SUMMARY
Non-homologous end-joining (NHEJ) is a major DNA double strand break repair pathway that is conserved in eukaryotes. In vertebrates, NHEJ further acquires end-processing capacities (e.g., hairpin opening) in addition to direct end-ligation. The catalytic subunit of DNA-PK (DNA-PKcs) is a vertebrate specific NHEJ factor that can be auto-phosphorylated or trans-phosphorylated by ATM kinase. Using a mouse model expressing a kinase-dead (KD) DNA-PKcs protein, we show that ATM-mediated trans-phosphorylation of DNA-PKcs regulates end-processing at the level of Artemis recruitment, while strictly auto-phosphorylation of DNA-PKcs is necessary to relieve the physical blockage on end-ligation imposed by the DNA-PKcs protein itself. Accordingly, DNA-PKcsKD/KD mice and cells show severe end-ligation defects and p53- and Ku-dependent embryonic lethality, but open hairpin-sealed ends normally in the presence of ATM kinase activity. Together, our findings identify DNA-PKcs as the molecular switch that coordinates end-processing and end-ligation at the DNA ends through differential phosphorylations.
INTRODUCTION
Non-homologous end-joining directly ligates two DNA ends and is a conserved DNA double strand break (DSB) repair pathway in eukaryotes. Defects in NHEJ leads to microcephaly, immunodeficiency, premature aging and cancer, underscoring the importance of this pathway in mammalian cells (Lieber, 2010). In vertebrates, NHEJ further evolved an end-processing capacity that allows for the repair of complex ends (e.g. hairpins). Among the seven known NHEJ factors, XRCC4, Lig4, XLF, and the Ku70/Ku80 dimer are conserved in all eukaryotes and each plays important roles in end-ligation. Meanwhile, end-processing is mediated by two vertebrate specific NHEJ factors: the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) and the Artemis endonuclease. While coordination between end-processing and end-ligation is likely critical for efficient and accurate NHEJ, they seem operate independently. As such hairpin ends are opened in the absence of Lig4 or XRCC4. Meanwhile, in the absence of DNA-PKcs or Artemis, direct ligation of simple DNA ends (e.g. blunt or cohesive) occurs efficiently, but hairpin ends cannot to be opened owing to a strict requirement for DNA-PKcs in the activation of the Artemis endonuclease (Davis et al., 2014). Yet, the mechanism underlying Artemis activation by DNA-PKcs is still not fully understood. Consistent with the fact that end-processing is required for a subset of NHEJ involving complex ends, DNA-PKcs- or Artemis- deficient cells display relatively moderate sensitivity to ionizing-radiation (IR) and proliferation defects in comparison to end-ligation defective XRCC4- or Lig4-deficient cells. Correspondingly, DNA-PKcs- or Artemis-null mice are viable and of normal size (Gao et al., 1998a, Taccioli et al., 1998, Rooney et al., 2002), while end-ligation defective XRCC4−/− and Lig4−/− mice invariably die during embryonic development with severe neuronal apoptosis (Barnes et al., 1998, Frank et al., 1998, Gao et al., 1998b, Gao et al., 2000, Frank et al., 2000).
NHEJ is also required for V(D)J recombination, the mechanism that assembles the functional antigen receptor gene products from germline V, D and J gene segments in developing lymphocytes (Lieber, 2010). The RAG endonuclease initiates V(D)J recombination by recognizing the recombination signaling sequence (RSS) and introducing DSBs between RSSs and the participating V, D or J gene segments. RAG cleavage generates two kinds of DNA ends: blunt, phosphorylated signal ends (SEs) and hairpin-sealed coding ends (CEs). The two SEs are directly ligated via NHEJ to form the signal joint (SJ). The two hairpin-sealed CEs must first be opened by DNA-PKcs and Artemis prior to ligation to form the coding joint (CJ). In this context, V(D)J recombination is a unique physiological system that readily distinguishes the end-processing and end-ligation steps of NHEJ. CJs encode the variable region exon of antigen receptor genes required for lymphocyte development, thus defects in either the end-ligation or the end-processing components of NHEJ abrogate V(D)J recombination and lymphocyte development, and lead to severe combined immunodeficiency in patients and animal models (Lieber, 2010, Franco et al., 2006).
On the molecular level, DNA-PKcs belongs to the PI3 Kinase related protein kinase (PI3KK) family that also includes ATM and ATR. Upon DNA damage, Ku70/80 heterodimer recognizes and binds DSBs. DNA-bound Ku recruits DNA-PKcs to the DNA ends to form the DNA-PK holo-enzyme and activates the kinase activity of DNA-PKcs. Activated DNA-PKcs phosphorylates partially overlapping substrates (e.g. H2AX, 53BP1) with ATM, which underlies the critical redundant functions of ATM and DNA-PKcs in DNA repair and embryonic developments (Zha et al., 2011b, Callen et al., 2009, Gapud et al., 2011). DNA-PKcs itself is also auto-phosphorylated as well as trans-phosphorylated by ATM during the DNA damage response (Meek et al., 2008, Davis et al., 2014). Both the ABCDE cluster flanking Thr2609 and the PQR cluster around the Ser2056 on human DNA-PKcs can be auto- phosphorylated, but the T2609 cluster is primarily phosphorylated by ATM or ATR under different cellular stresses (Chen et al., 2007, Davis et al., 2010, Meek et al., 2008). Mutagenesis studies revealed the importance of DNA-PKcs phosphorylation in DNA repair and also indicated that phosphorylation of S2056 limits end-resection whereas T2609 phosphorylation promotes resection (Cui et al., 2005). To distinguish the role of DNA-PKcs auto- vs trans-phosphorylation and identify the specific function of DNA-PKcs auto-phosphorylation in NHEJ, we generated and characterized a mouse model that expresses catalytically inactive DNA-PKcs protein. Our findings revealed a critical, yet previously overlooked structural function of the DNA-PKcs protein in coordinating end-ligation and end-processing and further identified distinct functions of trans- vs auto-phosphorylation of DNA-PKcs in vertebrate NHEJ.
RESULTS
Expression of Kinase Dead DNA-PKcs alone leads to embryonic lethality
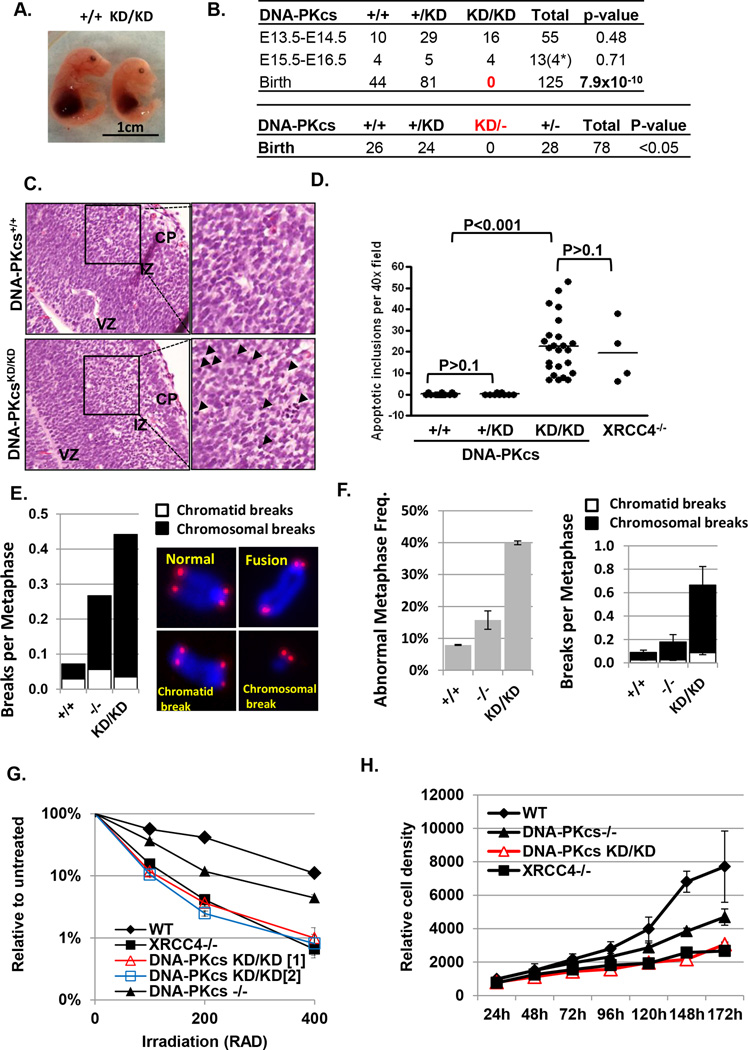
To interrogate the impact of DNA-PKcs auto- vs. trans-phosphorylation on the function of the DNA-PKcs protein itself, we generated a mouse model that expresses a catalytically inactive DNA-PKcs protein by mutating the conserved aspartic acid (D) 3922 in the putative catalytic loop of murine DNA-PKcs into alanine (A, D3922A) (Fig.S1A). This allele is thereafter referred to as “KD” for kinase dead DNA-PKcs (Fig.S1B–C). The D to A mutation of the corresponding residue in the related ATM kinase eliminates the kinase activity without significantly affecting protein stability (Yamamoto et al., 2012, Bakkenist and Kastan, 2003). The loss of DNA-PK kinase activity is confirmed by a modified DNA-PK kinase assay (see supplementary material for details) using DNA-PKcsKD/KD cell lysate in vitro (Fig.S1D). And the expression of DNA-PKcs KD protein was verified in DNA-PKcsKD/KD embryonic stem (ES) cells, murine embryonic fibroblasts (MEFs) and transformed B cells by Western blot (Fig.S1E). In contrast to the normal development of DNA-PKcs−/− mice (Gao et al., 1998a, Taccioli et al., 1998), DNA-PKcsKD/KD embryos were visibly small at embryonic day 14.5 (E14.5) (Fig.1A) and cannot be found beyond E16.5 (Fig.1B). DNA-PKcs+/KD mice are undistinguishable from DNA-PKcs+/+ littermates, but DNA-PKcs−/KD mice also cannot be found at birth (Fig.1B and Fig. S1F). Histological analyses of E14.5 brain revealed severe neuronal apoptosis in DNA-PKcsKD/KD embryos, at levels comparable to end-ligation deficient XRCC4−/− embryos (Fig.1C–D) (Gao et al., 2000). Consistent with normal development, neuronal apoptosis was not found in DNA-PKcs+/+ or DNA-PKcs+/KD littermates (Fig.1C–D). Notably, the apoptotic inclusions characterized by condensed nuclei are most prominent in the “post”-mitotic intermediate zone of the DNA-PKcsKD/KD embryonic brain and are relatively rare in the proliferating ventricular zone (Fig.1C). A similar pattern has been noted for XRCC4- or Lig4- deficient murine embryos (Gao et al., 2000, Frank et al., 2000, Barnes et al., 1998, Frank et al., 1998, Gao et al., 1998b) and is indicative of defects in the G0/G1 phase of the cell cycle, when NHEJ is more prominent.
Figure 1. Homozygous kinase dead (KD) mutation of DNA-PKcs leads to severe genomic instability and embryonic lethality with extensive neuronal apoptosis.
(A) DNA-PKcs+/+ and DNA-PKcsKD/KD embryos at E15.5. The targeting scheme and basic characterization of the DNA-PKcs KD allele can be found in Fig. S1. (B) Upper panel: Genotypes of embryos at different ages obtained by inter-crossing DNA-PKcs+/KD mice. * indicates post-mortem embryos. The p-values were calculated using the chi-squared test. Lower panel: The number of live birth mice obtained from intercrossing between DNA-PKcs+/− and DNA-PKcs+/KD mice. The p-value was calculated with the chi-square test. (C) H&E staining of E14.5 embryonic brain. Cortical plate (CP) and intermediate zone (IZ) contain post-mitotic neurons and ventricular zone (VZ) contains the proliferating cells. Black arrowheads denote apoptotic inclusions. (D) The frequency of apoptotic inclusions per 40× field in E14.5 brain. (E) The frequency of chromosomal (black box) or chromatid (white box) breaks per metaphase in ES cells. The panel on the right shows the examples of intact chromosome, chromosomal or chromatid breaks. The raw counts can be found in Fig.S2A. (F) Left panel: Frequency of metaphases with cytogenetic abnormalities among P3 primary DNA-PKcs+/+, DNA-PKcs−/− and DNA-PKcsKD/KD MEF cells. At least three independently derived lines were assayed for each genotype. Right panel: The frequency of chromatid (white box) and chromosomal (black box) breaks measured by T-FISH analyses in WT, DNA-PKcs−/− and DNA-PKcsKD/KD MEF cells. (G) IR sensitivity of WT, DNA-PKcs−/−, DNA-PKcsKD/KD and XRCC4−/− ES cells measured by colony formation assay. (H) Proliferation of P2 primary MEFs with different genotypes. One of two independent repeats is shown. The y-axis is the relative cell number measured by the fluorescence-nucleotide dye (CyQuant Cell Proliferation Assay, Invitrogen, CA).
Increased genomic instability and severe NHEJ defects in DNA-PKcsKD/KD cells
To determine whether DNA repair defects underlie the embryonic lethality of DNA-PKcs KD/KD mice, we performed telomere fluorescent in situ hybridization (T-FISH), which reveals that nearly 40% of metaphases from DNA-PKcsKD/KD ES cells or primary MEFs carry cytogenetic abnormalities, in contrast to <10% in DNA-PKcs+/+ cells (Fig. 1E–1F and Fig.S2A). T-FISH recognizes two kinds of chromosomal abnormalities: chromosome breaks that involve both sister-chromatids and chromatid breaks that involve only one of the two sister chromatids. While chromatid breaks likely occur after DNA replication in the S or G2 phase of the cell cycle, chromosomal breaks often result from the replication of a G1 phase DSB (Fig.1E). Notably, both DNA-PKcsKD/KD MEF and ES cells preferentially accumulate chromosomal breaks (Fig.1E–F and Fig.S2A), similar to previously characterized NHEJ deficient XRCC4 or XLF deficient cells (Li et al., 2008, Zha et al., 2007). While DNA-PKcs−/− ES cells are only moderately sensitive to IR, DNA-PKcsKD/KD ES cells are hypersensitive to IR, similar to XRCC4−/− ES cells (Fig.1G). DNA-PKcs KD/KD primary MEFs also display severe proliferation defects (Fig.1H) and immortalized DNA-PKcsKD/KD MEFs are hypersensitivity to Etoposide, a topoisomerase II inhibitor that generates DSBs (Fig.S2B), but they are not sensitive to PARP inhibition (Fig.S2C), which preferentially kills homologous recombination (HR) deficient cells (Bryant et al., 2005). HR functionality, as measured by an integrated DR-GFP reporter (Pierce et al., 2001), is also similar between DNA-PKcsKD/KD and DNA-PKcs+/+ ES cells (Fig.S2D). Together, these findings reveal that expression of catalytically inactive DNA-PKcs protein increases genomic instability by inhibiting NHEJ, but not HR. The much more severe genomic instability in DNA-PKcsKD/KD cells in comparison to the DNA-PKcs−/− controls further suggest that the DNA-PKcs protein has a role in NHEJ beyond end-processing.
P53 deficiency rescues the embryonic development, but not lymphocyte development, in DNA-PKcsKD/KD mice
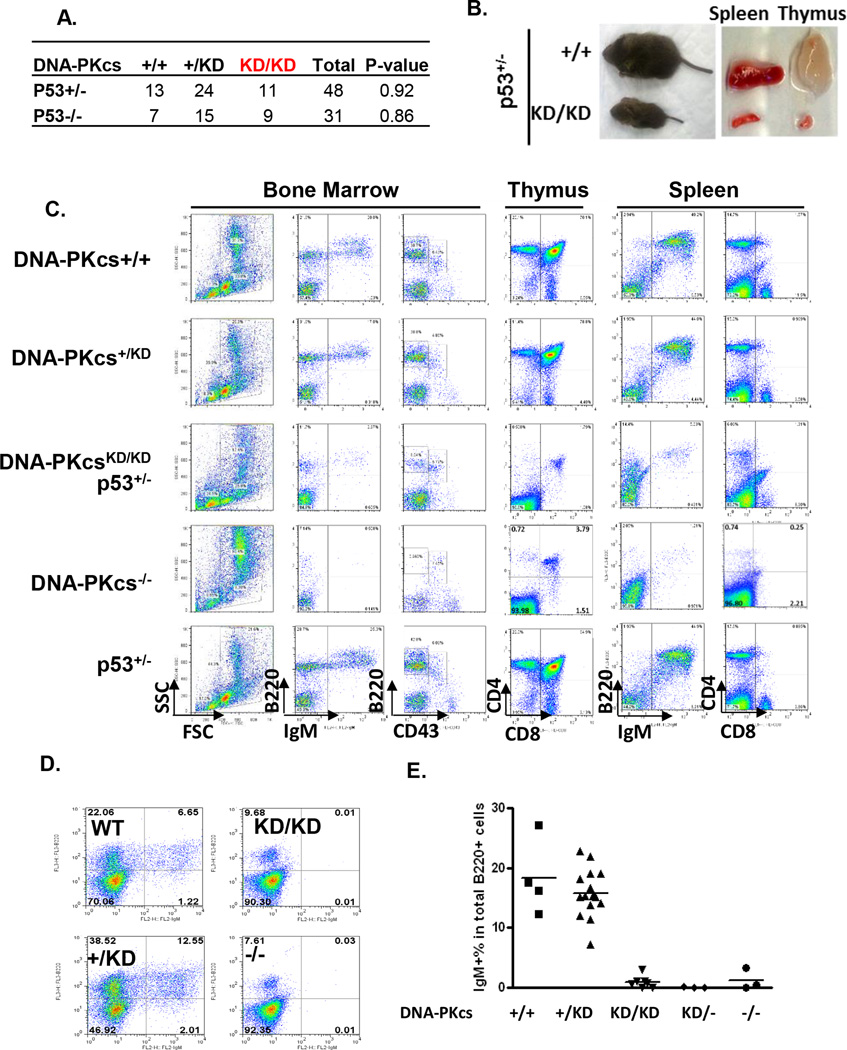
To further elucidate the role of DNA-PKcs in NHEJ, we examined lymphocyte development, where defects in end-processing and end-ligation can be readily distinguished (Lieber, 2010). Both homozygous (p53−/−) and heterozygous (p53+/−) p53 deficiency fully rescues the embryonic development of DNA-PKcsKD/KD mice (Fig.2A) and allows us to study lymphocyte development in live animals. The cellularity of the thymus and spleen from DNA-PKcsKD/KDp53+/− mice are reduced ~100 fold and ~30 fold, respectively, in comparison to the p53+/− littermates (Fig.2B and Fig. S3A). Flow cytometry analyses revealed that lymphocyte development in DNA-PKcsKD/KDp53+/−(−/−) mice is blocked at the progenitor stage, indicated by the lack of both naïve (B220+IgM+) and precursor (B220+CD43−IgM−) B cells in the bone marrow and the lack of both mature (CD4+CD8− or CD4−CD8+ single-positive) and precursor (CD4+CD8+ double positive) T cells in the thymus (Fig.2C and Fig.S3A-2B). Together with the markedly reduced ratio between precursor B cells (B220+CD43−IgM−) to progenitor B cells (B220+CD43+IgM−) (Fig.S3B), these observations are consistent with defects in V(D)J recombination. Progenitor B cells derived from p53-proficient DNA-PKcsKD/KD as well as DNA-PKcs−/KD fetal livers also failed to differentiate to IgM+B220+ naïve B cells in vitro (Fig.2D–2E), indicating that the V(D)J recombination defects are p53-independent and cell-autonomous. Consistent with the lack of dominant negative effects, progenitor B cells derived from DNA-PKcs+/KD fetal livers achieve similar differential capacities as those from DNA-PKcs+/+ littermates (Fig.2D–2E).
Figure 2. Cell autonomous and p53-indepndent developmental defects of DNA-PKcsKD/KD lymphocytes.
(A) Genotype distribution from DNA-PKcs+/KDp53+/− intercrossing. (B) Representative picture of p53+/−DNA-PKcsKD/KD and p53+/−DNA-PKcs+/+ littermates at 21 days of age. Spleen and thymus size are shown on the right. (C) Representative flow cytometry analyses of 4-week DNA-PKcs +/+, DNA-PKcs+/KD, DNA-PKcsKD/KDp53+/− DNA-PKcs−/− and p53 +/− mice. (D) Representative FACs analyses of day 10 fetal liver culture. Accumulation of naïve B cells (B220+IgM+) implies successful V(D)J recombination of both IgH and IgK loci. (E) The summary of IgM+% out of total B220+ B cells after 10 days of fetal liver culture as described before (Frank et al., 2000). Each dot represents an individual E14.5 embryo of the indicated genotypes. The dash lines represent the average of at least 3 independent embryos for each genotype.
End-ligation is abrogated in DNA-PKcsKD/KD Cells
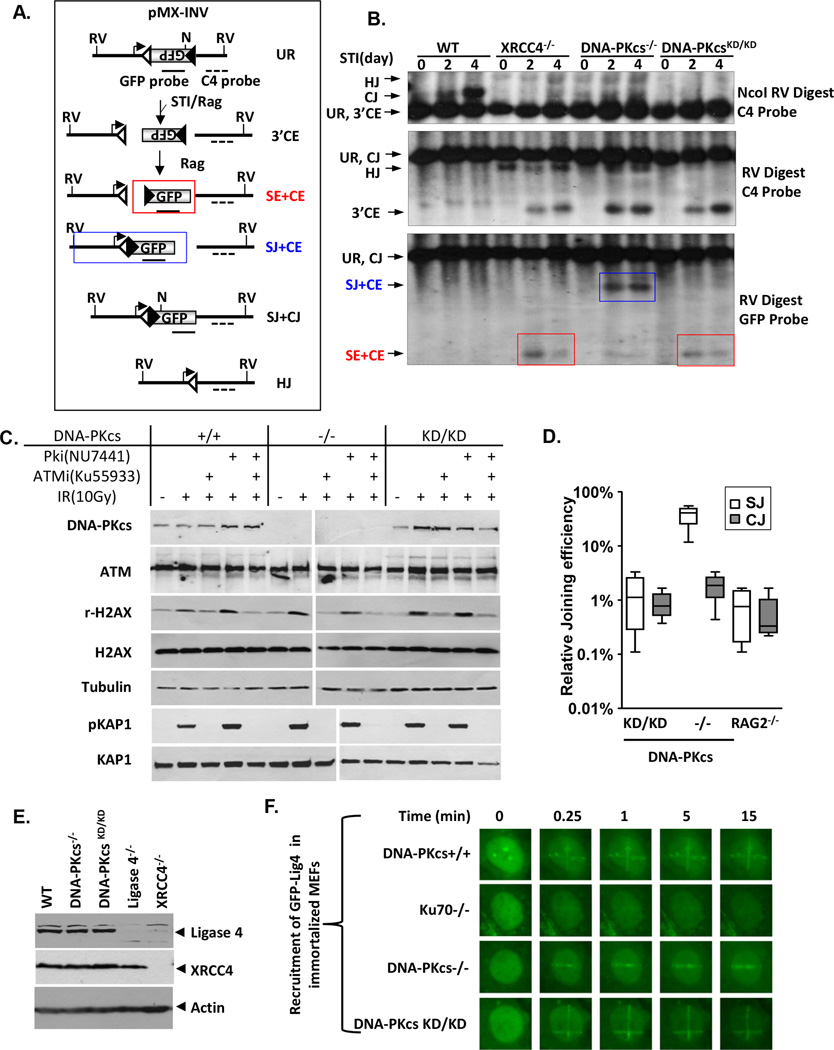
To ascertain whether the V(D)J recombination defects are caused by a failure of CE hairpin-opening (e.g., DNA-PKcs−/−) (Rooney et al., 2002, Gao et al., 1998a, Taccioli et al., 1998), end-ligation (e.g., XRCC4−/−) (Barnes et al., 1998, Frank et al., 1998, Gao et al., 1998b) or both, we generated v-abl kinase transformants from fetal liver-derived DNA-PKcsKD/KD B cells (Bredemeyer et al., 2006). Consistent with observations in MEFs and ES cells, DNA-PKcsKD/KD B cell lines are also hypersensitive to IR (Fig.S3C). In wild type v-abl-transformed B cells, the v-abl kinase inhibitor STI571 induces RAG expression and successful rearrangement of the chromosomally integrated inversional V(D)J recombination substrate pMX-INV (Fig.3A), as evidenced by GFP expression (Fig.S4A) and the accumulation of SJ and CJ products in Southern Blot analyses (Fig.3B- top panel) (Zha et al., 2011b). Consistent with the blockade in lymphocyte development in vivo, the formation of CJs is abrogated in DNA-PKcs−/−, XRCC4−/− as well as DNA-PKcsKD/KD cells, leading to the accumulation of unrepaired 3’CEs (Fig.3B, middle panel). End-ligation deficient XRCC4−/− cells cannot form SJs or CJs, and thus accumulate small SE-CE fragments (Fig.3A–3B, red box). DNA-PKcs−/− cells cannot form CJs owing to hairpin-opening defects, but form SJs efficiently, leading to the accumulation of the larger SJ-CE fragments (Fig.3A–3B, blue box). DNA-PKcsKD/KD cells accumulate SE-CE fragments only (Fig.3B, red box), similar to XRCC4−/− cells, indicating severe end-ligation defects. Consistent with the end-ligation defects, only 4.8% of residual SJs recovered from DNA-PKcsKD/KD cells are precise, in contrast to 93.8% from DNA-PKcs+/+ cells (Tab.S1). Similar results were obtained in several independent clones (Fig.S4B) and with a deletional SJ recombination substrate (pMX-Del-SJ, Fig.S4C). Together, these findings indicate that when DNA-PKcs protein is expressed, specifically DNA-PKcs kinase activity is necessary for direct end-ligation of NHEJ.
Figure 3. DNA-PKcsKD/KD lymphocytes have end-ligation defects.
(A) Schematic of pMX-INV V(D)J recombination substrates (Zha et al., 2011a). The un-rearranged substrate (UR), coding/signal end (CE/SE) intermediates and coding/signal joins (CJs/SJs) are diagramed. The recombination signal sequence (RSS, triangle), GFP probe (solid lines) and C4 probe (dash lines) are indicated. Positions of EcoRV (RV) sites and NcoI (N) sites are shown. The red box shows the isolated SE-CE fragments resulting from complete end-ligation defects. The blue box shows the SJ-CE fragments resulting from isolated CJ formation defects. (B) Southern blot analyses of inversional V(D)J rearrangements of representative DNA-PKcs+/+, XRCC4−/−, DNA-PKcs−/− and DNA-PKcsKD/KD cells with indicated digestion and probe. The red and blue boxes mark the SE-CE fragments and SJ-CE fragments, respectively. Flowcytometry analyses can be found in Fig. S4A. Additional clones can be found in Fig.S4B. (C) Western blot for DNA-PKcs, ATM, phosphorylated H2AX, total H2AX, phosphorylated KAP-1, total KAP-1, and α-tubulin in irradiated (10 Gy) MEFs with or without pre-incubation with 15 µM ATM kinase inhibitor (KU55933, Tocris Bioscience) or 5µM DNA-PKcs kinase inhibitor (NU7441, Tocris Bioscience) or both. Cell lysates were collected 2 hours after irradiation. Primary antibodies were used at the following dilutions: anti-DNA-PKcs (1:1000, Thermo), anti-ATM (1:500, MAT3, Sigma), anti-γH2AX (1:1,000, Millipore), anti-H2AX (1:1,000, Millipore), anti-KAP-1 (1:1,000, Cell Signaling), anti-phospho-KAP-1(S824) (1:1,000, Bethyl Laboratories) and anti-tubulin (1:1,000, Calbiochem). Additional experiments in v-abl kinse transformed pro-B cells can be found in Fig. S5A. (D) Extra-chromosomal V(D)J recombination efficiency. Each box represents the average and standard deviation of 4 independent repeats on two independent v-abl kinase transformed B cell lines of each genotype (see details in Fig.S5B). (E) Western blot for Lig4 and XRCC4 in DNA-PKcs+/+, DNA-PKcs−/−, DNA-PKcsKD/KD, Lig4−/−, and XRCC4−/− v-abl transformed B cell lines. Primary antibodies were used at the following dilutions: anti-XRCC4 (1:500, Santa Cruz), anti-Lig4 antibody (1:500, a generous gift from Dr. David Schatz) and anti-β-actin (1:10,000, Sigma). (F) Laser induced recruitment of GFP-Lig4 in immortalized MEFs.
ATM kinase activity is not affected in DNA-PKcsKD/KD cells
Next, we asked how DNA-PKcs kinase activity regulates end-ligation. ATM and DNA-PKcs share a similar consensus phosphorylation sequence (SQ/TQ) and have redundant functions in end-ligation, including SJ formation during V(D)J recombination (Callen et al., 2009, Zha et al., 2011b, Gapud et al., 2011), so we tested whether expression of catalytically inactive DNA-PKcs interferes with ATM kinase function. In this context, ATM protein levels and ATM-mediated phosphorylation of their shared substrates (e.g. H2AX) were not altered in DNA-PKcsKD/KD MEFs or B cells in comparison to the DNA-PKcs−/− control (Fig.3C and Fig.S5A). Specifically IR induced phosphorylation of H2AX is mediated by both ATM and DNA-PKcs in DNA-PKcs+/+ cells and is completely abolished by ATM kinase inhibitor alone in DNA-PKcsKD/KD and DNA-PKcs−/− cells, further confirming the loss of DNA-PKcs kinase activity in vivo (Fig.3C and Fig.S5A). Moreover, IR induced, ATM-mediated phosphorylation of KAP1 is also normal in DNA-PKcsKD/KD MEFs and B cells and is still sensitive to ATM kinase inhibition (Fig.3C and Fig.S5A), indicating that ATM function is not abrogated in DNA-PKcsKD/KD cells. This conclusion is further supported by the essential role of ATM kinase activity in end-processing in DNA-PKcs KD/KD cells (see below).
Extra-chromosomal end-ligation is also abrogated in DNA-PKcsKD/KD cells
ATM phosphorylates its chromatin-associated substrates (e.g. H2AX and 53BP1) to promote NHEJ, so ATM-deficiency preferentially affects chromosomal NHEJ, in comparison to extra-chromosomal NHEJ (Zha et al., 2011a). To determine whether this is also the case for DNA-PKcs, we measured NHEJ efficiency using plasmid-based extra-chromosomal V(D)J recombination substrates in v-abl kinase transformed DNA-PKcsKD/KD B cells. This assay is only semi-quantitative and is intended to measure severe NHEJ defects, so we performed four independent assays on cell lines from each genotype (Fig. 3D and Fig.S5B) and found that only CJ formation is reduced in DNA-PKcs−/− cells, but both CJ and SJ formation is reduced >20 fold in DNA-PKcsKD/KD cells (Fig.3D and Fig.S5B). Furthermore, the residual CJ/SJ junctions from DNA-PKcsKD/KD cells display large deletions and low fidelity (Tab.S2 and S3). Thus, we conclude that the DNA-PKcs protein regulates end-ligation in a manner independent of chromatin and independent of ATM.
Auto-phosphorylation of DNA-PKcs regulates end-ligation at the DNA ends by relieving the physical block imposed by the DNA-PKcs protein
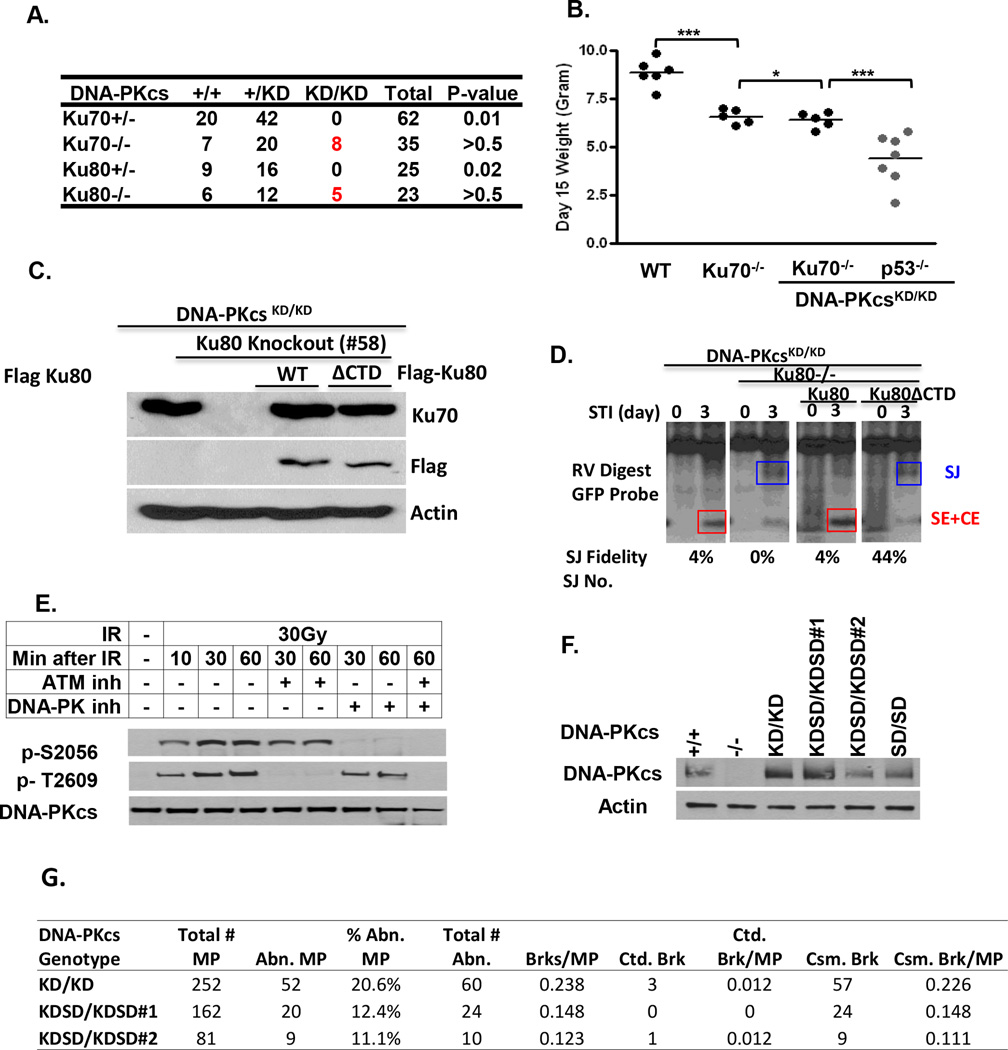
Defects in core NHEJ factors, especially Lig4 and XRCC4, abrogates both chromosomal and extra-chromosomal end-ligation, as observed in DNA-PKcsKD/KD cells. However, we found that the expression of Lig4 and XRCC4 (Fig.3E), as well as the DNA damage-induced recruitment of Lig4 to DSBs (Fig.3F) are not measurably affected in DNA-PKcs KD/KD cells, in comparison to DNA-PKcs−/− and DNA-PKcs+/+ controls. Given the kinase independent recruitment of DNA-PKcs to DSBs (Uematsu et al., 2007), we hypothesized that the DNA-PKcs protein might physically block end-ligation at the ends in the absence of auto-phosphorylation. To test this hypothesis, we deleted Ku, which is required for the DNA-binding of DNA-PKcs in vivo (Falck et al., 2005). Strikingly, complete deletion of Ku (Ku70−/− or Ku80−/−, but not Ku70+/− or Ku80+/−), fully rescues the embryonic development of DNA-PKcsKD/KD mice without rescuing the lymphocyte development (Fig.4A and FigS.5C). Moreover, DNA-PKcs KD/KDKu70−/− mice were identical to their Ku70−/− littermates and significantly bigger than DNA-PKcsKD/KDp53−/−(+/−) mice (Fig.4B), implying a rescue of the repair function beyond checkpoint removal.
Figure 4. DNA-PKcs regulates end-ligation at the DNA ends.
(A) Genotype distribution from inter-crossing Ku70+/− (or Ku80+/−) DNA-PKcs+/KD mice. Lymphocyte development of the Ku70−/−DNA-PKcs KD/KD mice and controls can be found in Fig. S5B. (B) The body weight (grams) of 15 day old mice of each genotype. The bar indicates the average and standard deviations. ***: p<0.01. *: p>0.5, using student t-test. (C) Western blot for Ku70 (1:1000, Abcam), Flag (M2, Sigma) and Actin (Sigma) in Ku80 deleted and reconstitute DNA-PKcsKD/KD pro-B cell lines. (D) Southern blot analyses of pMX-INV rearrangement in DNA-PKcs KD/KDKU80−/− cells with or without ectopic expression of full length (FL) or ΔCTD Ku80.The DNA was digested with EcoRV and probed with the GFP probe. The diagram can be found in Fig 3A. The summary and raw data of the SJ junctions can be found in Fig. S6 and Tab.S4 (E) Phosphorylation of S2056 and T2906 clusters in Hela cells after exposure to IR. The S2056 phosphorylation is selectively inhibited by DNA-PKcs kinase inhibitor, while the T2609 phosphorylation is abolished by ATM kinase inhibitor. The dose and the time after IR are listed in the table above. (F) Western blot for total DNA-PKcs protein in DNA-PKcsKD/KD, DNA-PKcsKDSD/KDSD and DNA-PKcsSD/SD ES cells. All ES cells were targeted homozygously. (G) Telomere-FISH analyses of two independent clones of DNA-PKcsKDSD/KDSD ES cells and their parental clone.
Ku70 and Ku80 depend on each other for protein stability and the Ku70/80 dimer is essential for end-ligation of NHEJ. In this context, Ku-deficiency also rescues the embryonic lethality of end-ligation defective Lig4−/− mice, presumably by increasing alternative end-joining (A-EJ), a mode of DSB repair that preferentially uses micro-homology (MH) at the junction (Karanjawala et al., 2002). To ascertain whether DNA-PKcs inhibits end-ligation of classical NHEJ specifically, we deleted Ku80 from DNA-PKcsKD/KD v-abl kinase transformed B cells using Crispr/Cas9 mediated gene editing. Homozygous Ku80 deleted cells was identified by locus specific genomic PCR and verified by loss of Ku70 protein stability by Western blot (Fig.4C). Ku80 deletion partially restored SJ formation in DNA-PKcsKD/KD cells as indicated by the accumulation of SJ-CE fragments (Fig.4D). However, all (100%) SJs from DNA-PKcsKD/KDKu80−/− cells are imprecise, including 60% (14/23) with MH (Fig.4D, Fig. S6A–6B and Tab. S4), consistent with the signature of A-EJ. Expression of WT Ku80 completely blocked SJ formation in DNA-PKcsKD/KDKu80−/− cells (Fig.4C–4D), confirming the specificity of Crispr/Cas9 mediated deletion of Ku80 in DNA-PKcsKD/KD cells. Expression of Ku80ΔCTD, which retains the Ku70-interacting domain but lacks the C-terminal sequences (Δ45aa) required for DNA-PKcs recruitment (Falck et al., 2005), restored the Ku70/80 dimer (Fig.4C) as well as SJ formation and importantly also SJ fidelity (~44% precise junctions with only 8% MH) in DNA-PKcs KD/KDKu80−/− cells (Fig.4D, and Fig. S6B). Together, these data support a model in which the DNA-PKcs protein physically blocks end-ligation at the DNA ends in the absence of auto-phosphorylation.
Among the phosphorylation sites on DNA-PKcs, the S2056 cluster is preferentially auto-phosphorylated, rather than phosphorylated by ATM in human cells (Fig.4E) (Davis et al., 2014, Meek et al., 2008). Unfortunately, numerous phospho-specific antibodies recognizing phosphorylated S2056 of human DNA-PKcs fail to recognize the phosphorylation of the corresponding S2053 in murine cells, presumably due to the divergence of nearby protein sequences between human and mouse (Fig.S7A, P vs R at +3 position). Nevertheless, we introduced the phospho-mimetic mutation S2053D (referred to as SD) into DNA-PKcsKD/KD ES cells (Fig.S7A–7C). The homozygous in-cis S2053D mutation partially rescues the genomic instability in DNA-PKcsKD/KD ES cells without altering DNA-PKcs protein levels (Fig.4F–4G), suggesting auto-phosphorylation of S2053 might promote end-ligation in the absence of DNA-PKcs kinase activity. The moderate rescue also suggests additional phosphorylation sites, including those within and beyond the S2053 cluster, are likely involved in end-ligation as well. Together, our findings reveal that the DNA-PKcs protein suppresses end-ligation at the DNA ends in a Ku -dependent, ATM-independent manner and that this suppression can be relieved through auto-phosphorylation, but not by ATM-mediated trans-phosphorylation.
DNA-PKcs protein, but not its kinase activity, is required for hairpin opening in vivo
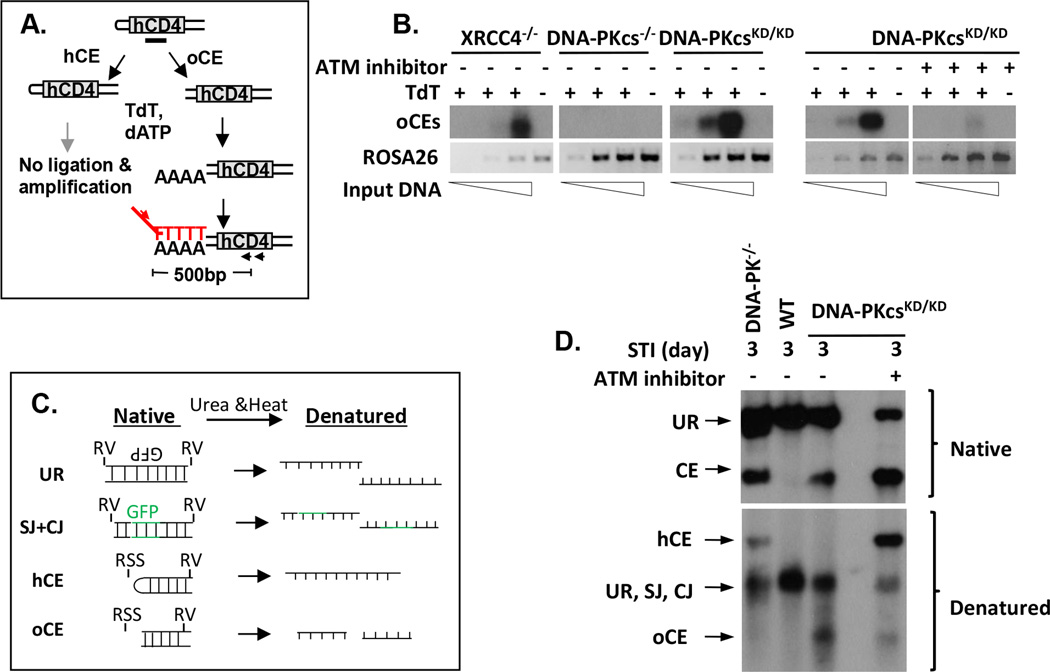
Since the hairpin opening is blocked in the absence of DNA-PKcs, we next tested whether DNA-PKcs kinase activity is required for end-processing. Two independent assays – TdT-mediated PCR and urea denaturing gel electrophoresis were employed to determine the 3’CE hairpin status as a measurement for end-processing (Zha et al., 2011a). TdT adds poly-A tracks to open CEs (oCEs), but not hairpin-sealed CEs (hCEs), and the poly-A track provides a template for subsequent PCR amplification (Fig.5A). Therefore, the hairpin opened 3’CEs in XRCC4−/− cells can be easily detected by TdT-mediated PCR, but the hairpin CEs in DNA-PKcs−/− cells cannot be amplified (Fig.5B). To our surprise, 3’CEs in DNA-PKcsKD/KD cells generated robust TdT-mediated PCR products indicative of open CEs (Fig.5B). Efficient hairpin opening in DNA-PKcsKD/KD cells was further validated using urea denaturing gel electrophoresis, which resolves hairpin (hCE; larger) and open (oCE; smaller) CE fragments by size (Fig.5C–D). At first glance, this appears to be inconsistent with in vitro reconstitution assays (with DNA-PKcs, KU and Artemis), which showed that ATP hydrolysis, and thus DNA-PK “kinase activity” is required for hairpin-opening (Ma et al., 2002). In this context, we found that hairpin opening in DNA-PKcs KD/KD cells can be completely blocked by ATM kinase inhibitor (Fig.5B and 5D). Together with the largely normal lymphocyte development in ATM deficient cells, our results suggest that while DNA-PKcs kinase activity is exclusively required for end-ligation, ATM and DNA-PKcs kinase activities are redundant for end-processing in vivo. This conclusion explains why DNA-PKcs kinase inhibitors alone fail to block CJ formation and hairpin opening in DNA-PKcs+/+ cells (Lee et al., 2013) (Fig. S7D). Together with the lack of hairpin opening in DNA-PKcs−/− cells, our finding indicates that hairpin opening requires DNA-PKcs protein and the kinase activity provided by either ATM or DNA-PKcs.
Figure 5. ATM-dependent hairpin opening in DNA-PKcsKD/KD lymphocytes.
Diagram (A) and results (B) from the TdT-mediated ligation PCR assay. Specifically, TdT adds a poly-A tail to open hairpins, which can be subsequently amplified with universal primers to poly T tails, and an internal primer within hCD4. The PCR product is then analyzed on a gel, transferred to a membrane and probed with P32 labeled oligo probes against hCD4. Two representative assays are shown in Fig 5B. PCR corresponding to the ROSA26 locus was used as loading control. Diagram (C) and results (D) of urea denaturing gel electrophoresis assay. DNA was digested with EcoRV. For denaturing gel electrophoresis, ½ of the DNA is subjected to 8M of Urea at 80°C to denature the double stranded DNA. RV=EcoRV. More detail of the experimental procedures can be found in (Zha et al., 2011a, Helmink et al., 2011).
ATM mediated phosphorylation of DNA-PKcs regulates end-processing through Artemis recruitment
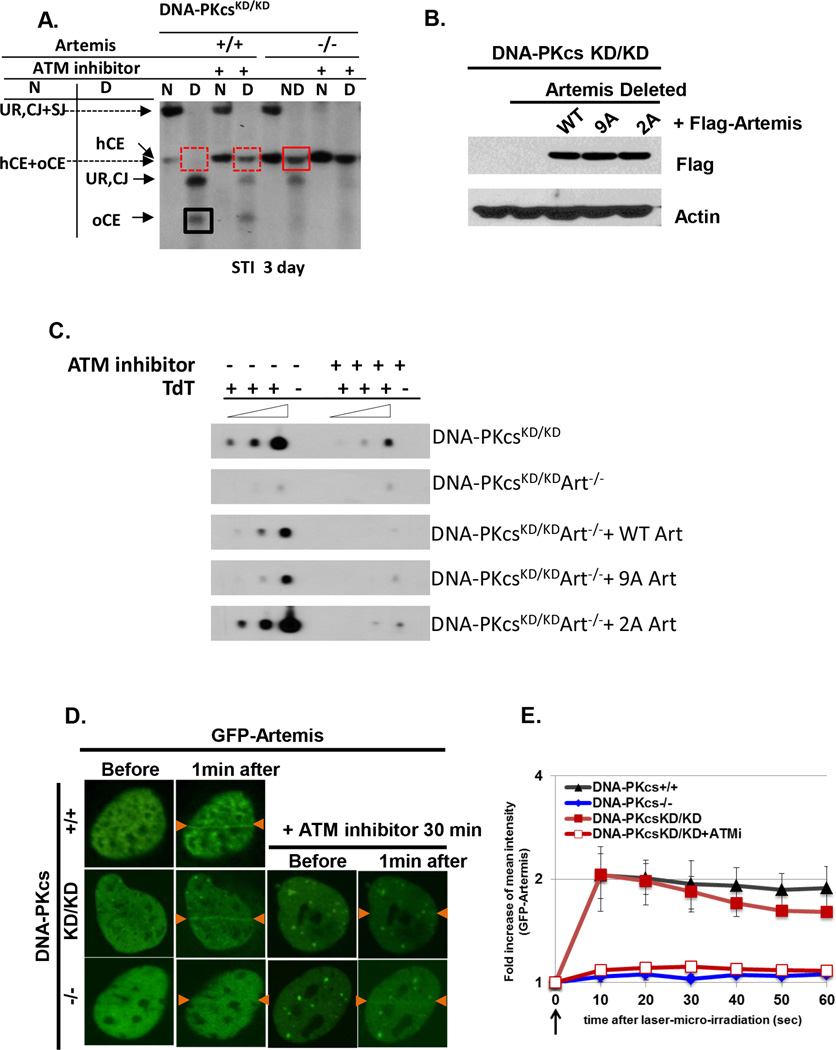
ATM kinase activity promotes many nuclease activities, including MRE11 and CtIP, which all have hairpin opening activity in vitro (Helmink et al., 2011, Zha et al., 2011a). Using Crispr-Cas9 mediated deletion (Fig.S7E), we showed that hairpin opening in DNA-PKcsKD/KD cells is exclusively mediated by the Artemis endonuclease. As such, deletion of Artemis completely blocked hairpin opening in DNA-PKcsKD/KD cells in the absence of ATM kinase inhibitor (Fig.6A). Yet surprisingly hairpin opening in DNA-PKcsKD/KD cells does not require any of the 9 canonical phosphorylation sites at the C-terminal of Artemis (Goodarzi et al., 2006) (Fig.6B–6C), implying that DNA-PKcs is the phosphorylation target required for end-processing.
Figure 6. Artemis, but not Artemis phosphorylation, is required for hairpin opening in DNA-PKcsKD/KD lymphocytes.
(A) Urea denaturing gel analyses of Artemis+/+ and Artemis−/− DNA-PKcsKD/KD cells with or without pretreatment with ATM kinase inhibitor (KU55933, 15uM).The scheme of Crispr mediated deletion of Artemis can be found in Fig. S7. (B) Ectopic expression of Flag tagged Artemis (WT or 2A or 9A) in DNA-PkcsKD/KD cells. Flag antibody (M2, Sigma) was used at 1:1000. (C) TdT-mediated PCR of DNA-PKcsKD/KDArtemis−/− cells without or with ectopic expression of WT, 9A or 2A-Artemis. (D) Representative imaging of GFP-Artemis recruitment in immortalized MEFs. (E) Quantification of increased florescence intensity of GFP-Artemis at the laser damaged area. The data represents the average and standard deviation of 10 cells at each time point.
Next, we asked how DNA-PKcs phosphorylation regulates Artemis activity. In DNA-PKcs+/+ MEFs, laser induced DSBs rapidly recruit GFP-Artemis to the site of damage and this recruitment is largely abolished in DNA-PKcs−/− MEFs (Fig.6D–E), suggesting the DNA-PKcs protein is required for the recruitment of Artemis. Consistent with normal hairpin opening, the recruitment of GFP-Artemis to laser-induced DSBs is largely normal in DNA-PKcsKD/KD cells (Fig.6D–E), but is completely blocked by ATM kinase inhibitor (Fig.6D–E). Together, these data suggest that the DNA-PKcs protein and ATM mediated phosphorylation of DNA-PKcs regulates end-processing by promoting the recruitment of Artemis endonuclease.
DISCUSSION
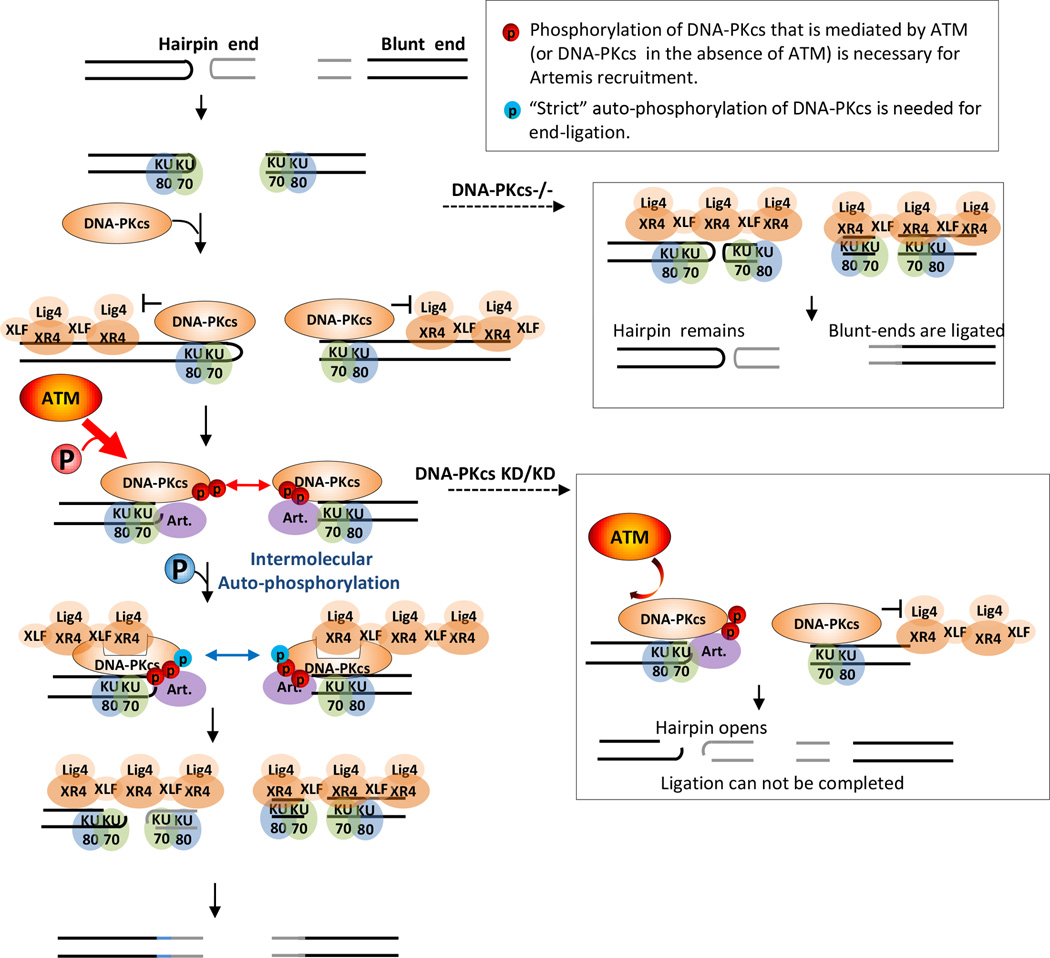
NHEJ is the most accessible DSB repair pathway in highly differentiated cells. In vertebrates, NHEJ entails end-processing in addition to end-ligation to repair DSBs with diverse chemical structures. In this study, we show that DNA-PKcs regulates both end-processing and end-ligation at DNA ends through phosphorylation (Fig. 7). At least two kinds of phosphorylations occur on DNA-PKcs: the ATM or DNA-PKcs mediated trans-phosphorylation of DNA-PKcs (red in Fig. 7) promotes end-processing by recruiting Artemis endonuclease, while distinct strict auto-phosphorylation of DNA-PKcs (blue in Fig. 7) is necessary for end-ligation by removing the physical blockade imposed by DNA-PKcs protein. In this context, the need for auto- and likely intermolecular phosphorylation of DNA-PKcs for end-ligation coupled with the “DNA” dependent kinase activity of DNA-PKcs ensure two ends are proximal before ligation. The temporal delay of end-ligation imposed by DNA-PKcs auto-phosphorylation likely provides a window for end-processing, where the redundant kinase activity of ATM and DNA-PKcs induces robust end-processing to avoid potential fruitless ligation attempts. Therefore, the DNA-PKcs protein serves as a molecular switch that coordinates end-processing with end-ligation through phosphorylation to maximize the NHEJ efficiency.
Figure 7. Distinct DNA-PKcs phosphorylations regulate end-ligation and end-processing.
In this diagram, we illustrate the normal vertebrate NHEJ mechanism on the left and the NHEJ mechanism in the context of DNA-PKcs−/− or DNA-PKcsKD/KD backgrounds on the right. In this model, we highlight two kinds of DNA-PKcs phosphorylation: red, ATM-mediated phosphorylation of DNA-PKcs, and blue, strict auto-phosphorylation of DNA-PKcs. First, in red, we emphasize the role of ATM-mediated DNA-PKcs phosphorylation in Artemis endonuclease recruitment for end processing (e.g. hairpin opening). Notably, in the absence of ATM, DNA-PKcs can also phosphorylate Artemis at such sites (thinner red arrows). Second, in blue, we note the sites on DNA-PKcs where strict auto-phosphorylation occurs are required for end-ligation. For the illustration purposes, we have shown that DNA-PKcs opens up to allow end-ligation by the Lig4-XRCC4-XLF complex, but DNA-PKcs could also simply dissociate from the DNA to allow end-ligation.
DNA-PKcs has previously been found to have, at most, a moderate role in the end-ligation phase of NHEJ. Here we show that catalytically inactive DNA-PKcs protein completely blocks end-ligation, leads to severe genomic instability in post-mitotic neurons, ES cells, MEFs, as well as in developing lymphocytes during V(D)J recombination, which is strikingly similar to those observed in cells lacking bona fide core NHEJ factors, such as XRCC4 or Lig4. We further show, unlike ATM kinase (Zha et al., 2011a), DNA-PKcs regulates NHEJ at the DNA ends independent of the chromatin context: extra-chromosomal and chromosomal end-ligation are both abrogated in DNA-PKcsKD/KD cells (Fig.3). Given the expression and recruitment of Lig4 is not affected in DNA-PKcsKD/KD cells, yet deletion of Ku and specifically the C-terminal domain of Ku80 rescue the end-ligation defects in DNA-PKcsKD/KD cells, it is clear that DNA-PKcs regulates end-ligation at DNA ends. One possibility would be that DNA-PKcs together with Ku forms a barrier at the DNA ends that prevents the XRCC4-XLF-Lig4 filament (Mahaney et al., 2013) from extending across the breaks (Fig.7) and auto-phosphorylation of DNA-PKcs relieve this blockade through large conformation changes or simple dissociation (Dobbs et al., 2010, Davis et al., 2014, Meek et al., 2008). In this context, Alanine substitution at 7 potential phosphorylation sites of DNA-PKcs inhibits T4 bacterial ligase in vitro (Block et al., 2004), consistent with the physical blocking rather than specific inhibitory interaction.
DNA-PKcsKD/− mice also died before birth (Fig.1B), but DNA-PKcs+/KD mice had no measurable defects in all assays including birth ratio, neuronal apoptosis, growth rate, lymphocyte development, fetal liver culture and even sensitivity IR sensitivity assays (Fig.1B–D and 2C, Fig.S1F and S3), suggesting that the kinase dead protein does not process dominant negative activity in the presence of the wild type DNA-PKcs. This absence of a classical dominant-negative effect has also been seen in previously characterized ATM+/KD mice and is consistent with “inter”-molecular, rather than “intra”-molecular auto-phosphorylation of DNA-PKcs, ATM and even ATR (Meek et al., 2008, Liu et al., 2011, Bakkenist and Kastan, 2003). As such, the kinase-proficient DNA-PKcs can trans-phosphorylate another kinase-dead DNA-PKcs protein to relieve the inhibition on end-ligation. Notably, expression of catalytically inactive ATM kinase also inhibits DNA repair, likely a subset of homologous dependent repair beyond the simple loss of ATM (Yamamoto et al., 2012, Daniel et al., 2012), suggesting that auto- and intermolecular phosphorylation dependent structural changes are a common means by which PI3KKs regulate “specific” repair pathways beyond their well-recognized signaling function. Consistent with this notion, several studies suggest that specific inhibitors for ATM or DNA-PKcs kinase inhibits DNA repair beyond that which is associated with the simple loss of ATM or DNA-PKcs (Allen et al., 2003, White et al., 2010). However, the precise molecular mechanism of DNA-PKcs kinase inhibitors would require further study, as DNA-PKcs kinase inhibitors do not block end-ligation, while catalytically inactive DNA-PKcs protein does (Fig. 3 and S7D) (Lee et al., 2013).
The requirement for DNA-PK kinase activity in end-ligation, but not in end-processing, further suggests that at least two distinct DNA-PKcs phosphorylation clusters exist. Among them, strict auto-phosphorylation of DNA-PKcs is necessary for end-ligation. In this context, we show that phospho-mimetic S2053D mutation, corresponding to the preferentially “auto”-phosphorylated S2056 cluster of human DNA-PKcs, moderately, but consistently, improves genomic stability of DNA-PKcsKD/KD cells (Fig.S7A and Fig. 4). Yet additional sites are likely involved. On the other hand, ATM-mediated phosphorylation of DNA-PKcs is sufficient for end processing. The T2609 cluster of DNA-PKcs is primarily phosphorylated by ATM (Fig.4E) (Davis et al., 2014) and can also be phosphorylated by DNA-PKcs. Alanine substitution at 3 of the 5 potential phosphorylation sites within the mouse T2609 cluster (referred to as the 3A allele) does not block hairpin opening, but renders hairpin-dependent CJ formation hypersensitive to both ATM and DNA-PK kinase inhibitors (Lee et al., 2013), consistent with redundancy between ATM and DNA-PKcs kinase activity in end-processing. Notably T2609 can also be phosphorylated by ATR after UV exposure (Meek et al., 2008, Davis et al., 2014). DNA-PKcs3A/3A mice also developed bone marrow failure, sharing molecular features with Faconia Anemia (FA) patients (Zhang et al., 2011), suggesting the T2609 cluster might regulate end-processing beyond hairpin opening. These distinct roles of DNA-PKcs auto- vs trans-phosphorylation in end-ligation and end-processing could potentially explain the opposite effects of phosphorylation of the S2056 and T2609 clusters on end-protection (Cui et al., 2005).
Finally while ATM is not absolutely required for hairpin opening, ATM inhibitor partially blocks hairpin opening in XLF−/− cells and selectively delays hairpin dependent CJ formation, but not SJ formation (Fig.S7D) (Zha et al., 2011a, Lee et al., 2013, Bredemeyer et al., 2006), Using the DNA-PKcs KD mouse model, we show that ATM mediated phosphorylation of DNA-PKcs regulates end-processing at the step of Artemis recruitment; as such ATM kinase inhibitors completely abolished hairpin opening and recruitment of Artemis to DSBs in DNA-PKcsKD/KD cells. These findings reveal a fundamental role of ATM in directly regulating Artemis and end-processing that is otherwise masked by the overlapping DNA-PKcs kinase activity and provides the molecular mechanism for the epistatic function of ATM and Artemis in ~10% of DNA repair (Riballo et al., 2004) and selective CJ defects in ATM kinase inhibitor treated cells.
EXPERIMENTAL PROCEDURES
Mice
DNA-PKcs−/−, Ku70+/−, Ku80+/−, XRCC4+/− and p53+/− mice and cells have been described before (Gao et al., 1998b, Gao et al., 1998a, Gu et al., 1997, Jacks et al., 1994, Nussenzweig et al., 1996). The DNA-PKcs KD mutation converts the conserved aspartic acid 3922 within the putative catalytic loop to alanine (D3922A) (Fig.S1). To generate the DNA-PKcs KD mutant allele, DNA sequences from the genomic DNA-PKcs (Prkdc) locus were generated via PCR from TC1 ES cell DNA (129 strain). A targeting construct was designed to insert a NeoR gene cassette oriented in the same transcriptional orientation from the endogenous DNA-PKcs (Prkdc) promoter into intron 83 next to the D3922A mutation in exon 83. A 2.6 kb 5’ arm and 4.9 kb 3’arm were PCR generated separately, cloned into the pBK vector and sequenced. The 3’arm was directly subcloned into pLNTK in the desired orientation. The mutation was introduced into the 5’ arm using site-directed mutagenesis and confirmed by sequencing (Fig.S1B). The mutated 5’ arm contains the exon with mutation (indicated by an open box in Fig.S1B) was then sub-cloned into pLNTK. The targeting construct was then electroporated into CSL3 ES cells (129 strain) and successful targeting was determined via Southern blot analyses using SpeI digested genomic DNA and a 5’ genomic probe as outlined in Fig.S1B. The WT band is ~7.0kb and the targeting introduced an additional SpeI site and reduced the band to ~4.5kb (Fig.S1C). The correct clones were confirmed with a 3’ probe. A total of five independent targeted clones were sequenced for the DNA-PKcs KD mutation, 3 were verified with correct mutations. Two were injected for germ line transmission and yielded identical phenotypes (hereafter referred to as the DNA-PKcs KD allele).
V(D)J recombination assays
Chromosomal V(D)J recombination with an integrated substrate was carried out as described (Zha et al., 2011a). Briefly, v-abl transformed pro-B-cell lines were isolated from various mouse lines that harbored an Eμ-Bcl2 transgene. EμBcl2+ is included to prevent apoptosis associated with STI571 treatment (Zha et al., 2011b). The pro-B lines were infected with the pMX-INV or pMX-Del-SJ retroviral vector and assayed for V(D)J recombination as described. STI571 is used at 3µM and ATM inhibitor KU55933 is used at 15µM final concentration throughout the paper. The diagram of pMX-INV is shown in Fig.3A and of pMX-SJ deletional substrates is shown in Fig.S4C. For sequence analyses, the chromosomal SJs were amplified by PCR and cloned into the pGEMTeasy vector (Promega). Precise SJs reconstitute a restriction site for ApaL1. Imprecise SJs were sequenced. Extra-chromosomal V(D)J recombination assays were performed in v-abl transformed cell lines as described before (Zha et al., 2011a). Please see supplementary experimental procedure for details.
Cas9 mediated deletion of Ku80 or Artemis
Guide RNA sequence targeting murine Ku80 (5’GTCCAGGACTCAGCTAGTTAAGG and 5’GATCTCGGCATCCCTCATGCAGG) or Artemis (5’GTTCCCGGAAGTGGCGCCCAGG and 5’GGTAAGACTAGACAGGGGTCTGG) were cloned into the pX330 Crispr plasmid (http://www.genome-engineering.org/crispr/?page_id=23). PAMs are underlined. A pair of gRNA containing plasmids were electroporated into v-abl kinase transformed DNA-PKcsKD/KD cells using the 4D Nucleofector apparatus (Buffer SF, program N100, Lonza, Walkersville, MD). One day later, the cells were plated for single clones and PCR was used to screen each single clone for the deletion. When necessary, heterozygous targeted clones were subjected to a 2nd round of gRNA targeted deletion to remove the 2nd allele. Complete deletion was verified by Western blot for Ku80 or by sequencing the deletion junctions for Artemis. Reliable antibodies for murine Ku80 and Artemis are not available. However, Ku80 is required for the protein stability of Ku70, so we verified the functional loss of Ku80 by the absence of Ku70 protein in DNA-PKcsKD/KDKu80−/− Abelson lines. For Artemis deletion, we cloned and sequenced the deletional junction as seen in Figure S7E and confirmed the loss of ATG and the first exon. The specificity of the deletion was further validated by reconstitution with wild type Ku80 (Fig.4C) or Artemis (Fig.6B).
Laser induced recruitment of Artemis and Ligase 4
The open reading frames of Artemis or Lig 4 were cloned into the pEGFP-C1 vector (Clontech) in frame with GFP. Immortalized MEFs were transiently transfected with GFP-Artemis or GFP-Lig4 and seeded into special plates. The recruitment of GFP-Artemis (Wei et al., 2013) or GFP-Lig4 (Li and Yu, 2013) was performed as previously described. Briefly, Olympus FV1000 confocal microscopy (F10PRDMYR-1, Olympus) equipped with FV1000 SIM Scanner, the 405 nm laser diode (F10OSIM405, Olympus) and FV1000 software package was employed to induce DNA damage (output power at the lens≅5 mW/scan) and follow the recruitment of GFP-Artemis. The recruitment of GFP-Artemis to the laser damaged area is quantified by calculating the ratio of the mean intensity between the sites of laser microirradiation/background area in the same nucleus. At least 10 cells were quantified at each time frame to obtain the average and standard deviation.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dr. Chyuan-Sheng Lin, Kenta Yamamoto and Frances Lee for technical assistance and advice. We thank Dr. David Schatz for providing the anti-Ligase 4 antibody, Dr. Andre Nussenzweig for providing the Ku80-deficient mice, Drs. Marin Jasin and Jeremy Stark for sharing the DR-GFP reporter system and Dr. Penny Jeggo for providing the Artemis-9A plasmid. We thank Drs. Richard Baer and Lorraine Symington for the critical reading of this manuscript. We apologize to colleagues, whose work could not be cited due to space limitations and was covered by reviews instead. This work is in part supported by NIH/NCI 5RO1CA158073, 5RO1CA184187, 1PO1CA174653-01 and American Cancer Society Research Scholar Grant (RSG-13-038-01 DMC) to SZ and NIH AG045545-01 to LL. SZ and XY are recipients of the Leukemia Lymphoma Society Scholar Award and XL is a Leukemia Lymphoma Society Fellow. WJ was in part supported by NIH/NCI T32-CA09503. JC is in part supported by NIH/NCI F31CA183504-01A1 and CTSA/NIH TL1 TR000082.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
WJ, JC, XL, LL, XY and SZ designed experiments. SZ, WJ, JC and XL wrote the paper. WJ, JC, XL, BJL, CLi and SZ analyzed the DNA-PKcs KD mice and cells, YW, SZ, and RLD generated the DNA-PKcs KD mice. SN and LL performed the Artemis recruitment experiment. CLiu and XY performed the Lig4 recruitment experiment.
REFERENCES
- Allen C, Halbrook J, Nickoloff JA. Interactive competition between homologous recombination and non-homologous end joining. Mol Cancer Res. 2003;1:913–920. [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- Block WD, Yu Y, Merkle D, Gifford JL, Ding Q, Meek K, Lees-Miller SP. Autophosphorylation-dependent remodeling of the DNA-dependent protein kinase catalytic subunit regulates ligation of DNA ends. Nucleic Acids Res. 2004;32:4351–4357. doi: 10.1093/nar/gkh761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, Sleckman BP. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Callen E, Jankovic M, Wong N, Zha S, Chen HT, Difilippantonio S, Di Virgilio M, Heidkamp G, Alt FW, Nussenzweig A, Nussenzweig M. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell. 2009;34:285–297. doi: 10.1016/j.molcel.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem. 2007;282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- Cui X, Yu Y, Gupta S, Cho YM, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol Cell Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Pellegrini M, Lee BS, Guo Z, Filsuf D, Belkina NV, You Z, Paull TT, Sleckman BP, Feigenbaum L, Nussenzweig A. Loss of ATM kinase activity leads to embryonic lethality in mice. J Cell Biol. 2012;198:295–304. doi: 10.1083/jcb.201204035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AJ, Chen BP, Chen DJ. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 2014;17:21–29. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AJ, So S, Chen DJ. Dynamics of the PI3K-like protein kinase members ATM and DNA-PKcs at DNA double strand breaks. Cell Cycle. 2010;9:2529–2536. doi: 10.4161/cc.9.13.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Franco S, Alt FW, Manis JP. Pathways that suppress programmed DNA breaks from progressing to chromosomal breaks and translocations. DNA Repair (Amst) 2006;5:1030–1041. doi: 10.1016/j.dnarep.2006.05.024. [DOI] [PubMed] [Google Scholar]
- Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- Frank KM, Sharpless NE, Gao Y, Sekiguchi JM, Ferguson DO, Zhu C, Manis JP, Horner J, Depinho RA, Alt FW. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- Gao Y, Chaudhuri J, Zhu C, Davidson L, Weaver DT, Alt FW. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 1998a;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, Depinho RA, Alt FW. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, Bronson RT, Malynn BA, Bryans M, Zhu C, Chaudhuri J, Davidson L, Ferrini R, Stamato T, Orkin SH, Greenberg ME, Alt FW. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998b;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- Gapud EJ, Dorsett Y, Yin B, Callen E, Bredemeyer A, Mahowald GK, Omi KQ, Walker LM, Bednarski JJ, Mckinnon PJ, Bassing CH, Nussenzweig A, Sleckman BP. Ataxia telangiectasia mutated (Atm) and DNA-PKcs kinases have overlapping activities during chromosomal signal joint formation. Proc Natl Acad Sci U S A. 2011;108:2022–2027. doi: 10.1073/pnas.1013295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, Lees-Miller SP. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Seidl KJ, Rathbun GA, Zhu C, Manis JP, Van Der SN, Davidson L, Cheng HL, Sekiguchi JM, Frank K, Stanhope-Baker P, Schlissel MS, Roth DB, Alt FW. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- Helmink BA, Tubbs AT, Dorsett Y, Bednarski JJ, Walker LM, Feng Z, Sharma GG, Mckinnon PJ, Zhang J, Bassing CH, Sleckman BP. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 2011;469:245–249. doi: 10.1038/nature09585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Karanjawala ZE, Adachi N, Irvine RA, Oh EK, Shibata D, Schwarz K, Hsieh CL, Lieber MR. The embryonic lethality in DNA ligase IV-deficient mice is rescued by deletion of Ku: implications for unifying the heterogeneous phenotypes of NHEJ mutants. DNA Repair (Amst) 2002;1:1017–1026. doi: 10.1016/s1568-7864(02)00151-9. [DOI] [PubMed] [Google Scholar]
- Lee BS, Gapud EJ, Zhang S, Dorsett Y, Bredemeyer A, George R, Callen E, Daniel JA, Osipovich O, Oltz EM, Bassing CH, Nussenzweig A, Lees-Miller S, Hammel M, Chen BP, Sleckman BP. Functional Intersection of ATM and DNA-PKcs in Coding End Joining During V(D)J Recombination. Mol Cell Biol. 2013 doi: 10.1128/MCB.00308-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Alt FW, Cheng HL, Brush JW, Goff PH, Murphy MM, Franco S, Zhang Y, Zha S. Lymphocyte-Specific Compensation for XLF/Cernunnos End-Joining Functions in V(D)J Recombination. Mol. Cell. 2008;31:631–640. doi: 10.1016/j.molcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23:693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Shiotani B, Lahiri M, Marechal A, Tse A, Leung CC, Glover JN, Yang XH, Zou L. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell. 2011;43:192–202. doi: 10.1016/j.molcel.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- Mahaney BL, Hammel M, Meek K, Tainer JA, Lees-Miller SP. XRCC4 and XLF form long helical protein filaments suitable for DNA end protection and alignment to facilitate DNA double strand break repair. Biochem Cell Biol. 2013;91:31–41. doi: 10.1139/bcb-2012-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek K, Dang V, Lees-Miller SP. DNA-PK: the means to justify the ends? Adv Immunol. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Chen C, Da C, S v, Sanchez M, Sokol K, Nussenzweig MC, Li GC. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature. 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, Devido J, Foy D, Chaudhuri J, Lombard D, Alt FW. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol. Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- Taccioli GE, Amatucci AG, Beamish HJ, Gell D, Xiang XH, Torres Arzayus MI, Priestley A, Jackson SP, Marshak RA, Jeggo PA, Herrera VL. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 1998;9:355–366. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, Van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Nakajima S, Hsieh CL, Kanno S, Masutani M, Levine AS, Yasui A, Lan L. Damage response of XRCC1 at sites of DNA single strand breaks is regulated by phosphorylation and ubiquitylation after degradation of poly(ADP-ribose) J Cell Sci. 2013;126:4414–4423. doi: 10.1242/jcs.128272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JS, Choi S, Bakkenist CJ. Transient ATM kinase inhibition disrupts DNA damage-induced sister chromatid exchange. Sci Signal. 2010;3:ra44. doi: 10.1126/scisignal.2000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Wang Y, Jiang W, Liu X, Dubois RL, Lin CS, Ludwig T, Bakkenist CJ, Zha S. Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. J Cell Biol. 2012;198:305–313. doi: 10.1083/jcb.201204098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S, Alt FW, Cheng HL, Brush JW, Li G. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proc Natl Acad Sci U S A. 2007;104:4518–4523. doi: 10.1073/pnas.0611734104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, Dubois RL, Alt FW. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 2011a;469:250–254. doi: 10.1038/nature09604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S, Jiang W, Fujiwara Y, Patel H, Goff PH, Brush JW, Dubois RL, Alt FW. Ataxia telangiectasia-mutated protein and DNA-dependent protein kinase have complementary V(D)J recombination functions. Proc Natl Acad Sci U S A. 2011b;108:2028–2033. doi: 10.1073/pnas.1019293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Yajima H, Huynh H, Zheng J, Callen E, Chen HT, Wong N, Bunting S, Lin YF, Li M, Lee KJ, Story M, Gapud E, Sleckman BP, Nussenzweig A, Zhang CC, Chen DJ, Chen BP. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J Cell Biol. 2011;193:295–305. doi: 10.1083/jcb.201009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.