multiple alignments can be produced.
• Amino acid substitution frequencies can be calculated based on fragments considered for alignment. This option has been implemented to calculate rate matrices as proposed by Devauchelle et al. (17).
ALIGNMENT COMPARISON USING AltAVist
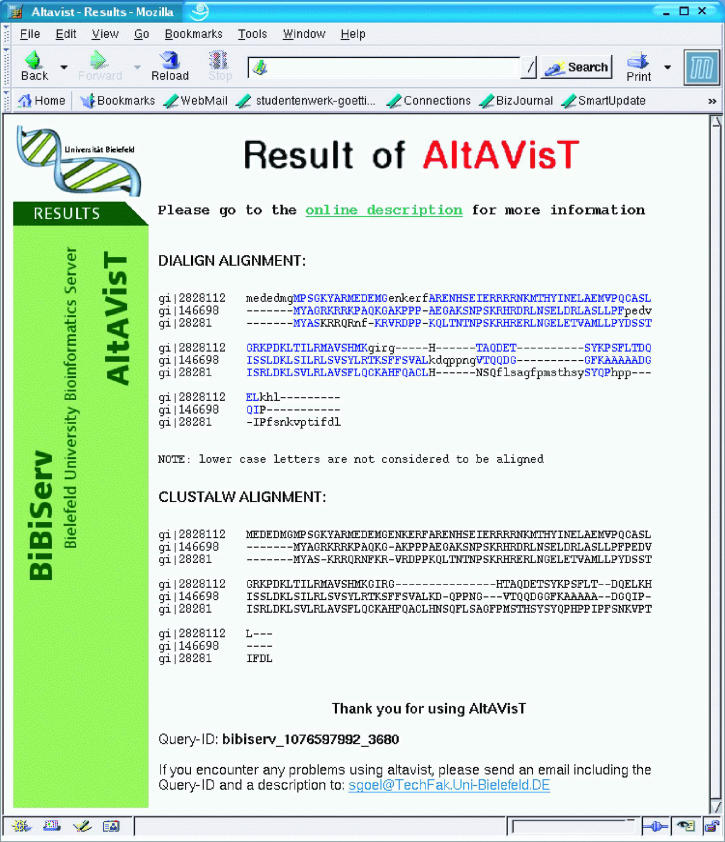
Another extension of the original DIALIGN algorithm is the possibility to compare alignments created by DIALIGN to alignments created by alternative programs. No automatic alignment procedure can be expected to produce biologically meaningful alignments under all possible conditions. Therefore, it is common practice to run more than one software program on a sequence set and to compare the different output alignments. Alignment regions where different tools agree are generally believed to be more reliable than regions where they disagree. The program AltAVisT (Alternative Alignment Visualization Tool) (18) compares two different multiple alignments of the same sequence set to each other. It searches for regions where these two alignments coincide and returns a graphical representation of those regions as shown in Figure 1. If a multiple sequence set is uploaded, AltAVisT runs both DIALIGN and CLUSTAL W (5) and compares the two resulting alignments. Alternatively, two pre-calculated multi-alignments of the same underlying sequence set can be uploaded and compared. AltAVisT is also available through BiBiServ (http://bibiserv.TechFak.Uni-Bielefeld.DE/altavist/).
Figure 1.
Comparison of multiple alignments by AltAVisT. A DIALIGN alignment is compared to a CLUSTAL W alignment of the same input data set. Those regions where both alignments coincide are printed in blue. (Local) agreement of different alignment methods usually indicates alignment reliability.
PROGRAM LIMITATIONS
In recent years, alignment of large genomic sequences has become a powerful tool for genome sequence analysis (19–21). In various research projects, DIALIGN has been found useful for this purpose. The standard version of the program, however, is far too slow to align sequences in the order of hundreds of kilobases or more. We therefore combined DIALIGN with Michael Brudno's rapid local alignment tool CHAOS to speed up the alignment procedure (14). A specialized WWW server for alignment of genomic sequences based on CHAOS and DIALIGN is introduced in a companion paper (22).
Acknowledgments
ACKNOWLEDGEMENTS
I would like to thank Saïd Abdeddaïm and Andreas Dress for their various contributions to the DIALIGN algorithm, Isabelle Schneider for programming assistance, Sachin Goel for his work on AltAVisT, Michael Brudno for many useful discussions, Alexander Sczyrba, Henning Mersch and Jan Krüger for their help at BiBiServ and Peter Meinicke, Isabelle Schneider, Jan Weyer-Menkhoff and Mario Stanke for critically reading the manuscript. Two unknown referees made helpful comments on the manuscript.
REFERENCES
- 1.Notredame C. (2002) Recent progress in multiple sequence alignment: a survey. Pharmacogenomics, 3, 131–144. [DOI] [PubMed] [Google Scholar]
- 2.Morgenstern B., Dress,A. and Werner,T. (1996) Multiple DNA and protein sequence alignment based on segment-to-segment comparison. Proc. Natl Acad. Sci. USA, 93, 12098–12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgenstern B. (1999) DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics, 15, 211–218. [DOI] [PubMed] [Google Scholar]
- 4.Thompson J.D., Plewniak,F. and Poch,O. (1999) BAliBASE: a benchmark alignment database for the evaluation of multiple sequence alignment programs. Bioinformatics, 15, 87–88. [DOI] [PubMed] [Google Scholar]
- 5.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson J.D., Plewniak,F. and Poch,O. (1999) A comprehensive comparison of protein sequence alignment programs. Nucleic Acids Res., 27, 2682–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Notredame C., Higgins,D. and Heringa,J. (2000) T-COFFEE: a novel algorithm for multiple sequence alignment. J. Mol. Biol., 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 8.Lassmann T. and Sonnhammer,E.L. (2002) Quality assessment of multiple alignment programs. FEBS Lett., 529, 126–130. [DOI] [PubMed] [Google Scholar]
- 9.Lee C., Grasso,C. and Sharlow,M.F. (2002) Multiple sequence alignment using partial order graphs. Bioinformatics, 18, 452–464. [DOI] [PubMed] [Google Scholar]
- 10.Abdeddaim S. (1997) Incremental computation of transitive closure and greedy alignment. Proceedings of the Eigth Annual Symposium on Combinatorial Pattern Matching, Lecture Notes in Computer Science. Vol. 1264, Springer Verlag, Heidelberg, pp. 167–179. [Google Scholar]
- 11.Morgenstern B., Hahn,K., Atchley,W.R. and Dress,A.W.M. (1998) Segment-based scores for pairwise and multiple sequence alignments. In Glasgow,J., Littlejohn,T., Major,F., Lathrop,R., Sankoff,D. and Sensen,C. (eds), Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology, AAAI Press, Menlo Parc, CA, pp. 115–121. [PubMed] [Google Scholar]
- 12.Abdeddaim S. and Morgenstern,B. (2001) Speeding up the DIALIGN multiple alignment program by using the ‘greedy alignment of biological sequences library’ (GABIOS-LIB). Lecture Notes in Comput. Sci., 2066, 1–11. [Google Scholar]
- 13.Morgenstern B., Rinner,O., Abdeddaim,S., Haase,D., Mayer,K., Dress,A. and Mewes,H.-W. (2002) Exon discovery by genomic sequence alignment. Bioinformatics, 18, 777–787. [DOI] [PubMed] [Google Scholar]
- 14.Brudno M., Chapman,M., Gottgens,B., Batzoglou,S. and Morgenstern,B. (2003) Fast and sensitive multiple alignment of large genomic sequences. BMC Bioinformatics, 4, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenhof H.-P., Morgenstern,B. and Reinert,K. (1999) An exact solution for the segment-to-segment multiple sequence alignment problem. Bioinformatics, 15, 203–210. [DOI] [PubMed] [Google Scholar]
- 16.Sammeth M., Morgenstern,B. and Stoye,J. (2003) Divide-and-conquer alignment with segment-based constraints. Bioinformatics, 19, ii189–ii195. [DOI] [PubMed] [Google Scholar]
- 17.Devauchelle C., Grossmann,A., Henaut,A., Holschneider,M., Monnerot,M., Risler,J. and Torresani,B. (2001) Rate matrices for analyzing large families of protein sequences. J. Comput. Biol., 8, 381–399. [DOI] [PubMed] [Google Scholar]
- 18.Morgenstern B., Goel,S., Sczyrba,A. and Dress,A. (2003) AltAVisT: a WWW server for comparison of alternative multiple sequence alignments. Bioinformatics, 19, 425–426. [DOI] [PubMed] [Google Scholar]
- 19.Miller W. (2001) Comparison of genomic DNA sequences: solved and unsolved problems. Bioinformatics, 17, 391–397. [DOI] [PubMed] [Google Scholar]
- 20.Chain P., Kurtz,S., Ohlebusch,E. and Slezak,T. (2003) An applications-focused review of comparative genomics tools: capabilities, limitations, and future challenges. Brief. Bioinform., 4, 105–123. [DOI] [PubMed] [Google Scholar]
- 21.Pollard D.A., Bergman,C.M., Stoye,J., Celniker,S.E. and Eisen,M.B. (2004) Benchmarking tools for the alignment of functional noncoding DNA. BMC Bioinformatics, 5, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brudno M., Steinkamp,R. and Morgenstern,B. (2004) The CHAOS/DIALIGN WWW server for multiple alignment of genomic sequences. Nucleic Acids Res., 32, W41–W44. [DOI] [PMC free article] [PubMed] [Google Scholar]